Abstract
Documentation of maternal uniparental disomy of chromosome 7 in 10% of patients with Russell-Silver syndrome (RSS), characterized by prenatal and postnatal growth retardation and dysmorphic features, has suggested the presence of an imprinted gene on chromosome 7 whose mutation is responsible for the RSS phenotype. Human GRB10 on chromosome 7, a homologue of the mouse imprinted gene Grb10, is a candidate, because GRB10 has a suppressive effect on growth, through its interaction with either the IGF-I receptor or the GH receptor, and two patients with RSS were shown to have a maternally derived duplication of 7p11-p13, encompassing GRB10. In the present study, we first demonstrated that the GRB10 gene is also monoallelically expressed in human fetal brain tissues and is transcribed from the maternally derived allele in somatic-cell hybrids. Hence, human GRB10 is imprinted. A mutation analysis of GRB10 in 58 unrelated patients with RSS identified, within the N-terminal domain of the protein, a P95S substitution in two patients with RSS. In these two cases, the mutant allele was inherited from the mother. The fact that monoallelic GRB10 expression was observed from the maternal allele in this study suggests but does not prove that these maternally transmitted mutant alleles contribute to the RSS phenotype.
Russell-Silver syndrome (RSS [MIM 180860]) is characterized by prenatal and postnatal growth retardation accompanied by dysmorphic features, such as triangular facies and fifth-finger clinodactyly (Silver et al. 1953; Russell 1954; Price et al. 1999). On the basis of familial occurrence (Robichaux et al. 1981; Duncan et al. 1990; Teebi 1992; Al-Fifi et al. 1996) and occasional chromosomal rearrangements (Christensen and Nielsen 1978; Wilson et al. 1985; Butler et al. 1988; Roback et al. 1991; Ramirez-Duenas et al. 1992; Tamura et al. 1993; Schinzel et al. 1994; Rogan et al. 1996; Eggermann et al. 1998; Monk et al. 2000), a genetic etiology for RSS has been suggested. However, the gene responsible for RSS has not yet been identified.
The recent discovery of maternal uniparental disomy of chromosome 7 [mUPD7] in 10% of patients with RSS has suggested the presence, on chromosome 7, of an imprinted gene whose mutation is responsible for the RSS phenotype (Kotzot et al. 1995; Eggermann et al. 1997; Preece et al. 1997; Bernard et al. 1999). The predicted function of this putative RSS gene is regulation of growth, since prenatal and postnatal growth retardation is the hallmark of RSS. Mutations in either a maternally repressed (i.e., paternally expressed) gene or a paternally repressed (i.e., maternally expressed) gene could account for the RSS phenotype.
If we assume that the putative RSS gene is paternally expressed, the gene would not be expressed in mUPD7 cells. Therefore, the predicted function of the putative gene would be facilitation of growth. Loss-of-function mutations in the gene, when transmitted paternally, should lead to the RSS phenotype. The human PEG1 (paternally expressed gene-1) gene on 7q31 was considered to be an excellent candidate gene, because (1) human PEG1 and mouse Peg1 are imprinted and are expressed from the paternal allele (Kobayashi et al. 1997; Riesewijk et al. 1997) and (2) a loss-of-function mutation in mouse Peg1, when paternally transmitted, is associated with severe growth retardation (Kaneko-Ishino et al. 1995). However, analysis of PEG1 in 49 patients with RSS did not reveal any mutations (Riesewijk et al. 1998).
Alternatively, we could assume that the putative RSS gene is maternally expressed. In this scenario, the gene would be expressed in excess in mUPD7 cells. Hence, the predicted function of the putative RSS gene would be suppression rather than facilitation of growth. A gain-of-function mutation in such a gene, when transmitted maternally, would contribute to the RSS phenotype. The human growth factor receptor–bound protein 10/maternally expressed gene-1 (GRB10/MEG1) gene, on 7p11.2-p12 (Jerome et al. 1997), is considered to be a prime candidate, because (1) mouse Grb10 is subject to imprinting and is maternally expressed (Miyoshi et al. 1998), (2) maternal uniparental disomy of the proximal region of mouse chromosome 11 encompassing Grb10 leads to prenatal growth retardation (Cattanach et al. 1998), and (3) in vitro studies indicate that, by interacting with either the insulin-like growth factor I (IGF-I) receptor (O'Neill et al. 1996; Morrione et al. 1997) or the growth hormone receptor (Moutoussamy et al. 1998), GRB10 has a suppressive effect on growth.
