Abstract
The results of genetic linkage studies for autism have suggested that a susceptibility locus for the disease is located on the long arm of chromosome 7 (7q). An autistic individual carrying a translocation, t(7;13)(q31.3;q21), with the chromosome 7 breakpoint located in the region of 7q implicated by genetic studies was identified. A novel gene known as “RAY1” (or “FAM4A1”) was found to be directly interrupted by the translocation breakpoint. The gene, which was found to be encoded by 16 exons with evidence of alternative splicing, spanned ⩾220 kb of DNA at 7q31.3. Mutation screening of the entire coding region in a set of 27 unrelated autistic individuals failed to identify phenotype-specific variants, suggesting that coding region mutations are unlikely to be involved in the etiology of autism. Apparent homologues of RAY1 have also been identified in mouse, rat, pig, chicken, fruit fly, and nematode. The human and mouse genes share similar splicing patterns, and their predicted protein products are 98% identical.
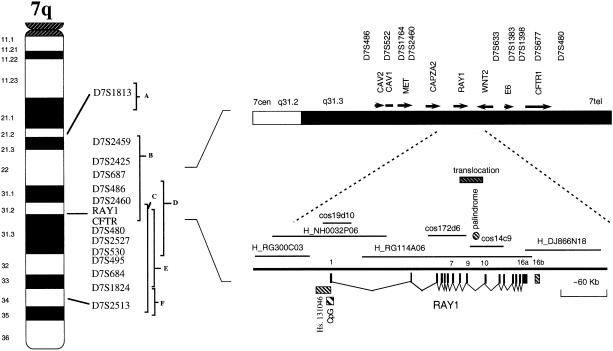
Autism, or autistic disorder (AD [MIM 209850]), a severe and debilitating developmental disorder with onset in childhood, is characterized by impaired communication and social interaction coupled with preoccupations, repetitive and ritualistic behaviors, and a restricted pattern of interests and activities. The prevalence of typical autism has been estimated to be 5.5/10,000 (Fombonne 1999), whereas a broader phenotype of autism-spectrum disorders has a population prevalence of 18.7/10,000 (Fombonne 1999). The contribution of genetic factors to autism is well established, but the mode of genetic transmission is unclear. Results from the first genomewide scan for linkage to autism were published (International Molecular Genetic Study of Autism Consortium [IMGSAC] 1998), and regions on six chromosomes (chromosomes 4, 7, 10, 16, 19, and 22) with multipoint maximum LOD scores (MLS) of >1 were identified. As shown in figure 1, the most significant MLS (3.55) was found with genetic markers residing on the long arm of chromosome 7 (7q). Results from other genome scans have also suggested that 7q might contain a putative autism-susceptibility locus (fig. 1; Ashley-Koch et al. 1999; Barrett et al. 1999; Philippe et al. 1999), with some evidence to suggest maternal imprinting at the locus (Ashley-Koch et al. 1999). Linkage results for a severe speech and language disorder have also implicated a locus (SPCH1 [MIM 602081]) within the same general region of 7q (Fisher et al. 1998).
Figure 1.
Ideogram of the long arm of chromosome 7 indicating the relative positions of positive linkage results from genome scans for autism and severe speech and language disorder (from Barrett et al. 1999 [A]; Fisher et al. 1998 [B; for SPCH1]; IMGSAC 1998 [C]; Philippe et al. 1999 [D]; Ashley-Koch et al. 1999 [E]; and Barrett et al. 1999 [F]). The position of RAY1 and the translocation breakpoint for t(7;13)(q31.2;q21), relative to neighboring genes and DNA markers, is also shown. RAY1 contains 16 exons and spans two BAC clones and one PAC clone. Exon 7 is alternatively spliced, and alternate 3′ terminal exons (16a or 16b), are employed in different transcripts. The translocation breakpoint probably occurs between exons 9 and 10 (cosmid clone 14c9 is translocated and 172d6 is not translocated). The relative positions of the CpG island, antisense transcript Hs. 131046, and the 139-bp palindrome are also shown. The end sequences of cosmids cos14c9, cos 19d10, cos172d6, and BAC H_NH0032P06 were generated, which allowed them to be aligned to the existing genomic sequence.
