Abstract
The MICs of cationic, hydrophobic peptides of the prototypic sequence KKAAAXAAAAAXAAWAAXAAAKKKK-amide (where X is one of the 20 commonly occurring amino acids) are in a low micromolar range for a panel of gram-negative and gram-positive bacteria, with no or low hemolytic activity against human and rabbit erythrocytes. The peptides are active only when the average segmental hydrophobicity of the 19-residue core is above an experimentally determined threshold value (where X is Phe, Trp, Leu, Ile, Met, Val, Cys, or Ala). Antimicrobial activity could be increased by using peptides that were truncated from the prototype length to 11 core residues, with X being Phe and with 6 Lys residues grouped at the N terminus. We propose a mechanism for the interaction between these peptides and bacterial membranes similar to the “carpet model,” wherein the Lys residues interact with the anionic phospholipid head groups in the bacterial membrane surface and the hydrophobic core portion of the peptide is then able to interact with the lipid bilayer, causing disruption of the bacterial membrane.
Antimicrobial peptides have been found in all living organisms (4, 10). In the past 20 years, more than 700 antibacterial peptides have been identified (http://www.bbcm.univ.trieste.it/∼tossi/pag1.htm). These peptides have a broad spectrum of activities, killing or neutralizing many gram-negative and gram-positive bacteria, including some antibiotic-resistant strains, in vitro and also many fungi, viruses, and parasites (13). They are usually made up of between 12 and 50 residues, and while they differ widely in sequence and structure, they do share some common features in that they are polycationic, ca. 50% of their amino acids are hydrophobic, and they are generally amphipathic (i.e., with systematically alternating hydrophobic and polar residues along the primary sequence) (for reviews, see references 8, 12, 15, and 31). Cationic antimicrobial peptides have been divided into major classes based on their three-dimensional structures, including β-sheet structures stabilized by two or three disulfide bonds, linear α-helices, extended coils, and loop structures resulting from a single disulfide bond (4, 8). The most abundant—and the most widely studied—naturally occurring type is the linear, amphipathic, α-helical peptide (see, e.g., reference 28).
Despite the large numbers of antibiotics currently available (36), antibiotic resistance has been of great concern during the last decades due to the extensive clinical use of classical antibiotics (5, 33, 34). As most well-known antibiotics act by interfering in a specific manner with bacterial homeostasis, the bacteria can evolve resistance through mechanisms such as preventing the antibiotic from binding to or entering the organism, producing an enzyme that inactivates the antibiotic, and/or changing the internal binding site of the antibiotic. Therefore, it is of considerable interest to find, or develop, antibiotics with a new mechanism(s) of action which can potentially evade the emergence of resistance.
In the search for such new antibiotics, the group of linear α-helical peptides have attracted increasing research and clinical interest during the last decade (9, 11, 29), as the microenvironment of the lipid bilayer has been shown to have a stabilizing effect on the α-helical structure of peptides (17). Structural analyses by nuclear magnetic resonance spectroscopy indicate that peptide antibiotics strongly interact with lipids, and several models have been proposed to explain the antibiotic and membrane-disrupting activities of these peptides (2, 27), although some details of their mechanism(s) of action remain under investigation. One widely postulated hypothesis is that of the “self-promoted uptake” of the peptides across the outer membranes of gram-negative bacteria (13), which suggests that the peptides interact with the negatively charged outer leaflet of the cytoplasmic membranes (of both gram-negative and gram-positive bacteria) and disrupt the membranes via either a “barrel-stave” or a “carpet” mechanism (2, 24, 27).
