Abstract
The opportunistic fungal pathogen Candida albicans is the major causative agent of oropharyngeal candidiasis (OPC) in AIDS. The development of azoles, such as fluconazole, for the treatment of OPC has proven effective except in cases where C. albicans develops resistance to fluconazole during the course of treatment. In the present study, we used microarray technology to examine differences in gene expression from a fluconazole-susceptible and a fluconazole-resistant well-characterized, clinically obtained matched set of C. albicans isolates to identify genes which are differentially expressed in association with azole resistance. Among genes found to be differentially expressed were those involved in amino acid and carbohydrate metabolism; cell stress, cell wall maintenance; lipid, fatty acid, and sterol metabolism; and small molecule transport. In addition to CDR1, which has previously been demonstrated to be associated with azole resistance, the drug resistance gene RTA3, the ergosterol biosynthesis gene ERG2, and the cell stress genes CRD2, GPX1, and IFD5 were found to be upregulated. Several genes, such as the mitochondrial aldehyde dehydrogenase gene ALD5, the glycosylphosphatidylinositol synthesis gene GPI1, and the iron transport genes FET34 and FTR2 were found to be downregulated. Further study of these differentially regulated genes is warranted to evaluate how they may be involved in azole resistance. In addition to these novel findings, we demonstrate the utility of microarray analysis for studying the molecular mechanisms of drug resistance in pathogenic organisms.
Candida albicans is an opportunistic human fungal pathogen that causes mucosal, cutaneous, and systemic infections, including oropharyngeal candidiasis (OPC), the most frequent opportunistic infection among patients with AIDS (9, 18). Fluconazole and other azole antifungal agents have proven effective in the management of OPC; however, with increased use of these agents treatment failures have occurred. Such failures have been associated with the emergence of azole-resistant strains of C. albicans (24, 31, 32, 42).
Several mechanisms of resistance to the azole antifungal agents have been described in C. albicans (11, 12, 24, 35, 36, 40, 41). Point mutations in the gene encoding the target of the azoles, lanosterol demethylase (ERG11), result in conformational changes that prevent effective binding between the azoles and their target (36, 41). Increased expression of the ERG11 gene has also been associated with azole resistance (40). This presumably results in increased production of lanosterol demethylase, which overwhelms the capacity of the azole antifungal agent. Two classes of efflux pumps have been implicated in azole resistance; the energy-dependent ATP binding cassette (ABC) transporters and the proton motive force-dependent major facilitators (11, 12, 24, 35, 40). The genes encoding two of the ABC transporters in C. albicans, CDR1 and CDR2, as well as that encoding a major facilitator, CaMDR1, have been shown to be overexpressed in resistant isolates. Overexpression of these efflux pumps is presumed to prevent the accumulation of sufficient effective concentrations of the azole antifungal agent in the fungal cell. Overexpression of the CDR genes confers resistance to several azoles, whereas overexpression of CaMDR1 is specific to fluconazole (42). High-level azole resistance appears to accumulate gradually over time and to involve multiple mechanisms (30).
Due to the clonal nature of C. albicans, it is critical that matched sets of sensitive and resistant isolates of a single strain be used when characterizing molecular mechanisms of resistance (42). For this reason, several series of clinical isolates have been collected and initially characterized (11, 12, 24, 26, 32, 35, 40). One particular series of 17 isolates was obtained from an AIDS patient with recurrent episodes of OPC who required progressively higher doses of fluconazole due to multiple treatment failures. This series has been extensively characterized by several research groups (31, 32, 40). Earlier examination by using Ca3 typing revealed that all but one of the isolates in this set were isogenic (40). Reported MICs for these isolates ranged from 0.25 to >64 μg/ml for the initial and final isolates in this series, respectively. It has been demonstrated previously that the final isolate in this series overexpresses the ERG11, CDR1, CDR2, and CaMDR1 genes (25, 40).
Although increased gene expression is a common theme in azole antifungal resistance in C. albicans, studies of the molecular mechanisms involved in this process have focused on only a limited number of genes. Additionally, little is known about the transcriptional regulation of genes known to be involved in azole resistance. Finally, impaired drug import is a common mechanism of drug resistance; however, the mechanism by which the azole antifungal agents enter the fungal cell is poorly understood.
In the present study we used cDNA microarray analysis to identify differentially expressed genes associated with azole resistance by comparing the transcriptional profile of an azole-resistant C. albicans isolate to its matched azole-susceptible parent isolate. A number of genes were found to be differentially expressed between these isolates, including several involved in amino acid and carbohydrate metabolism; cell stress; cell wall maintenance; lipid, fatty acid, and sterol metabolism; and small molecule transport. The present study broadens the characterization of these isolates to reveal genes associated with fluconazole resistance that previously have not been identified and lays the groundwork for the application of functional genomics to this problem in pathogenic fungi.
(Preliminary results from the present study were presented at the Sixth American Society for Microbiology Conference on Candida and Candidiasis held on 13 to 17 January 2002 in Tampa, Fla.)
MATERIALS AND METHODS
Candida isolates and growth conditions.