A patient with autism who had a translocation, t(7;13)(q31.2;q21), involving the implicated region of 7q was identified. The translocation was transmitted by an unaffected mother. Diagnosis of the patient, a 12-year-old boy, was made by use of DSM-IIIR diagnostic criteria for autism, using the Autism Diagnostic Interview (Le Couteur et al. 1989) and the Autism Diagnostic Observational Schedule (Lord et al. 1989). Following the hypothesis that the translocation might disrupt an autism-susceptibility gene, we mapped the site of the breakpoint by use of FISH analysis of metaphase spreads from a lymphoblastoid cell line from the patient. A bacterial-artificial-chromosome (BAC) clone (H_RG114A06 [GenBank accession number AC002542]) that spanned the breakpoint was identified. By use of exon-specific cosmid probes for the gene described below, the translocation breakpoint could be positioned between exons 9 and 10. Of interest is the fact that this region contains an almost perfectly symmetrical palindromic sequence that is 139 bp long (nucleotides 126287–126425). Nonhomologous recombination at a high frequency has been observed for palindromic transgenes in mice (Akgun et al. 1997). Thus, it is conceivable that this sequence may be the actual position of the translocation breakpoint in this individual. Spectral karyotyping was also performed, and the results lent further support to the conclusion that additional rearrangements were not present (data not shown). Clone H_RG114A06 was localized within the 7q critical region for linkage for autism, just proximal to CFTR and between genes WNT2 and CAPZA2 (fig. 1).
A combination of DNA sequence analysis and cDNA library screening was used to clone a gene that spanned the breakpoint. In brief, the expressed-sequence-tag (EST) clone IMAGE #724082 (Research Genetics), which mapped to H_RG114A06, was used to screen a human colon carcinoma cDNA library and a human fetal brain cDNA library. Through alignment of the cDNA sequence to the genomic DNA sequence, it was possible to identify a gene that consisted of 16 exons and spanned ⩾220 kb. This gene has been named “RAY1” (“FAM4A1” [approved by the Human Genome Organization nomenclature committee]; GenBank accession numbers AF234882 and AF234883). Alternatively spliced transcripts differing at exon 7 and having two different exon 16s, to give transcripts of 2,047–2,760 bp with corresponding predicted open reading frames (ORFs) of 554–585 amino acids, respectively, have been identified. Two additional exons, which have been identified in ESTs (IMAGE #1840889 [GenBank accession number AI221163] and IMAGE #1628386 [GenBank accession number AI018574]) but not in full-length clones, lie at nucleotides 162185–162390 and 162922–162994 (H_RG114A06) between exons 12 and 13. Exon 1 was found to be upstream of clone H_RG114A06, on BAC clone H_NH0032P06, which bridges the gap between BAC clones H_RG114A06 and H_RG300C03. This positions exon 1 at 90–150 kb upstream of exon 2 (fig. 1). A cosmid clone (cos19d10) containing exon 1 was isolated, and, through DNA sequencing of subclones, 2.1 kb of sequence flanking exon 1 (GenBank accession number AF234886) were determined, including a CpG island, a TATA box, and putative promoter sequences.
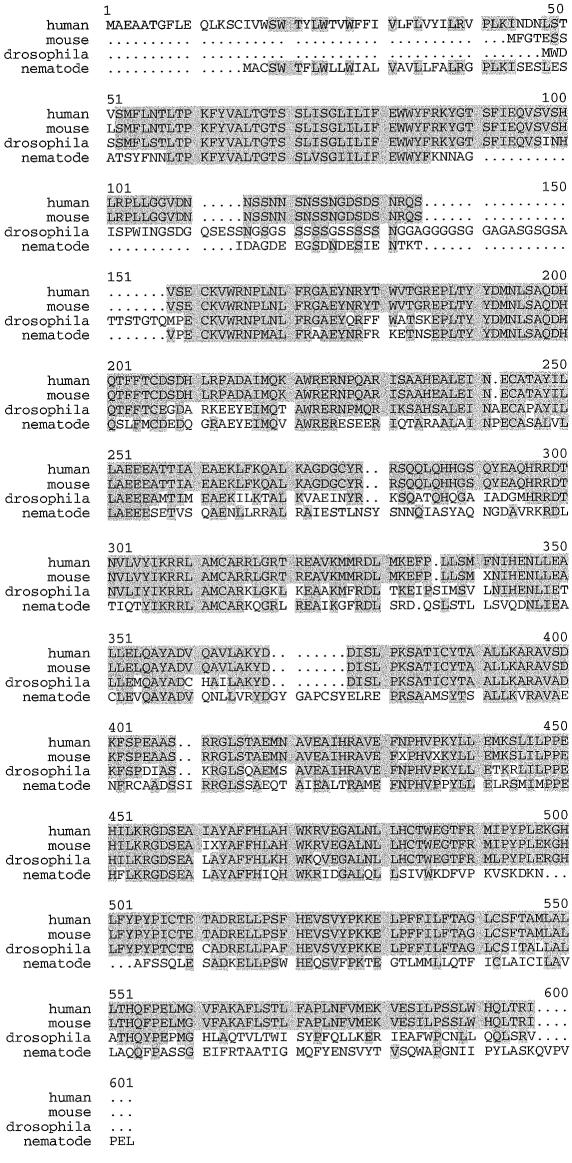
The human gene was also used to screen a mouse brain cDNA library, from which a mouse homologue, mray (Fam4a1 [GenBank accession numbers AF234884 and AF234885]), was cloned. The mouse homologue, which also demonstrates alternative splicing of exon 7 (assuming the same exon structure as human RAY1), has transcript lengths of 2,165 and 2,180 bp with corresponding predicted ORFs of 511 and 534 amino acids. Homology with the human protein is >98%. RAY1 and mray also correspond to a putative Caenorhabditis elegans gene, which encodes an ORF with 534 amino acids, and to a putative Drosophila gene, which has an ORF of 538 amino acids. The predicted C. elegans protein is 48% homologous to RAY1 and 46% homologous to mray, whereas the predicted Drosophila protein is 68% homologous to RAY1 and 66% homologous to mray (fig. 2).