Recent documentation of two patients with RSS who had a maternally derived interstitial duplication of 7p11-p13 encompassing GRB10 (Joyce et al. 1999; Monk et al. 2000) further strengthens arguments for GRB10 involvement in the pathogenesis of RSS. In the present study, we determined whether human GRB10 is imprinted, and we then screened 58 patients with RSS for GRB10 mutations.
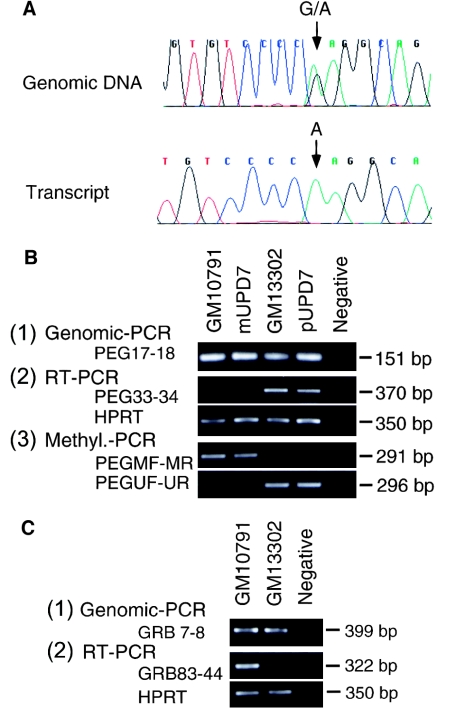
First, monoallelic expression of human GRB10 was investigated by reverse transcriptase (RT)–PCR analysis with a primer pair designed to amplify across a known G/A single-nucleotide polymorphism in exon 3 (Angrist et al. 1998). Human brain tissues for use in this analysis were obtained from the Research Resource Bank (Tokyo), with permission of the Ethics Committee at the National Center of Neurology and Psychiatry. Total RNA was extracted from brain by a commercial kit (QIAGEN) and was treated with RNase-free DNase. Then, cDNA was directly synthesized using the SuperScript II amplification kit (GIBCO/BRL) with an oligo-dT primer. RT-PCR analysis was done with nested primer pairs GRB76 (5′-GCAGAAGGAACCCATGGCTTTAG-3′) and GRB78 (5′-AGAGATGAGGTTCTAAACTGCTGGTC-3′) and nested primer pairs GRB83 (5′-CATCCGTACTACCAGGACAAGG-3′) and GRB44 (5′-TGACAGCGAGGATGTGCACAG-3′), which were designed on the basis of the published cDNA sequence of human GRB10 (GenBank accession number AF001534). Amplification was carried out in a 20-μl reaction volume containing 1 μl of reverse-transcription product, 1 × reaction buffer, 2.5 mM dNTP mix, 1.5 mM MgCl2, 10 nmol each primer, and 0.25 U of Taq Gold polymerase (PE Biosystems). Reaction conditions were denaturation at 95°C for 10 min; followed by 20 cycles at 95°C for 60 s, at 58°C for 45 s, and at 72°C for 90 s; and then a final extension step at 72°C for 10 min. A second round of PCR was performed under the same PCR conditions, with 1/20 of the first reaction product. The PCR products were directly sequenced by the dideoxy sequencing method (BigDye Dideoxy sequencing kit; PE Biosystems). Five fetal brain tissues (19–27 wk gestation), shown to be heterozygous for the G/A single-nucleotide polymorphisms within exon 3, were examined. Only one allele—g in two samples and a in three samples—was present in each of the five RT-PCR products (fig. 1A). Therefore, human GRB10 is monoallelically expressed in the fetal brain.
Figure 1.