C. albicans isolates 2-79 and 12-99 were obtained from Spencer W. Redding (32) and Theodore C. White (40). A compilation of MICs and alternate names for this set of isolates is presented in Table 1. An aliquot of glycerol stock from each isolate was diluted in YPD broth (1% yeast extract, 2% peptone, 1% dextrose) and grown overnight at 30°C in an environmental shaking incubator. Two independent sets of cultures were diluted to an optical density at 600 nm (OD600) of 0.1 to 0.2 in fresh YPD and grown as described above to early logarithmic phase to an equivalent OD for subsequent RNA isolation.
TABLE 1.
Compilation of previously reported and verified MICs for fluconazole-susceptible and -resistant C. albicans isolates
| Isolate | Alternate isolate namea | MIC (μg/ml)
|
|
|---|---|---|---|
| White (40) | The present study | ||
| 2-79 | 2 | 2 | 0.25 |
| 12-99 | 17 | >64 | >64 |
As reported by White (40).
Susceptibility testing of Candida isolates.
The MICs of fluconazole were determined by using broth microdilution techniques as described by the National Committee for Clinical Laboratory Standards (19). The cells were cultured in RPMI 1640 buffered at pH 7.0 with morpholinepropanesulfonic acid, with a starting inoculum of ca. 0.5 × 103 to 2.5 × 103 CFU/ml. Microtiter trays were incubated at 35°C in a moist, dark chamber, and the MICs were recorded after 48 h of incubation. The susceptibility endpoint was defined as the lowest concentration of antifungal agent that resulted in visible growth that was reduced by 80% compared to growth of the control. Determinations of the MICs for the isolates were performed prior to experimental use.
Total RNA isolation.
RNA was isolated by the hot phenol method (11). Briefly, cells were collected by centrifugation and snap-frozen in liquid nitrogen. Frozen cells were resuspended in 12 ml of AE buffer (50 mM sodium acetate [pH 5.2], 10 mM EDTA) at room temperature, after which 800 μl of 25% sodium dodecyl sulfate (SDS) and 12 ml of acid phenol (Fisher Scientific, Houston, Tex.) were added. The cell lysate was then incubated 10 min at 65°C with vortexing each minute, cooled on ice 5 min, and subjected to centrifugation for 15 min at 11,952 × g. Supernatants were transferred to new tubes containing 15 ml of chloroform, mixed, and subjected to centrifugation at 200 × g for 10 min. RNA was precipitated from the resulting aqueous layer by transferring that portion to new tubes containing 1 volume of isopropanol and 0.1 volume of 2 M sodium acetate (pH 5.0), thorough mixing, and then subjecting the mixture to centrifugation at 17,211 × g for 35 min at 4°C. The supernatants were removed, the pellet was resuspended in 10 ml of 70% ethanol, and the RNA collected by centrifugation at 17,211 × g for 20 min at 4°C. Supernatants were again removed, and RNA was resuspended in diethyl pyrocarbonate-treated water. The ODs were measured at 260 and 280 nm, and the integrity of the RNA was visualized by subjecting 2 to 5 μl of the sample to electrophoresis through a 1% agarose-morpholinepropanesulfonic acid gel.
Microarray preparation.
Microarrays were prepared by Incyte Genomics (St. Louis, Mo.) microarray facility by first generating DNA fragments by PCR, purifying the fragments by gel filtration, drying the DNA, and rehydrating the fragments in distilled water. Each DNA fragment was spotted by robotics onto modified glass slides and chemically bonded to the glass; the slides were then washed in distilled water, treated with 0.2% I-Block (Tropix, Bedford, Mass.) dissolved in 1× Dulbecco phosphate-buffered saline (Gibco/Invitrogen Corp., Carlsbad, Calif.) at 60°C for 30 min, and rinsed in 0.2% SDS for 2 min, followed by three 1-min washes with distilled water.
Probe preparation.
Fluorescently labeled cDNA probes were generated from duplicate sets of 2-79 and 12-99 isolate RNA samples. Total RNA was reverse transcribed with 5′ Cy3- or Cy5-labeled random 9-mers (Operon Technologies, Inc., Alameda, Calif.). The 25 μl-volume reaction mixtures, consisting of 1 μg of total RNA, 200 U of Moloney murine leukemia virus reverse transcriptase (Gibco/Invitrogen), 4 mM dithiothreitol, 1 U of RNase inhibitor (Ambion, Austin, Tex.), 0.5 mM concentrations of deoxynucleotide triphosphates, and 2 μg of labeled 9-mers in an enzyme buffer, were incubated at 37°C for 1 h and terminated by incubation for 5 min at 85°C. The paired reactions were combined and purified by using a TE-30 column (Clontech, Palo Alto, Calif.). The eluate volume was adjusted to 90 μl with distilled water and precipitated with 2 μg of glycogen, 300 μl of ethanol, and ammonium acetate at a final concentration of 0.6 M. After centrifugation the pellet was dissolved in hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.2% SDS, 1 mM dithiothreitol).
Microarray hybridization and scanning.