Figure 2.
Amino acid sequence alignment for cloned human RAY1 (FAM4A1), mouse mray (Fam4a1), and the deduced D. melanogaster (fruit fly) and C. elegans (nematode) genes dray and nray. The D. melanogaster gene was deduced by analysis of genomic clone CDM:10211043 (GenBank accession number AC014350), by use of GENSCAN, and from EST clones (GenBank accession numbers AA438606 and AA949923). The C. elegans gene sequence was deduced by analysis of genomic clone CEF11A10 (GenBank accession number Z68297), by use of FGENES, and from EST sequences (GenBank accession numbers C45487, C40736, C34292 and M80024). 5′ mRNAs (GenBank accession numbers AW360118 and AW436819) for a pig homologue were also identified; taken together, they encode for 241 N-terminal amino acids, which are identical to 240 of the first 242 amino acids of the human sequence. A rat EST clone (GenBank accession number AW141348) encodes the 3′ end of the gene, which is 100% identical to the human C-terminal 120 amino acids.
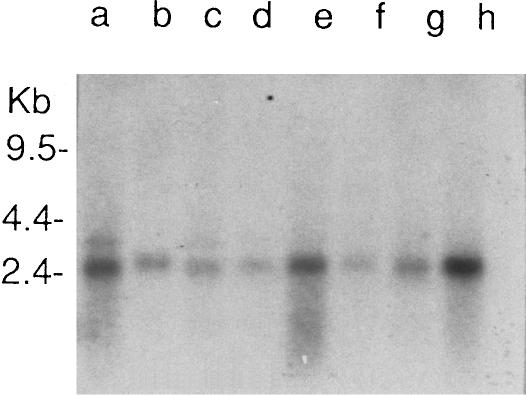
RAY1 is expressed in a wide range of tissues but is most abundant in the heart, liver, and pancreas (transcript ∼2.4 kb; fig. 3). The amino acid sequences of RAY1 and mray show no significant homology with any proteins in the database (SWISSPROT and PDB), and no consistent motifs were observed when protein analysis programs (Pfam, BLOCKS, ProDom, PRINTS, prosite pattern, and prosite profile) were used.
Figure 3.
Multiple-tissue northern blot (Clontech) for human RAY1, for selected tissues: heart (a), brain (b), placenta (c), lung (d), liver (e), skeletal muscle (f), kidney (g), and pancreas (h). A 2.4-kb transcript is abundant in most tissues—in particular, the heart, liver, and pancreas. A less abundant but slightly larger transcript can be seen for brain (∼2.5 kb), and larger transcripts (∼2.8 kb) are also present in the heart and the placenta, in addition to the 2.4-kb transcript.
A second gene, identified from ESTs from Unigene cluster Hs. 131046, is situated adjacent to RAY1 and is transcribed on the opposite strand, overlapping exon 1, just upstream of the AUG start site (nucleotide 1423; GenBank accession number AF234886). The 5′ end of the transcript starts at nucleotide 1252, and polyadenylation starts after either nucleotide 328 or nucleotide 456 (GenBank accession number AF234886). No significant ORF occurs in this transcript, possibly suggesting that this represents the 3′ UTR of a coding gene, the coding portion of which is situated in the unsequenced portion of intron 1. Alternatively, this transcript may be a noncoding RNA—possibly a riboregulator—involved in control of translation of the gene on the sense strand—namely, RAY1.
The reciprocal half of the translocation breakpoint on 13q21 was localized within CEPH YACs 925e11 and 857h4, which encompass the genetic markers CHLC.GATA42A09 and AFM311wd5 (to date, no transcripts are known to map within this BAC clone). Interestingly, an MLS of 3.0 was demonstrated for the marker D13S800 in a study by the Collaborative Linkage Study of Autism (Barrett et al. 1999), which also implicates this region in autism. D13S800 resides 15.5 cM distal to the translocation breakpoint on the female recombination map but is as close as 0.44 cM on the male recombination map (Genetic Location Database). The serotonin receptor HTR2A (MIM 182135) lies ∼3 cM proximal to the translocation (1.54 cM on the female map). Involvement of the serotonin system has long been suspected in autism, and secondary serotonin reuptake inhibitors are commonly used to treat a number of symptoms of autism.