Imprinting of the human GRB10 gene. A, Monoallelic expression of human GRB10 in fetal brain. The genomic sequence of a normal fetal tissue heterozygous for a G/A polymorphism in exon 3 (top) and the corresponding cDNA sequence (bottom). Note that only the “A” allele is expressed in this sample. B, Determination of the parental origin of the human chromosome 7 in somatic cell hybrids GM10791 and GM13302, which carry a whole chromosome 7 and a derivative chromosome der(7)t(7;17)(q36;q22), respectively: (1) both hybrids harbor PEG1, as evidenced by positive amplification with primer pair 5′-caatagtccacttcttacc-3′ and 5′-ggccgagatcttttaatc-3′ (Riesewijk et al. 1998), corresponding to exon 6 of the human PEG1 genomic sequence; (2) expression of the imprinted PEG1 isoform 1 in GM10791, mUPD7 lymphoblastoid cell line GM11496, GM13302, and a paternal upd(7) (pUPD7) lymphoblastoid cell line (Pan et al. 1998) analyzed by RT-PCR with GRB83 and GRB44, where PEG1 is expressed in GM13302 and pUPD7 but not in GM10791 or mUPD7 and the housekeeping gene, HPRT, is used as a control; (3) methylation status of the promoter of human PEG1 in each cell line, where genomic DNA, treated with bisulfite, is amplified with PEGMF-PEGMR pair and PEG1UF-PEG1UR, specific for the methylated and the unmethylated allele, respectively, and the promoter is methylated in GM10791 and in mUPD7 but is unmethylated in GM13302 and pUPD7. Collectively, the human chromosome 7 is paternally derived in GM13302 and maternally derived in GM10791. C, Expression of GRB10 in GM10791 and GM13302, where (1) both hybrids harbor GRB10, as evidenced by positive amplification, with primer pair GRB7 and GRB8, and (2) GRB10 is expressed in GM10791, which carries a maternally derived chromosome 7, but not in GM13302, which carries a paternally derived chromosome 7.
The parental origin of the expressed allele of human GRB10 was determined by our taking advantage of the recent finding that imprinting of human genes is maintained in human-rodent somatic hybrid cells (Gabriel et al. 1998). Somatic hybrid cells produced by crossing CHO cells and human lymphocytes retaining a single human chromosome 7 (GM10791 and GM13302; Coriell Cell Repositories) were investigated by RT-PCR and methylation-specific PCR (Herman et al. 1996; Kosaki et al. 1997, in press; Kubota et al. 1997). GM10791 and GM13302 carry a single chromosome 7 and a derivative chromosome der(7)t(7;17)(q36;q22), respectively (fig. 1B). Standard tissue-culture techniques were used to propagate the hybrid cell lines according to the supplier's recommendations, and genomic DNA and cDNA were extracted from the cells by commercial kits (QIAGEN). Expression of PEG1, a human imprinted gene on chromosome 7, known to be transcribed from the paternal allele (Kobayashi et al. 1997; Riesewijk et al. 1997; Kosaki et al. 2000), was evaluated by RT-PCR with the primer pair PEG33 (5′-ATGGGATAACGCGGCCATGGTG-3′) and PEG34 (5′-ATAGTGATGTGGTCTCGGTTTGTCACTG-3′) (Kosaki et al. 2000). The cycling conditions were denaturation at 95°C for 10 min; followed by 40 cycles at 95°C for 1 min, at 58°C for 1 min, and at 72°C for 2 min; and then a final extension step at 72°C for 10 min. PEG1 isoform 1 (Kosaki et al. 2000), the imprinted isoform, was expressed in GM13302 but not in GM10791 (fig. 1C). Thus, the human chromosome 7 in the somatic hybrid cells was derived paternally in GM13302 and maternally in GM10791.
To confirm this assignment of parental origin, the methylation status of the CpG island in the human PEG1 gene, which undergoes parent-of-origin–specific methylation (Kobayashi et al. 1997; Riesewijk et al. 1997), was evaluated by methylation-specific PCR, as described elsewhere (Kosaki et al., in press). PEGMF (5′-TAGTTGCGTTTCGTAAGCGTAGTGTC-3′) and PEGMR (5′-ACACAATCCTCCGCTCGCCTA-3′) were used to amplify the methylated allele, and PEG1UF (5′-GTGGTAGTTGTGTTTTGTAAGTGTAGTGTT-3′) and PEG1UR (5′-CACACAATCCTCCACTCACCTACA-3′) were used for the unmethylated allele. In GM13302 the promoter of PEG1 isoform 1 (Kosaki et al. 2000), was exclusively unmethylated, whereas in GM10791 the promoter was exclusively methylated. Hence, expression of PEG1 and methylation of the promoter of PEG1 collectively indicated that the human chromosome 7 was derived paternally in GM13302 and maternally in GM10791.
Last, expression of GRB10 was evaluated by RT-PCR, with the GRB83-GRB44 primers under PCR conditions the same as those used for amplification of PEG1. A GRB10 amplicon was present in GM10791, which contains a maternally derived chromosome 7, but not in GM13302, which contains a paternally derived chromosome 7. Hence, it can be concluded that human GRB10 is imprinted and expressed solely from the maternal allele.