Each set of samples was used for separate array hybridizations with cDNA from isolate 2-79 labeled with Cy3 and cDNA from isolate 12-99 labeled with Cy5 in the first hybridization and cDNA from isolate 2-79 labeled with Cy5 and cDNA from isolate 12-99 labeled with Cy3 in the second hybridization. A total of two hybridization experiments were performed. Probes were incubated at 65°C for 5 min, applied to a glass slide, and then covered with a 22-mm2 glass coverslip. Hybridization took place at 60°C for 6.5 h in a humidified chamber; the slides were then washed three times in solutions of decreasing ionic strength. The microarrays were scanned in both Cy3 and Cy5 channels by using a GenePix scanner (Axon, Foster City, Calif.) with a 10-μm resolution. The signal was converted into a resolution of 16 bits per pixel, yielding a 65,536-count dynamic range.
Microarray data analysis.
GEMTools software (Incyte Genomics) was used for image analysis and data visualization. Each element area was determined by a gridding and detection algorithm. In order to be considered for data analysis, each GEM element was required to pass each of the following criteria in at least one channel: (i) each element must have a minimum area of 40% of the average area calculated for all elements on the slide, and (ii) the element's signal-to-background ratio must be at least 2.5. Additionally, elements for which the homogeneity of the spotted DNA could not be confirmed by PCR analysis were excluded.
The local background values were calculated from the area surrounding each element and subtracted from the total element signal values. These adjusted values were used to determine differential gene expression (Cy3/Cy5 ratio) for each element. A correction factor was applied to account for systematic differences in the probe labels by using the differential gene expression ratio to balance the Cy5 signals. In the present study, only elements with a balanced differential expression greater than or equal to 2.5 or less than or equal to −2.5 were considered in further analyses. DNA sequences were annotated on the basis of results of BLASTN searches by using GenBank (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi), the Proteome Bioknowledge library (http://www.incyte.com/sequence/proteome/index.shtml), the Stanford University sequencing database (http://www-sequence.stanford.edu/group/candida), and the CandidaDB database (http://www.pasteur.fr/Galar_Fungail/CandidaDB/).
cDNA synthesis and reverse transcription-PCR (RT-PCR).
To synthesize cDNA, ca. 2 μg of total RNA was placed in a 0.5-ml reaction tube with 1 μg of oligo(dT) primer stock (ResGen/Invitrogen Corp., Carlsbad, Calif.), and the volume was adjusted to 15.5 μl with diethyl pyrocarbonate-treated water. The mixture was incubated for 10 min at 70°C and chilled on ice 1 min, after which the remainder of the reaction mixture was added from a master mix to the reaction tube in order for each reaction to contain a 1.25 mM concentration each of dATP, dCTP, dGTP, and TTP; 40 U of RNase inhibitor (Stratagene, LaJolla, Calif.); and 25 U of Moloney murine leukemia virus reverse transcriptase (Gibco/Invitrogen) in a buffer consisting of 0.05 M Tris-HCl (pH 8.3), 75 mM KCl, and 3 mM MgCl2. After brief mixing, the reaction was incubated for 10 min at room temperature, followed by incubation at 37°C for 1 h. Finally, the reaction was heated at 90°C for 5 min and either cooled on ice for 10 min or immediately stored at −20°C until use.
PCR was performed by mixing 1 μl of the appropriate dilution of cDNA (empirically determined for each gene to give a product in the linear range), 0.5 μg of each forward and reverse primer, 2.5 U of Taq polymerase (Sigma, St. Louis, Mo.), and 0.1% Triton X-100 in EasyStart Micro50 PCR tubes and then subjecting the reaction mixture to the following reaction conditions: one cycle of 94°C for 5 min; 32 cycles of 94°C for 1 min, a gene-specific annealing temperature for 1 min, and 72°C for 2 min; and 1 cycle of 72°C for 5 min. Equivalent volumes of PCR product were applied to a 3% agarose gel and separated by gel electrophoresis in 1× TAE. Primer sequences used for amplification of specific genes by RT-PCR are shown in Table 2.
TABLE 2.