We describe here a new category of nonamphipathic hydrophobic antimicrobial peptides; in this context, the hydrophobic core of the peptides is regarded as nonamphipathic because hydrophilic residues are absent. Meanwhile, N- and C-terminal Lys residues flanking the core act to solubilize these otherwise hydrophobic peptides in aqueous media to facilitate purification and characterization (22), as well as to provide a locus for attraction to bacterial membranes. These peptides, of the prototypic sequence KKAAAXAAAAAXAAWAAXAAAKKKK-amide (where X is one of the 20 commonly occurring amino acids) (20), were originally designed as transmembrane mimetic model peptides. Peptides of this design have several key features. (i) Ala is chosen as the background residue, reflecting its midrange hydropathy and frequent occurrence in membrane protein transmembrane domains. (ii) The hydrophobic core segment of the peptide comprises 19 residues, which is approximately the minimum number required to span a lipid bilayer (16). (iii) Distribution of the “guest” residues (X) in the hydrophobic segment has been designed to preserve both angular and longitudinal symmetry around the helix. Three guest residues amplify the effect of these residues, ensuring their structural impact. A Trp residue is inserted into the hydrophobic segment as a fluorescent probe to monitor characteristics of the local environment. It has been shown that these peptides spontaneously insert themselves into membranes when the average hydrophobicity of their core segment is above an experimentally determined threshold value. The threshold value is based on the Liu-Deber hydrophobicity scale, in which a hydrophobicity value for each guest residue is determined from the retention time of the corresponding X peptide as found by reverse-phase high-performance liquid chromatography (RP-HPLC) (19). Peptides with hydrophobicities above a threshold value (i.e., those in which X is Phe, Trp, Leu, Ile, Met, Val, Cys, or Ala) spontaneously insert themselves into membranes (19, 20). The latter observations, in combination with the cationic nature of these peptides, prompted us to investigate the potential activities of these peptides in bacterial membranes. In the present work, we report the antibiotic activities noted for these compounds and for further iterated structural analogs.
MATERIALS AND METHODS
Materials.
Reagents for peptide synthesis, cleavage, and purification included 9-fluorenylmethoxy carbonyl (Fmoc)-protected amino acids (Novabiochem), [5-(4-Fmoc-aminomethyl-3,5-dimethoxyphenoxy)valeric acid]-polyethylene glycol-polystyrene resin (Fmoc-PAL-PEG-PS resin; Applied Biosystems, Foster City, Calif.), N,N-dimethylformamide (peptide grade; Caledon Laboratories Ltd., Georgetown, Ontario, Canada), piperidine (Applied Biosystems or Acros), methanol (Caledon), N,N-diisopropylethylamine (DIEA; Aldrich), O-(7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU; Applied Biosystems or GL Biochem Ltd., Shanghai, China), diethyl ether (Caledon), triisopropylsilane (TIPS; Aldrich), phenol (Gibco), and acetonitrile (Caledon).
Reagents for a micro-bicinchoninic acid protein assay were obtained from Pierce (Rockford, Ill.). Mueller-Hinton broth and Bacto Agar were purchased from Difco Laboratories. All other reagents were of analytical grade.
Peptide synthesis.
Peptides were synthesized by using standard Fmoc chemistry on a PerSeptive Biosystems Pioneer peptide synthesizer. The synthesis utilized the Pioneer's standard (45-min) cycle. A low load (>0.15 mmol/g) of PAL-PEG-PS resin was used to produce an amidated C terminus. The HATU-DIEA activator pair was used with amino acids at a fourfold excess. Deprotection and cleavage of the peptides were carried out in a mixture of 95% trifluoroacetic acid (TFA), 2.5% water, and 2.5% TIPS (vol/vol/vol) or of 88% TFA, 5% phenol, 5% water, and 2% TIPS (vol/vol/vol/vol), under nitrogen, for 2 h at room temperature. Cleaved and deprotected peptides were precipitated with ice-cold diethyl ether. Centrifuged pellets were dried, redissolved in water, and lyophilized.
Purification of the peptides was performed on a C4 preparative RP-HPLC column (21.2 by 250 mm, 300 Å, 10-μm particle size) with a linear gradient of acetonitrile in 0.1% TFA. Crude peptide samples (5 to 12 mg) were dissolved in water and applied to the column. The fraction from the major peak was collected manually and lyophilized. Purified peptides were characterized by analytical RP-HPLC, mass spectrometry, and amino acid analysis. The RP-HPLC analyses were performed on a Vydac C4 column (4.6 by 250 mm, 300 Å, 5-μm particle size) with linear gradients of water-0.1% TFA (gradient A) and acetonitrile-0.1% TFA (gradient B) at a flow rate of 1 ml/min and with 1% gradient B/min, starting at a concentration of 10% gradient B. Peptide concentration was determined by amino acid analysis and micro-bicinchoninic acid protein assay.