After demonstrating maternal allele–specific expression, we undertook a mutation analysis of GRB10 in two groups of patients with RSS, one from Japan and the other from Europe. The first group consisted of 30 unrelated Japanese patients with RSS who fulfilled at least three of the following diagnostic criteria (Price et al. 1999): low birth weight (<−2SD); short stature at the time of diagnosis (<−2SD); characteristic facial features; and facial, limb, or trunk asymmetry. Clinical information and material for analysis were provided by clinicians throughout Japan. Appropriate informed consent was obtained from all subjects. Karyotypes were normal, and mUPD7 was excluded by either genotyping analysis or methylation analysis of PEG1 (Riesewijk et al. 1997; Kosaki et al., in press). Cell lines from the second group of patients with RSS were derived from the European Collection of Cell Cultures. A total of 28 RSS cell lines deposited by Richard Trembath, who defined the diagnostic criteria for RSS (Price et al. 1999), were analyzed.
Using primers based on the reported genomic sequence of human GRB10 (GenBank accession numbers AF073363–AF073378; Angrist et al. 1998), we amplified each of the 16 exons of GRB10 from genomic DNA. The PCR conditions were denaturation at 95°C for 10 min; followed by 35 cycles at 95°C for 1 min, at 58°C for 1 min, and at 72°C for 1 min; and a final extension step at 72°C for 10 min. The primer sequences can be obtained from the corresponding author, on request. PCR products were subjected to denaturing high-performance liquid chromatography (WAVE; Transgenomic) (Liu et al. 1998; Wagner et al. 1999), and, when abnormal chromatographic patterns were detected, the PCR products were sequenced on an automated sequencer (ABI310; PE Biosystems).
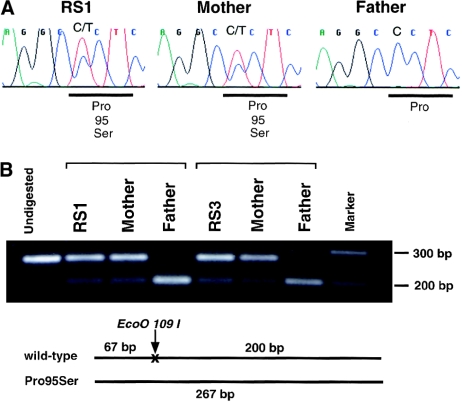
The mutation analysis revealed that two patients with RSS—RS1 and RS3—had a heterozygous C→T transition that was not present in 100 ethnically matched controls (200 chromosomes) (fig. 2). The base change was confirmed by EcoO109I digestion. This transition leads to a proline→serine substitution at codon 95 (P95S), which is located in the amino-terminal region of GRB10. Patient RS1, delivered at 39 wk gestation after an uneventful pregnancy, had a birth weight of 1.80 kg (−3.5 SD) and a length of 42.5 cm (−3.5 SD). At age 3.5 years, her weight was 10.5 kg and her height was 86.5 cm (both −3.5 SD). She had facial features that were characteristic of RSS, including triangular facies, downturned corners of the mouth, and fifth-finger clinodactyly. The P95S substitution was derived from the mother, who had normal height (163 cm) and no features of RSS. The unaffected mother received the mutant allele from her father (i.e., the maternal grandfather). RS3, who was unrelated to RS1 and was delivered at 39 wk gestation after an uneventful pregnancy, had a birth weight of 2.55 kg (−2SD) and a length of 45.5 cm (−2SD). He had the same characteristic physical features as patient RS1. The P95S substitution was derived from his mother, who had a height of 153 cm (−1SD). DNA samples from the maternal grandparents were not available.
Figure 2.
Identification of the GRB10 mutation. Exon 3 of GRB10 was amplified from genomic DNA of patients affected with Russell-Silver syndrome. Two patients, RS1 and RS3, had a heterozygous missense mutation (cct→tct, Pro95Ser) that was maternally derived. Primers GRB7 and GRB8 were used. A, Automated sequencing of the PCR products. B, Restriction enzyme digest of the PCR products. The missense mutation ablates an Eco O-109 I site. The mutant allele is demonstrated as a band at 267 bp.
Both the identification of the P95S substitution in the proline-rich amino-terminal region of GRB10 in two of 58 patients with RSS and the absence of similar mutations in 100 normal individuals suggest that the amino acid substitution may result in functionally significant change in the critical RSS-related gene on chromosome 7. The documented paternal origin of the mutated P95S allele in the mother of patient RS1 is consistent with the normal phenotype observed in this individual, in spite of the inheritance of the mutant allele. Although she possesses an allele with the same mutation as her affected daughter, this paternally derived allele would not be expressed in the mother. Ultimately, direct assessment of the effects of the mutation on the functional properties of the GRB10 protein will be required in order to verify their causal role in RSS.