DNA sequences and melting temperatures of primers used in RT-PCR
| Primer | Sequence | Tm (°C) |
|---|---|---|
| ald5-f | GGT TCT ACT GCC ACC GGT AA | 57.6 |
| ald5-r | TTG GGC ACC CAT AAA AGT GT | 56.1 |
| cdr1-f | GGC AAT TAG TCA AGA CTC TTC TTC AG | 57.2 |
| cdr2-f | CAG CTA GAC GAA AAG CCA TGG | 57.1 |
| cdr-ra | CAC CTG GTC TCA TAA TGG CAT C | 56.7 |
| cwh8-f | AAA AAT TCC GGT CGT GAT GG | 52.7 |
| cwh8-r | TCC AAC ATC TCT AGC CAC TGA A | 57.0 |
| erg2-f | GGT GCC ATG GGT ACA ATG TT | 56.8 |
| erg2-r | CCA GCC TTG AGC CAA TTC TA | 55.9 |
| erg11-f | TAC TGC TGC TGC CAA AGC TA | 58.0 |
| erg11-r | CCC AAA TGA TTT CTG CTG GT | 54.5 |
| fet34-f | TTC AAT CCA ACT GGT GCT GA | 55.7 |
| fet34-r | ACA CGT TGC ATG AAG GCA TA | 56.3 |
| ftr2-f | CAA ACA ATC AAT GGG CTC CT | 54.5 |
| ftr2-r | AAA CCT TCA CGC AAG CAA GT | 57.0 |
| gpi1-f | TAG AGG CAG CCA ACA TTT CA | 55.5 |
| gpi1-r | ACC CAA ACC CAT CAA TAG CA | 55.6 |
| gpx1-f | TCG TCA ATG TTG CTT CCA AA | 54.2 |
| gpx1-r | GGT CAA TCC CAA AAC ACC AC | 55.2 |
| ifd5-f | TAT TGG TGC TTC CTC CAT GA | 54.7 |
| ifd5-r | CAC CAG CAA TTG GAA TCA CA | 54.7 |
| lys21-f | CGA TAC TGG TTG TGC CAT TG | 55.0 |
| lys21-r | GAT TGG CCA AGA TGG CTT TA | 54.3 |
| mdh1-f | GCT TCC AAC GCC TAC AAA GT | 56.8 |
| mdh1-r | CAA TTG AGG CAT TGG TGT TG | 53.7 |
| mdr1-f | TTT TTG GGT GGA TTC TTT GC | 53.0 |
| mdr1-r | CGG TGA TGG CTC TCA ATC TT | 55.8 |
| met3-f | AAA CGG AGG GTT TTC TCC AT | 55.5 |
| met3-r | TTC TGA ATC ACC ACG GAA CA | 55.3 |
| mir1-f | GAT TTG TCC ACT GCT GCT CA | 56.6 |
| mir1-r | GGC TGG TGG ACA GTT CAA AG | 57.1 |
| rta3-f | CGA AGG CAA ACC AAG TCC AT | 54.9 |
| rta3-r | TAC CAA TCA TTG CTG CAT CC | 54.3 |
| 18S-f | GCC AGC GAG TAT AAG CCT TG | 56.7 |
| 18S-r | AGG CCT CAC TAA GCC ATT CA | 57.3 |
The cdr-r primer was used as the reverse primer for both the cdr1-f and the cdr2-f primers.
RESULTS AND DISCUSSION
Experimental design and broad categorization of findings.
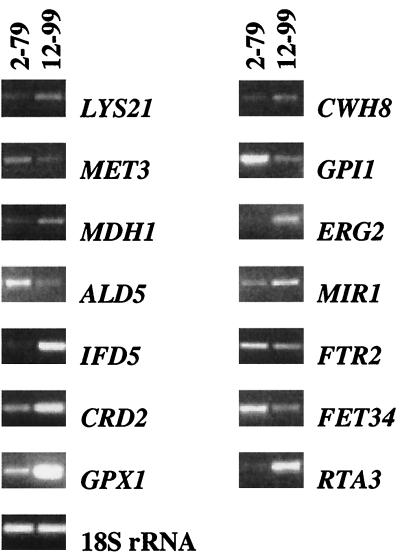
The development of microarray technology and the sequencing of the C. albicans genome makes possible a more complete identification of genes associated with drug resistance in C. albicans (Stanford C. albicans Genome Project; Stanford University, Palo Alto, Calif. [http://www-sequence.stanford.edu/group/candida]). In the present study, a portion of the C. albicans genome was examined simultaneously by using microarray technology to compare differences in gene expression between isogenic clinical isolates of fluconazole-susceptible and -resistant C. albicans. This was achieved by using the Incyte Genomics C. albicans GEM microarray described in detail previously (5). Fourteen genes were, for the first time, found to be differentially expressed in association with the azole-resistant phenotype (Table 3). Differential expression for these genes was then confirmed independently by RT-PCR (Fig. 1).
TABLE 3.
Genes that are differentially expressed in fluconazole-resistant C. albicans
| Role and gene name | Accession no. | Function | Mean fold change in gene expression ± SD |
|---|---|---|---|
| Amino acid metabolism | |||
| LYS21 | Z74230 | Lysine biosynthesis | 3.15 ± 1.25a |
| MET3 | AF164103 | Methionine biosynthesis (ATP sulfurylase) | −6.65 ± 2.899 |
| Carbohydrate metabolism | |||
| MDH1 | J02841 | Malate dehydrogenase | 3.525 ± 0.225a |
| ALD5 | U56605 | Mitochondrial aldehyde dehydrogenase | −3.825 ± 0.459a |
| Cell stress | |||
| IFD5 | U43281 | Alcohol dehydrogenase (lignocellulose degradation) | 12.45 ± 0.309a |
| CRD2 | AF268099 | Copper-binding metallothionein | 4.6 ± 0.0 |
| GPX1 | Z50123 | Glutathione peroxidase (oxidative stress response) | 2.85 ± 0.212 |
| Cell wall maintenance | |||
| CWH8 | AL033502 | Involved in mannoprotein layer generation | 3.6 ± 0.707 |
| GPI1 | Z73001 | Glucosaminyl-phosphatidylinositol anchoring | −3.0 ± 1.838 |
| Lipid, fatty acid, and sterol metabolism | |||
| ERG2 | M74037 | Ergosterol biosynthesis | 2.65 ± 1.626 |
| Small molecule transport | |||
| CDR1 | X77589 | ABC transporter in drug resistance | 3.67 ± 0.197a |
| MIR1 | M54879 | Mitochondrial phosphate transporter | 2.85 ± 0.778 |
| FTR2 | AF195776 | High-affinity iron permease | −5.9 ± 4.243 |
| FET34 | Y09329 | Multicopper oxidase | −9.4 ± 2.939a |
| Other | |||
| RTA3 | Z74757 | Unknown | 5.05 ± 2.192 |
The standard error was used due to the presence of multiple spots on each array for the gene in question.