Hydrophobicity.
The hydrophobicity of each peptide—given as the average hydropathy of the core hydrophobic segment (with flanking Lys or Arg residues excluded)—was calculated by using the Liu-Deber hydrophobicity scale (19).
CD spectroscopy.
Circular dichroism (CD) spectra were recorded at room temperature using a Jasco J-720 spectropolarimeter. Spectral scans were performed from 250 to 190 nm, with a step resolution of 0.1 nm and a bandwidth of 1.0 nm and at a speed of 50 nm/min. Values from three scans were averaged per sample. A 1-mm-path-length quartz cuvette was used for the measurements. Samples were measured at peptide concentrations of 30 to 40 μM in 20 mM Tris-HCl-20 mM NaCl, pH 7.4, with or without 20 mM sodium dodecyl sulfate (SDS).
Bacterial strains.
The Escherichia coli strain UB1005 and its antibiotic-supersusceptible derivative DC2 (25), a clinical isolate of Staphylococcus epidermidis (C621), and the coryneform bacterial strain Corynebacterium xerosis C875 were a kind gift from R. E. W. Hancock (University of British Columbia). E. coli ATCC 25922, Staphylococcus aureus ATCC 25923, Enterococcus faecalis ATCC 29212, Bacillus subtilis ATCC 6633, and Pseudomonas aeruginosa ATCC 27853, all from the American Type Culture Collection, were a kind gift from Anne Matlow (Infectious Diseases, Hospital for Sick Children).
Antimicrobial activity.
The MIC (measured in micromolar units) was determined by using a broth microdilution assay modified from the method of Amsterdam (1) as described in reference 35. Twofold serial dilutions of peptides, with concentrations ranging from 320 to 2.5 mM, were diluted in 0.2% bovine serum albumin with 0.01% acetic acid in polypropylene test tubes. Bacteria were grown overnight in Mueller-Hinton broth and diluted to approximately 4 × 105 CFU/ml. Bacteria (90 μl) and peptide (10 μl) were added to the wells of sterile 96-well polypropylene microtiter plates (cat. no. 3790; Costar). Plates were incubated at 37°C overnight. Plates were read visually and at 600 nm in a microplate reader (Molecular Devices). In the present study, the MIC was taken to be the lowest concentration that completely inhibited growth.
Hemolytic activity.
Freshly collected rabbit or human blood with heparin was centrifuged to remove the buffy coat, and the erythrocytes obtained were washed three times with phosphate-buffered saline (PBS), centrifuged for 10 min at 1,000 × g, and resuspended in PBS to 4% (vol/vol). Peptides were diluted in PBS to 400 and 100 μM, and 100 μl of the erythrocyte suspension and 100 μl of the peptide solution were added to the wells of a 96-well round-bottomed polypropylene microtiter plate. PBS and 0.1% Triton X-100 were used as agents for 0 and 100% hemolysis, respectively. Plates were incubated for 1 h at 37°C and centrifuged at 1,000 × g for 5 min. Supernatant (100 μl) was transferred to a 96-well flat-bottomed polystyrene plate, and the release of hemoglobin was monitored by measuring the absorbance at 540 nm in a microplate reader.
RESULTS
Antimicrobial activity of peptides.
The MICs of several KKAAAXAAAAAXAAWAAXAAAKKKK-amide peptides for selected gram-negative and gram-positive bacteria are shown in Table 1. The peptides with the guest residue Phe, Trp, Leu, Ile, Met, Cys, or Tyr each displayed antimicrobial effects on both gram-negative and gram-positive bacteria. The peptides with Ala or Val as a guest residue were active only against the gram-positive bacterium C. xerosis. Peptides with hydrophobicities below the threshold value (ca. 0.4), i.e., below the hydrophobicity on the Liu-Deber scale of the peptide in which X is Ala (e.g., those in which X is Ser) (19) (Table 1), generally showed no antimicrobial activity; peptides in this series in which X is Thr, Glu, Asp, Gln, Gly, Asn, or Pro (not shown in Table 1) similarly displayed no activity (MIC > 64) against this panel of organisms. Peptides of increased cationic character (where X is Lys, Arg, or His) did display activity against C. xerosis and E. coli DC2 at higher peptide concentrations.