Since GRB10 is a maternally expressed gene, it is expected to be overexpressed in patients with RSS who have maternal UPD7 or maternally derived duplication of 7p12 (Joyce et al. 1999; Monk et al. 2000). By analogy, the GRB10 mutations that we have identified are likely to result in a gain of function of the GRB10 protein. Functional studies are warranted to evaluate how the proline→serine substitution actually affects the activity of GRB10. Although several reports have provided evidence for inhibitory effects of GRB10 on insulin, IGF-I, and GH receptor signaling (O’Neill et al. 1996; Morrione et al. 1997; Moutoussamy et al. 1998), a recent study demonstrated positive growth-regulatory effects of GRB10 in cultured fibroblasts (Wang et al. 1999). The investigation of both the functional significance of the GRB10 mutations that we have identified and their relevance to growth failure in patients with RSS should help to define the actions and physiological importance of GRB10.
It is intriguing to note that the amino acid substitution is present in the proline-rich amino-terminal region of GRB10, which determines binding interactions with SH3-domain–containing proteins (Wojcik et al. 1999). Although amino acids corresponding precisely to P95 are not evident in mouse Grb10 (Ooi et al. 1995; Laviola et al. 1997), the absence of sequence identity at this position does not rule out a functionally significant effect of the P95S mutation in humans. Indeed, multiple known disease-causing substitutions have been identified involving amino acid residues that are not identical in other homologous human and mouse genes.
In the present study, we have demonstrated GRB10 coding-sequence mutations in only a small subset of patients with RSS. Such a low prevalence of GRB10 mutations is not unexpected, because the etiology of RSS is most likely genetically heterogeneous (Wakeling et al. 1998). Studies have shown that the RSS phenotype is associated not only with chromosome 7 abnormalities but also with other chromosomal aberrations, including balanced translocation involving 17q (Midro et al. 1993) and deletion of 15q26-qter (Wilson et al. 1985; Rogan et al. 1996).
It has been proposed that the RSS phenotype in patients with 15q26-qter deletions results from hemizygosity for the IGF-I receptor (IGF1R) (Peoples et al. 1995; Siebler et al. 1995). Because GRB10 may negatively regulate IGF-I receptor–mediated growth and mitogenesis (O'Neill et al. 1996; Morrione et al. 1997), it is conceivable that overexpression of GRB10 (e.g., mUPD7 and duplication of 7p11-13) and haploinsufficiency of the IGF-I receptor (e.g., 15q deletion) would confer a similar phenotype. Since hemizygosity for IGF-1 also has been associated with a phenotype similar to that of RSS (Woods et al. 1996), we suggest that loss-of-function mutations in IGF1 or its receptor and, most probably, gain-of-function mutations in GRB10, as a negative modulator of the IGF-I receptor, all lead to a common RSS phenotype. The documentation of GRB10 mutations in two patients with RSS in this study further supports the concept that defects in the insulin-like growth-factor axis all lead to the common RSS phenotype.
In addition to its effects on the IGF-I receptor–signaling pathway, it recently has been shown that GRB10 can associate with the growth hormone (GH) receptor and inhibit transcription of a GH reporter gene (Moutoussamy et al. 1998). This in vitro finding suggests that GRB10 mutations may affect GH as well as IGF-I signaling. Since some patients with RSS respond to GH therapy and others do not (Stanhope et al. 1998), it will be important ultimately to determine whether the presence of mutations in GRB10 correlates with the effectiveness of GH therapy.
In summary, we have shown that the human GRB10 gene is subject to genomic imprinting. A mutation analysis of GRB10 in 58 unrelated patients with RSS identified two individuals with a P95S substitution in the amino-terminal region of the GRB10 protein. In both cases, the P95S mutant allele was inherited from the mother. The fact that monoallelic GRB10 expression was observed from the maternal allele in this study suggests but does not prove that these maternally transmitted mutant alleles may contribute to the RSS phenotype.