FIG. 1.
Evaluation of differential expression by RT-PCR of genes found to be associated with azole antifungal resistance in this set of isolates. RT-PCR of C. albicans 18S rRNA was performed as a control.
The two isolates used in the present study are from a set of 17 isogenic clinical isolates that have been extensively characterized (25, 40, 43) (Table 1). It should be noted that two genes, CDR2 and MDR1, known to be differentially expressed in association with azole resistance in this set of isolates (and others) were not represented on the array used in the present study. Confirmation of differential expression of genes previously associated with azole resistance in these isolates was therefore performed by RT-PCR (Fig. 2). Consistent with previous findings by others (25, 40), we observed the genes encoding the transporters CDR1, CDR2, and CaMDR1 to be upregulated in the resistant isolate (Fig. 2). We did not, however, observe a change in expression for ERG11 between the two isolates. This could be due to loss of this genetic change between the initial characterization of these isolates and the present study, or it may be reflective of subtle changes in expression between these isolates that are not readily detectible by semiquantitative RT-PCR. Indeed, a recent analysis showed only a 1.2- to 2.3-fold change in expression of ERG11 between isolates 1 and 17 of this series (25). Yet another explanation may be the choice of controls used for normalization of gene expression. Previous studies have used ACT1 as a control compared to 18S rRNA used in the present study. It is possible that differences in the expression of ACT1 between resistant and susceptible isolates could account for this discrepancy.
FIG. 2.
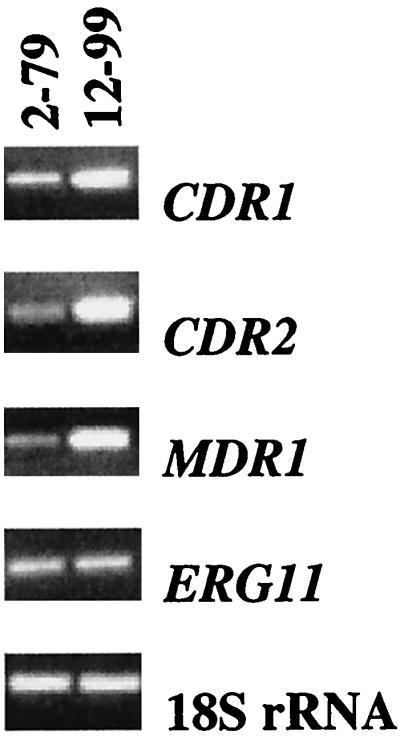
Evaluation of differential expression by RT-PCR of genes previously associated with azole antifungal resistance in this set of isolates. RT-PCR of C. albicans 18S rRNA was performed as a control.
Ergosterol biosynthesis, carbohydrate metabolism, cell wall maintenance, and iron transport genes.
ERG2 was the only gene involved in ergosterol biosynthesis found to be differentially expressed. ERG2 encodes C-8 sterol isomerase, an enzyme in the late stages of the ergosterol biosynthesis pathway responsible for the conversion of fecosterol to episterol. A relationship between the late stages of ergosterol biosynthesis and the activity of the Saccharomyces cerevisiae homolog of Cdr1p, Pdr5p, has been reported. Disruptions in ERG6, ERG2, ERG3, and ERG4 each reduced the ability of Pdr5p to confer drug resistance (17). Upregulation of ERG2 could therefore serve to provide membrane conditions that are optimal for Cdr1p and Cdr2p activity in C. albicans. Furthermore, the upregulation of ERG2 could potentially compensate for a partial inhibition of lanosterol demethylase by an azole antifungal agent. Theoretically, any lanosterol successfully converted to ignosterol could more efficiently be further converted to ergosterol.
ALD5 encodes mitochondrial aldehyde dehydrogenase which is critical to the regulation and/or biosynthesis of electron transport chain components (21). Impairment or loss of mitochondrial function has been associated with increased expression of Pdr5p in S. cerevisiae (15). Downregulation of ALD5 may therefore contribute to overexpression of CDR1 and CDR2 in the resistant isolate.