TABLE 1.
MICs of selected 25-residue KKAAAXAAAAAXAAWAAXAAAKKKK-amide peptides for gram-negative and gram-positive bacteria
| Peptide X residue | Core segment hydrophobicitya | Molecular mass (Da) | MIC (μM)b
|
|||
|---|---|---|---|---|---|---|
| Gram-negative bacteria
|
Gram-positive bacteria
|
|||||
| E. coli DC2 | E. coli UB1005 | C. xerosis C875 | S. epidermidis C621 | |||
| F | 1.18 | 2,480 | 4 | 16 | <0.25 | 2 |
| W | 1.16 | 2,597 | 8 | 16 | <0.25 | <0.25 |
| L | 1.14 | 2,378 | 16 | 32 | <0.25 | 2 |
| I | 1.09 | 2,378 | 16 | >32 | <0.25 | 4 |
| M | 0.90 | 2,432 | 32 | >32 | 0.5 | 16 |
| V | 0.87 | 2,336 | >32 | >32 | 0.5 | >32 |
| C | 1.02 | 2,368 | 8 | 16 | 1 | >32 |
| Y | 0.71 | 2,328 | 16 | 32 | <0.25 | 8 |
| A | 0.42 | 2,252 | >64 | >64 | 2 | >64 |
| R | −0.05 | 2,507 | 32 | >64 | 8 | >64 |
| S | −0.06 | 2,300 | >64 | >64 | >64 | >64 |
| H | −0.34 | 2,450 | 64 | >64 | 32 | >64 |
| K | −0.40 | 2,423 | 64 | >64 | 16 | >64 |
Values correspond to the hydrophobicity of the core segment determined by the Liu-Deber scale (20): Lys residues are not included in the calculation. Core segment hydrophobicities of greater than +0.4 are above the threshold for membrane insertion (7).
Values are representative of results from three or more separate experiments. MICs may be converted to micrograms per milliliter by dividing molecular mass (in daltons) by 1,000 and then multiplying by the value in micromolar units.
The peptide in which X is Phe (designated F25) was among those with the highest antimicrobial activity and, interestingly, is also the most hydrophobic on the Liu-Deber hydropathy scale. Accordingly, starting from the Phe peptide (25 residues), a series of further peptides was synthesized in order to investigate the relevance to antibiotic activity of two key variables: the length of the hydrophobic core and the position(s) of the X residue(s). In addition, we synthesized peptides in which flanking Lys residues were interchanged with Arg residues, as the latter amino acid is known for its excellent membrane-surface binding properties (26). These peptides are listed in Table 2 together with their relative hydrophobicities as calculated by the Liu-Deber scale. MICs of the latter group of peptides for gram-negative and gram-positive bacteria are shown in Tables 3 and 4, respectively.
TABLE 2.
Amino acid sequences and hydrophobicities of antimicrobial peptides
| Peptide | Amino acid sequencea | Core segment hydrophobicityb | Molecular mass (Da) |
|---|---|---|---|
| F25 | KKAAAFAAAAAFAAWAAFAAAKKKK-NH2 | 1.18 | 2,480 |
| F25-6K | KKKKKKAAAFAAAAAFAAWAAFAAA-NH2 | 1.18 | 2,480 |
| 4F | KKAAAAFAAFAAWFAAFAAAAKKKK-NH2 | 1.43 | 2,556 |
| F21 | KKAFAAAAAFAAWAAFAKKKK-NH2 | 1.45 | 2,196 |
| F17 | KKKAAAFAAWAAFAKKK-NH2 | 1.47 | 1,835 |
| F17-R | RRRAAFAAWAAFAARRR-NH2 | 1.47 | 2,003 |
| F17-6K | KKKKKKAAFAAWAAFAA-NH2 | 1.47 | 1,835 |
| Al1-d F17-6K | kkkkkkaafaawaafaa-NH2 | 1.47 | 1,835 |
| F17-6R | RRRRRRAAFAAWAAFAA-NH2 | 1.47 | 2,003 |
| KAFW | KKKKKKAAAAFWAAAAF-NH2 | 1.47 | 1,835 |
| 3F17-6K | KKKKKKAAFAAFAAFAA-NH2 | 1.49 | 1,796 |
| W17-6K | KKKKKKAAWAAWAAWAA-NH2 | 1.45 | 1,913 |
For clarity, the Phe and Trp residues are in bold. Amino acids in lowercase are d-enantiomers.