Acknowledgments
We thank Taichi Suzuki from Tokyo Technical College for excellent laboratory assistance. This work was supported in part by a grant from the Pharmacia-Upjohn Fund for Growth & Development Research and by the Foundation for Growth Science in Japan.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Coriell Cell Repository, http://locus.umdnj.edu/nigms
- European Collection of Cell Cultures, http://fuseiv.star.co.uk/camr/
- GenBank, http://www.ncbi.nlm.nih.gov/GenBank/ (for sequences of cDNA [accession number AF001534] and genomic DNA [accession numbers AF073363–AF073378] of human GRB10)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for RSS [MIM 180860])
References
- Al-Fifi S, Teebi AS, Shevell M (1996) Autosomal dominant Russell-Silver syndrome. Am J Med Genet 61:96–97 [DOI] [PubMed] [Google Scholar]
- Angrist M, Bolk S, Bentley K, Nallasamy S, Halushka MK, Chakravarti A (1998) Genomic structure of the gene for the SH2 and pleckstrin homology domain-containing protein GRB10 and evaluation of its role in Hirschsprung disease. Oncogene 17:3065–3070 [DOI] [PubMed] [Google Scholar]
- Bernard LE, Penaherrera MS, Van Allen MI, Wang MS, Yong SL, Gareis F, Langlois S, et al (1999) Clinical and molecular findings in two patients with Russell-Silver syndrome and UPD7: comparison with non-UPD7 cases. Am J Med Genet 87:230–236 [PubMed] [Google Scholar]
- Butler MG, Fogo AB, Fuchs DA, Collins FS, Dev VG, Phillips JA III (1988) Two patients with ring chromosome 15 syndrome. Am J Med Genet 29:149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattanach BM, Shibata H, Hayashizaki Y, Townsend KM, Ball S, Beechey CV (1998) Association of a redefined proximal mouse chromosome 11 imprinting region and U2afbp-rs/U2af1-rs1 expression. Cytogenet Cell Genet 80:41–47 [DOI] [PubMed] [Google Scholar]
- Christensen MF, Nielsen J (1978) Deletion short arm 18 and Silver-Russell syndrome. Acta Paediatr Scand 67:101–103 [DOI] [PubMed] [Google Scholar]
- Duncan PA, Hall JG, Shapiro LR, Vibert BK (1990) Three-generation dominant transmission of the Silver-Russell syndrome. Am J Med Genet 35:245–250 [DOI] [PubMed] [Google Scholar]
- Eggermann T, Eggermann K, Mergenthaler S, Kuner R, Kaiser P, Ranke MB, Wollmann HA (1998) Paternally inherited deletion of CSH1 in a patient with Silver-Russell syndrome. J Med Genet 35:784–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermann T, Wollmann HA, Kuner R, Eggermann K, Enders H, Kaiser P, Ranke MB (1997) Molecular studies in 37 Silver-Russell syndrome patients: frequency and etiology of uniparental disomy. Hum Genet 100:415–419 [DOI] [PubMed] [Google Scholar]
- Gabriel JM, Higgins MJ, Gebuhr TC, Shows TB, Saitoh S, Nicholls RD (1998) A model system to study genomic imprinting of human genes. Proc Natl Acad Sci USA 95:14857–14862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 93:9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome CA, Scherer SW, Tsui LC, Gietz RD, Triggs-Raine B (1997) Assignment of growth factor receptor-bound protein 10 (GRB10) to human chromosome 7p11.2-p12. Genomics 40:215–216 [DOI] [PubMed] [Google Scholar]
- Joyce CA, Sharp A, Walker JM, Bullman H, Temple KI (1999) Duplication of 7p12.1-p13, including GRB10 and IGFBP1, in a mother and daughter with features of Silver-Russell syndrome. Hum Genet 105:273–280 [DOI] [PubMed] [Google Scholar]
- Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, et al (1995) Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat Genet 11:52–59 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Kohda T, Miyoshi N, Kuroiwa Y, Aisaka K, Tsutsumi O, Kaneko-Ishino T, et al (1997) Human PEG1/MEST, an imprinted gene on chromosome 7. Hum Mol Genet 6:781–786 [DOI] [PubMed] [Google Scholar]
- Kosaki K, Kosaki R, Craigen WJ, Matsuo N (2000) Isoform-specific imprinting of the human PEG1/MEST gene. Am J Hum Genet 66:309–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki K, Kosaki R, Robinson W, Shaffer LS, Craigen WJ, Sato S, Matsuo N. Diagnosis of maternal UPD 7 by methylation specific PCR. J Med Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki K, McGinniss MJ, Veraksa AN, McGinnis WJ, Jones KL (1997) Prader-Willi syndrome and Angelman syndromes: diagnosis with a bisulfite treated–methylation specific PCR method. Am J Med Genet 73:308–313 [PubMed] [Google Scholar]