We found two genes involved in metal ion transport that were downregulated in the present study. FET34, downregulated 9.4-fold, is required for FTR1-mediated iron uptake in S. cerevisiae (1), whereas the high-affinity iron permease FTR2 was downregulated 5.9-fold. Previous studies have indicated a relationship between iron accessibility and azole resistance in C. albicans. In addition to studies that have demonstrated a synergistic effect of lactoferrin (or related compounds) and fluconazole on fungal cell killing (20, 28), Wakabayashi et al. showed that clinically isolated azole-resistant strains of C. albicans were more susceptible to lactoferrin treatment than azole-susceptible strains (39). Whether downregulation of iron transport genes such as FTR2 and FET34 in fluconazole-resistant isolates is responsible for enhanced lactoferrin susceptibility remains uncertain. Further studies should be performed to determine what role downregulation of these iron transport genes has in fluconazole resistance.
Glycosylphosphatidylinositol (GPI)-anchored proteins are critical for cell wall maintenance and integrity (33). GPI1 encodes a product that is intimately involved in GPI synthesis (23). CWH8 encodes a dolichyl pyrophosphate (Dol-P-P) phosphatase capable of converting Dol-P-P to dolichyl monophosphate (Dol-P) and Pi on the luminal surface of the endoplasmic reticulum in yeast. This process is needed for the reutilization of the dolichyl moiety in further rounds of lipid intermediate biosynthesis. Both Dol-P and Dol-P-P are released on the luminal surface of the endoplasmic reticulum during GPI anchor synthesis (10). Downregulation of GPI1 concomitant with upregulation of CWH8 may reflect altered regulation of GPI-anchored proteins that, through changes in cell wall composition, may alter susceptibility to azole antifungal agents.
RTA3 is a gene of unknown function. However, it is homologous to S. cerevisiae RTA1, which has been implicated in resistance to 7-aminocholesterol (37). Upregulation of RTA3 may also contribute to the azole-resistant phenotype.
Role of oxidative stress response genes in azole resistance.
Isolates of C. glabrata lacking the ERG11 gene have been shown to be more susceptible to neutrophils and H2O2 than isolates with ERG11 intact (16). It is therefore possible that chemical inhibition of the ERG11 gene product, lanosterol demethylase, with an azole antifungal agent may produce a similar effect. This suggests that imparting to the fungal cell such enhanced susceptibility to oxidative damage may be part of the mechanism of action of the azole antifungals. In support of this hypothesis, we found several genes involved in cell stress responses to be upregulated in association with azole resistance in this set of isolates.
LYS21 encodes one of the two homocitrate synthase enzyme isoforms that catalyze the first step in the alpha-aminoadipate pathway for the biosynthesis of lysine (14). LYS7 encodes the enzyme that catalyzes the second step in this pathway, the dehydration of homocitrate to cis-homocitrate. LYS7 has been implicated in oxidative stress protection. In S. cerevisiae, deletion of the LYS7 gene resulted in sensitivity to superoxide-generating drugs. This was associated with impaired superoxide dismutase activity (13). It is therefore possible that upregulation of LYS21 may contribute to this oxidative stress response.
CRD2 encodes a copper-binding metallothionein that plays a role in reducing oxidative stress (38). Copper is essential for enzymes involved in a multitude of biological processes such as respiration, free radical destruction, and iron homeostasis (34). GPX1 encodes glutathione peroxidase, a significant component of the glutathione and glutathione-dependent enzyme system. This system protects cells from environmental toxins and has been implicated in the resistance of tumor cells to anticancer agents, particularly those whose effects are mediated by free radicals (3, 8). Increased activity of this enzyme system is often observed in conjunction with increased activity of the ABC transporter Mdr1p in drug-resistant human cancer cells (4, 22). IFD5 is a member of a family of homologs of S. cerevisiae YPL088W, a putative alcohol dehydrogenase/oxidoreductase, and was recently shown to contain a drug response element in its promoter that leads to induction of mRNA expression upon estradiol treatment of C. albicans (27). This drug response element was shared by only three other genes, including CDR1 and CDR2. Furthermore, YPL088W was 1 of 26 genes in S. cerevisiae upregulated in the pdr1-3 mutant strain, indicating its responsiveness to PDR1 and PDR3 (7, 29).
Given that the resistant isolate in this set exhibits a stably resistant phenotype after multiple serial passages (25, 40), these changes in gene expression are likely due to a stable change in transcriptional regulation. Indeed, in S. cerevisiae, the loss of transcriptional control of azole resistance genes is often associated with single amino acid substitutions in the transcriptional regulators Pdr1p and Pdr3p (2, 6). Further examination of the gene expression profiles of azole-resistant isolates of C. albicans will likely assist in the identification of the transcription factors and regulatory elements involved in this phenotype.
The studies described here were carried out by using a cDNA microarray representing only a portion of the C. albicans genome. As this technology improves and more comprehensive arrays, such as those currently under development by Eurogentec and the Biotechnology Research Institute, National Research Council of Canada, become widely available, it will be possible to monitor changes in expression for every gene in the genome. Furthermore, as a consensus is reached on the annotation of the C. albicans genome, expanded utility of the tools of bioinformatics will allow greatly expanded functional genomics analyses of the phenomenon of azole antifungal resistance in this pathogenic fungus.
Conclusions.