The RAY1 gene was screened for mutations in DNA from 33 individuals with autism who were from 23 multiplex families and from 4 unrelated probands (primer sequences and PCR conditions are posted at The Chromosome 7 Project Web site or are available from the authors on request). Ascertainment and diagnostic criteria for the families were those described by Bolton et al. (1992). Each exon was screened using cycle sequencing (USB) in forward and reverse. No patient-specific variants were found, although several polymorphisms were observed, including a T→C polymorphism in intron 10, located 7 bp downstream from the donor-splice site (GenBank accession number AC002542; nucleotide 134833), and a T→C polymorphism in intron 11, located 16 bp downstream from the exon 11 donor-splice site (GenBank accession number AC002542; nucleotide 153259).
PCR followed by restriction digestion with StyI was used to analyze the frequency of the intron 10 polymorphism, and allele-specific oligonucleotide hybridization (T allele, 5′-AAT AGT TTG CGC G-3′; C allele, 5′-AAT AGC TTG CGC G-3′) was used to determine allele frequencies for the intron 11 variant in the probands with autism and a population of unrelated, unaffected individuals (N=88). No allelic association with autism was observed (exon 10 χ2=0, P=1; exon 11 χ2=0.26, P=.61), and there was no apparent segregation with autism in the 23 families genotyped.
The findings presented here suggest that disruption or mutation at RAY1 may not be directly involved in the etiology of autism and that the translocation in this patient is purely coincidental. However, it would be premature to exclude this region, because other transcripts may either overlap RAY1 or be transcribed from the opposite strand. Also, further mutation screening of RAY1 may be required in additional patients, to exclude it as a rare cause of autism. There exists the further possibility that a mutation or variant at RAY1 may have a more subtle effect as a risk factor for autism, requiring the action of other genes for phenotypic expression. To demonstrate this, disequilibrium studies in a much larger sample with autism may be required.
Acknowledgments
We would like to thank Michael Rutter, for the clinical samples used for the mutation screening set. Much gratitude is owed to Shelley Stregger, Jennifer Skaug, Xiao-Mei Shi, and Barbara Kellam at The Centre for Applied Genomics, The Hospital for Sick Children, for help with the sequencing and mapping work. S.W.S. is a scholar of the Medical Research Council of Canada.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- The Chromosome 7 Project, http://www.genet.sickkids.on.ca/chromosome7/ray1primers/ (for primer sequences and PCR sequences for mutation analysis of individuals exons on RAY1 [FAM4A1])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BAC clone H_RG114A06 [accession number AC002542], RAY1 and mray [FAM4A1 and Fam4a1; accession numbers AF234882–AF234886], and H_N0032P06, cos14c9, and cos19d10 [accession numbers AZ081238 and AZ254572–AZ254575])
- Genetic Location Database, The, http://cedar.genetics.soton.ac.uk/public_html/ldb.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AD [MIM 209850], SPCH1 [MIM 602081], and HTR2A [MIM 182135])
References
- Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, et al (1997) Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol 17:5559–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, et al (1999) Genetic studies of autistic disorder and chromosome 7. Genomics 61:227–236 [DOI] [PubMed] [Google Scholar]
- Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, et al (1999) An autosomal genomic screen for autism: Collaborative Linkage Study of Autism. Am J Med Genet (Neuropsychiatr Genet) 88:609–615 [DOI] [PubMed] [Google Scholar]
- Bolton P, Pickles A, Butler L, Summers D, Webb T, Lord C, Le Couteur A, et al (1992) Fragile X in families multiplex for autism and related phenotypes: prevalence and criteria for cytogenetic diagnosis. Psychiatr Genet 2:277–300 [Google Scholar]
- Fisher SE, Vargha-Khadem F, Watkins KE, Monaco AP, Pembrey ME (1998) Localisation of a gene implicated in a severe speech and language disorder. Nat Genet 18:168–170 [DOI] [PubMed] [Google Scholar]
- Fombonne E (1999) The epidemiology of autism: a review. Psychol Med 29:769–786 [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (1998) A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum Mol Genet 7:571–578 [DOI] [PubMed] [Google Scholar]
- Le Couteur A, Rutter M, Lord C, Rios P, Robertson S, Holdgrafer M, MacLennan J (1989) Autism diagnostic interview: a standardized investigator-based instrument. J Autism Dev Disord 19:363–387 [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, Schopler E (1989) Autism diagnostic observational schedule: a standardized observation of communicative and social behaviour. J Autism Dev Disord 19:505–524 [DOI] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, et al (1999) Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet 8:805–812 [DOI] [PubMed] [Google Scholar]