- Kotzot D, Schmitt S, Bernasconi F, Robinson WP, Lurie IW, Ilyina H, Mehes K, et al (1995) Uniparental disomy 7 in Silver-Russell syndrome and primordial growth retardation. Hum Mol Genet 4:583–587 [DOI] [PubMed] [Google Scholar]
- Kubota T, Das S, Christian SL, Baylin SB, Herman JG, Ledbetter DH (1997) Methylation-specific PCR simplifies imprinting analysis. Nat Genet 16:16–17 [DOI] [PubMed] [Google Scholar]
- Laviola L, Giorgino F, Chow JC, Baquero JA, Hansen H, Ooi J, Zhu J, et al (1997) The adapter protein Grb10 associates preferentially with the insulin receptor as compared with the IGF-I receptor in mouse fibroblasts. J Clin Invest 99:830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Smith D, Rechtzigel KJ, Thibodeau SN, James DC (1998) Denaturing high performance liquid chromatography (DHPLC) used in the detection of germline and somatic mutations. Nucleic Acids Res 26:1396–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midro AT, Debek K, Sawicka A, Marcinkiewicz D, Rogowska M (1993) Second observation of Silver-Russell syndrome in a carrier of a reciprocal translocation with one breakpoint at site 17q25. Clin Genet 44:53–55 [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Kuroiwa Y, Kohda T, Shitara H, Yonekawa H, Kawabe T, Hasegawa H, et al (1998) Identification of the Meg1/Grb10 imprinted gene on mouse proximal chromosome 11, a candidate for the Silver-Russell syndrome gene. Proc Natl Acad Sci USA 95:1102–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk D, Wakeling EL, Proud V, Hitchins M, Abu-Amero SN, Stanier P, Preece MA, et al (2000) Duplication of 7p11.2-p13, including GRB10, in Silver-Russell syndrome. Am J Hum Genet 66:36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrione A, Valentinis B, Resnicoff M, Xu S, Baserga R (1997) The role of mGrb10alpha in insulin-like growth factor I-mediated growth. J Biol Chem 272:26382–26387 [DOI] [PubMed] [Google Scholar]
- Moutoussamy S, Renaudie F, Lago F, Kelly PA, Finidori J (1998) Grb10 identified as a potential regulator of growth hormone (GH) signaling by cloning of GH receptor target proteins. J Biol Chem 273:15906–15912 [DOI] [PubMed] [Google Scholar]
- O'Neill T, Rose DW, Pillay TS, Hotta K, Olefsky JM, Gustafson TA (1996) Interaction of a GRB-IR splice variant (a human GRB10 homolog) with the insulin and insulin-like growth factor I receptors: evidence for a role in mitogenic signaling. J Biol Chem 271:22506–22513 [DOI] [PubMed] [Google Scholar]
- Ooi J, Yajnik V, Immanuel D, Gordon M, Moskow JJ, Buchberg AM, Margolis B (1995) The cloning of Grb10 reveals a new family of SH2 domain proteins. Oncogene 10:1621–1630 [PubMed] [Google Scholar]
- Pan Y, McCaskill CD, Thompson KH, Hicks J, Casey B, Shaffer LG, Craigen WJ (1998) Paternal isodisomy of chromosome 7 associated with complete situs inversus and immotile cilia. Am J Hum Genet 62:1551–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples R, Milatovich A, Francke U (1995) Hemizygosity at the insulin-like growth factor I receptor (IGF1R) locus and growth failure in the ring chromosome 15 syndrome. Cytogenet Cell Genet 70:228–234 [DOI] [PubMed] [Google Scholar]
- Preece MA, Price SM, Davies V, Clough L, Stanier P, Trembath RC, Moore GE (1997) Maternal uniparental disomy 7 in Silver-Russell syndrome. J Med Genet 34:6–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SM, Stanhope R, Garrett C, Preece MA, Trembath RC (1999) The spectrum of Silver-Russell syndrome: a clinical and molecular genetic study and new diagnostic criteria. J Med Genet 36:837–842 [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Duenas ML, Medina C, Ocampo-Campos R, Rivera H (1992) Severe Silver-Russell syndrome and translocation (17;20) (q25;q13). Clin Genet 41:51–53 [DOI] [PubMed] [Google Scholar]