Our study is the first to use microarray hybridization techniques to identify genes associated with a drug-resistant phenotype in clinical isolates of a pathogenic fungus. The ability to examine thousands of genes simultaneously allowed for not only the identification of the differential expression of genes previously shown to be associated with drug resistance but also that of a variety of other genes. Among these are the drug resistance gene RTA3; the ergosterol biosynthesis gene ERG2; the cell stress genes CRD2, GPX1, and IFD5; the iron transport genes FTR2 and FET34; and the cell wall maintenance genes CWH8 and GPI1. Each of these genes possesses putative ties to antifungal drug resistance, but they have been studied little if at all as to their specific function in this process. The present study identifies new potential targets in the development of pharmacological approaches to circumvent azole antifungal resistance and to improve the therapeutic index of this valuable class of antifungal agents. Further investigation of the functional genomics of azole antifungal resistance in this and other pathogenic fungi is warranted.
Acknowledgments
This work was supported by a 2001-2002 Society of Infectious Diseases Pharmacists/Pfizer Research Award.
We thank Spencer Redding and Theodore White for providing the isolates used in this study and Russ Lewis for performing susceptibility testing.
REFERENCES
- 1.Andrews, N. C., M. D. Fleming, and H. Gunshin. 1999. Iron transport across biologic membranes. Nutr. Rev. 57:114-123. [DOI] [PubMed] [Google Scholar]
- 2.Balzi, E., W. Chen, S. Ulaszewski, E. Capieaux, and A. Goffeau. 1987. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J. Biol. Chem. 262:16871-16879. [PubMed] [Google Scholar]
- 3.Black, S. M., and C. R. Wolf. 1991. The role of glutathione-dependent enzymes in drug resistance. Pharmacol. Ther. 51:139-154. [DOI] [PubMed] [Google Scholar]
- 4.Buser, K., F. Joncourt, H. J. Altermatt, M. Bacchi, A. Oberli, and T. Cerny. 1997. Breast cancer: pretreatment drug resistance parameters (GSH-system, ATase, P-glycoprotein) in tumor tissue and their correlation with clinical and prognostic characteristics. Ann. Oncol. 8:335-341. [DOI] [PubMed] [Google Scholar]
- 5.De Backer, M. D., T. Ilyina, X. J. Ma, S. Vandoninck, W. H. Luyten, and H. Vanden Bossche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaveau, T., A. Delahodde, E. Carvajal, J. Subik, and C. Jacq. 1994. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol. Gen. Genet. 244:501-511. [DOI] [PubMed] [Google Scholar]
- 7.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 8.Doroshow, J. H., S. Akman, F. F. Chu, and S. Esworthy. 1990. Role of the glutathione-glutathione peroxidase cycle in the cytotoxicity of the anticancer quinones. Pharmacol. Ther. 47:359-370. [DOI] [PubMed] [Google Scholar]
- 9.Feigal, D. W., M. H. Katz, D. Greenspan, J. Westenhouse, W. Winkelstein, Jr., W. Lang, M. Samuel, M., S. P. Buchbinder, N. A. Hessol, and A. R. Lifson, et al. 1991. The prevalence of oral lesions in HIV-infected homosexual and bisexual men: three San Francisco epidemiological cohorts. AIDS 5:519-525. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez, F., J. S. Rush, D. A. Toke, G. S. Han, J. E. Quinn, G. M. Carman, J. Y. Choi, D. R. Voelker, M. Aebi, and C. J. Waechter. The CWH8 gene encodes a dolichyl pyrophosphate phosphatase with a luminally oriented active site in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 276:41455-41464. [DOI] [PubMed]
- 11.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhäuser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franz, R., M. Ruhnke, and J. Morschhäuser. 1999. Molecular aspects of fluconazole resistance development in Candida albicans. Mycoses 42:453-458. [DOI] [PubMed] [Google Scholar]
- 13.Gamonet, F., and G. J. Lauquin. 1998. The Saccharomyces cerevisiae LYS7 gene is involved in oxidative stress protection. Eur. J. Biochem. 251:716-723. [DOI] [PubMed] [Google Scholar]
- 14.Garrad, R. C., and J. K. Bhattacharjee. 1992. Lysine biosynthesis in selected pathogenic fungi: characterization of lysine auxotrophs and the cloned LYS1 gene of Candida albicans. J. Bacteriol. 174:7379-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallstrom, T. C., and W. S. Moye-Rowley. 2000. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275:37347-37356. [DOI] [PubMed] [Google Scholar]
- 16.Kan, V. L., A. Geber, and J. E. Bennett. 1996. Enhanced oxidative killing of azole-resistant Candida glabrata strains with ERG11 deletion. Antimicrob. Agents Chemother. 40:1717-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur, R., and A. K. Bachhawat. 1999. The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae. Microbiology 145:809-818. [DOI] [PubMed] [Google Scholar]
- 18.Klein, R. S., C. A. Harris, C. B. Small, B. Moll, M. Lesser, and G. H. Friedland. 1984. Oral candidiasis in high-risk patients as the initial manifestation of the acquired immunodeficiency syndrome. N. Engl. J. Med. 311:354-358. [DOI] [PubMed] [Google Scholar]
- 19.Klepser, M. E., D. Malone, R. E. Lewis, E. J. Ernst, and M. A. Pfaller. 2000. Evaluation of voriconazole pharmacodynamics using time-kill methodology. Antimicrob. Agents Chemother. 44:1917-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuipers, M. E., H. G. de Vries, M. C. Eikelboom, D. K. Meijer, and P. J. Swart. 1999. Synergistic fungistatic effects of lactoferrin in combination with antifungal drugs against clinical Candida isolates. Antimicrob. Agents Chemother. 43:2635-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurita, O., and Y. Nishida. 1999. Involvement of mitochondrial aldehyde dehydrogenase ALD5 in maintenance of the mitochondrial electron transport chain in Saccharomyces cerevisiae. F. E. M. S. Microbiol. Lett. 181:281-287. [DOI] [PubMed] [Google Scholar]
- 22.Lee, W. P., C. L. Lee, and H. C. Lin. 1996. Glutathione S-transferase and glutathione peroxidase are essential in the early stage of adriamycin resistance before P-glycoprotein overexpression in HOB1 lymphoma cells. Cancer Chemother. Pharmacol. 38:45-51. [DOI] [PubMed] [Google Scholar]
- 23.Leidich, S. D., and P. Orlean. 1996. Gpi1, a Saccharomyces cerevisiae protein that participates in the first step in glycosylphosphatidylinositol anchor synthesis. J. Biol. Chem. 271:27829-27837. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Ribot, J. L., R. K. McAtee, L. N. Lee, W. R. Kirkpatrick, T. C. White, D. Sanglard, and T. F. Patterson. 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 42:2932-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyons, C. N., and T. C. White. 2000. Transcriptional analyses of antifungal drug resistance in Candida albicans. Antimicrob. Agents Chemother. 44:2296-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marr, K. A., T. C. White, J. A. van Burik, and R. A. Bowden. 1997. Development of fluconazole resistance in Candida albicans causing disseminated infection in a patient undergoing marrow transplantation. Clin. Infect. Dis. 25:908-910. [DOI] [PubMed] [Google Scholar]
- 27.Micheli, M. M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 28.Minn, Y., E. Brummer, and D. A. Stevens. 1997. Effect of iron on fluconazole activity against Candida albicans in presence of human serum or monocyte-derived macrophages. Mycopathologia 138:29-35. [DOI] [PubMed] [Google Scholar]
- 29.Nawrocki, A., S. J. Fey, A. Goffeau, P. Roepstorff, and P. M. Larsen. 2001. The effects of transcription regulating genes PDR1, pdr1-3, and PDR3 in pleiotropic drug resistance. Proteomics 1:1022-1032. [DOI] [PubMed] [Google Scholar]
- 30.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfaller, M. A., J. Rhine-Chalberg, S. W. Redding, J. Smith, G. Farinacci, A. W. Fothergill, and M. G. Rinaldi. 1994. Variations in fluconazole susceptibility and electrophoretic karyotype among oral isolates of Candida albicans from patients with AIDS and oral candidiasis. J. Clin. Microbiol. 32:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Redding, S., J. Smith, G. Farinacci, M. Rinaldi, A. Fothergill, J. Rhine-Chalberg, and M. Pfaller. 1994. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS: documentation by in vitro susceptibility testing and DNA subtype analysis. Clin. Infect. Dis. 18:240-242. [DOI] [PubMed] [Google Scholar]
- 33.Richard, M., S. Ibata-Ombetta, F. Dromer, F. Bordon-Pallier, T. Jouault, and C. Gaillardin. 2002. Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol. Microbiol. 44:841-853. [DOI] [PubMed] [Google Scholar]
- 34.Riggle, P. J., and C. A. Kumamoto. 2000. Role of a Candida albicans P1-type ATPase in resistance to copper and silver ion toxicity. J. Bacteriol. 182:4899-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soustre, I., Y. Letourneux, and F. Karst. 1996. Characterization of the Saccharomyces cerevisiae RTA1 gene involved in 7-aminocholesterol resistance. Curr. Genet. 30:121-125. [DOI] [PubMed] [Google Scholar]
- 38.Viarengo, A., B. Burlando, N. Ceratto, and I. Panfoli. 2000. Antioxidant role of metallothioneins: a comparative overview. Cell. Mol. Biol. 46:407-417. [PubMed] [Google Scholar]
- 39.Wakabayashi, H., T. Okutomi, S. Abe, H. Hayasawa, M. Tomita, and H. Yamaguchi. 1998. Enhanced anti-Candida activity of neutrophils and azole antifungal agents in the presence of lactoferrin-related compounds. Adv. Exp. Med. Biol. 443:229-237. [DOI] [PubMed] [Google Scholar]
- 40.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White, T. C. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, T., K. Wright, S. F. Hurst, and C. J. Morrison. 2000. Enhanced extracellular production of aspartyl proteinase, a virulence factor, by Candida albicans isolates following growth in subinhibitory concentrations of fluconazole. Antimicrob. Agents Chemother. 44:1200-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]