- Riesewijk AM, Blagitko N, Schinzel AA, Hu L, Schulz U, Hamel BC, Ropers HH, et al (1998) Evidence against a major role of PEG1/MEST in Silver-Russell syndrome. Eur J Hum Genet 6:114–120 [DOI] [PubMed] [Google Scholar]
- Riesewijk AM, Hu L, Schulz U, Tariverdian G, Hoglund P, Kere J, Ropers HH, et al (1997) Monoallelic expression of human PEG1/MEST is paralleled by parent-specific methylation in fetuses. Genomics 42:236–244 [DOI] [PubMed] [Google Scholar]
- Roback EW, Barakat AJ, Dev VG, Mbikay M, Chretien M, Butler MG (1991) An infant with deletion of the distal long arm of chromosome 15 (q26.1-qter) and loss of insulin-like growth factor I receptor. Am J Med Genet 38:74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaux V, Fraikor A, Favara B, Richer M (1981) Silver-Russell syndrome: a family with symmetric and asymmetric siblings. Arch Pathol Lab Med 105:157–159 [PubMed] [Google Scholar]
- Rogan PK, Seip JR, Driscoll DJ, Papenhausen PR, Johnson VP, Raskin S, Woodward AL, et al (1996) Distinct 15q genotypes in Russell-Silver and ring 15 syndromes. Am J Med Genet 62:10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A (1954) A syndrome of intrauterine dwarfism recognizable at birth with craniofacial dysostosis, disproportionate short arms and other anomalies. Proc R Soc Med 47:1040–1044 [PubMed] [Google Scholar]
- Schinzel AA, Robinson WP, Binkert F, Fanconi A (1994) An interstitial deletion of proximal 8q (q11-q13) in a girl with Silver-Russell syndrome-like features. Clin Dysmorphol 3:63–69 [PubMed] [Google Scholar]
- Siebler T, Lopaczynski W, Terry CL, Casella SJ, Munson P, De Leon DD, Phang L, et al (1995) Insulin-like growth factor I receptor expression and function in fibroblasts from two patients with deletion of the distal long arm of chromosome 15. J Clin Endocrinol Metab 80:3447–3457 [DOI] [PubMed] [Google Scholar]
- Silver H, Kiyasu W, George J, Deamer W (1953) Syndrome of congenital hypertrophy, shortness of stature and elevated gonadotropins. Pediatrics 12:356–368 [PubMed] [Google Scholar]
- Stanhope R, Albanese A, Azcona C (1998) Growth hormone treatment of Russell-Silver syndrome. Horm Res 49 Suppl 2:37–40 [PubMed] [Google Scholar]
- Tamura T, Tohma T, Ohta T, Soejima H, Harada N, Abe K, Niikawa N (1993) Ring chromosome 15 involving deletion of the insulin-like growth receptor gene in a patient with features of Silver-Russell syndrome. Clin Dysmorphol 2:106–113 [PubMed] [Google Scholar]
- Teebi AS (1992) Autosomal recessive Silver-Russell syndrome. Clin Dysmorphol 1:151–156 [PubMed] [Google Scholar]
- Wagner TMU, Hirtenlehner K, Shen P, Moeslinger R, Muhr D, Fleischmann E, Concin H, et al (1999) Global sequence diversity of BRCA2: analysis of 71 breast cancer families and 95 control individuals of worldwide populations. Hum Mol Genet 8:413–423 [DOI] [PubMed] [Google Scholar]
- Wakeling EL, Abu-Amero S, Price SM, Stanier P, Trembath RC, Moore GE, Preece MA (1998) Genetics of Silver-Russell syndrome. Horm Res 49 Suppl 2:32–36 [PubMed] [Google Scholar]
- Wang J, Dai H, Yousaf N, Moussaif M, Deng Y, Boufelliga A, Swamy OR, et al (1999) Grb10, a positive, stimulatory signaling adapter in platelet-derived growth factor BB-, insulin-like growth factor I-, and insulin-mediated mitogenesis. Mol Cell Biol 19:6217–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GN, Sauder SE, Bush M, Beitins IZ (1985) Phenotypic delineation of ring chromosome 15 and Russell-Silver syndromes. J Med Genet 22:233–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik J, Girault JA, Labesse G, Chomilier J, Mornon JP, Callebaut I (1999) Sequence analysis identifies a ras-associating (RA)–like domain in the N-termini of band 4.1/JEF domains and in the Grb7/10/14 adapter family. Biochem Biophys Res Commun 259:113–120 [DOI] [PubMed] [Google Scholar]
- Woods KA, Camacho-Hubner C, Savage MO, Clark AJ (1996) Intrauterine growth retardation and postnatal growth failure associated with deletion of the insulin-like growth factor I gene. N Engl J Med 335:1363–1367 [DOI] [PubMed] [Google Scholar]