Calculated by the Liu-Deber scale (20). Each value is the mean residue hydrophobicity of the core segment; Lys or Arg was not included.
TABLE 3.
MICs of peptides for gram-negative bacteria
| Peptide | MIC (μM)a
|
|||
|---|---|---|---|---|
| E. coli DC2 | E. coli UB1005 | E. coli ATCC 25922 | P. aeruginosa ATCC 27853 | |
| F25 | 4 | 16 | >32 | 8 |
| F25-6K | 8 | 16 | 16 | 32 |
| 4F | ND | 8 | 16 | 8 |
| F21 | 4 | 8 | 32 | >32 |
| F17 | 16 | 32 | >32 | >32 |
| F17-R | 1 | 2 | 8 | 16 |
| F17-6K | 0.5 | 1 | 8 | 16 |
| A11-d F17-6K | ND | 0.5 | 2 | 8 |
| F17-6R | 0.5 | 1 | 4 | 8 |
| KAFW | 0.5 | 2 | 16 | 16 |
| 3F17-6K | ND | 2 | 16 | 16 |
| W17-6K | ND | 1 | 8 | 8 |
TABLE 4.
MICs of peptides for gram-positive bacteria
| Peptide | MIC (μM)a
|
||||
|---|---|---|---|---|---|
| C. xerosis C875 | S. epidermidis C621 | S. aureus ATCC 25923 | E. faecalis ATCC 29212 | B. subtilis ATCC 6633 | |
| F25 | <0.25 | 2 | >32 | >32 | 1 |
| F25-6K | 1 | 4 | ND | ND | ND |
| 4F | ND | 4 | 32 | >32 | 2 |
| F21 | 0.5 | 8 | >32 | >32 | 2 |
| F17 | 4 | >32 | >32 | >32 | 16 |
| F17-R | 1 | 4 | >32 | >32 | 2 |
| F17-6K | 0.5 | 4 | >32 | >32 | 8 |
| All-d F17-6K | ND | 4 | 32 | >32 | 4 |
| F17-6R | 0.5 | 4 | ND | ND | ND |
| KAFW | 0.5 | 8 | ND | ND | ND |
| 3F17-6K | ND | 8 | >32 | >32 | 16 |
| W17-6K | ND | 8 | 32 | >32 | 4 |
By shortening the Phe peptide by 2 residues at each end of the hydrophobic core (to create peptide F21), we found that antimicrobial activity was not significantly affected. Upon further shortening of the peptide by 4 residues at the N-terminal side of the hydrophobic core (to create peptide F17), the activity was decreased fourfold against gram-negative bacteria and 8- to 16-fold against gram-positive bacteria. Changing the flanking Lys residues in the F17 peptide to Arg residues significantly increased the antimicrobial activity. Notably, after grouping the 6 Lys or Arg residues at the N-terminal side of the molecule, with none at the C-terminal side (as in F17-6K; its enantiomer, all-d F17-6K; and F17-6R), we observed higher antimicrobial activity from the new peptide than from the F17 peptide itself. We further found that 17-residue peptides with six Lys residues at the N terminus but either with no Trp (3F17-6K) or with Trp instead of Phe (W17-6K) showed antimicrobial activities similar to those of the F17-6K, F17-R, and F17-6R peptides.
Finally, we noted that 25-residue peptides that were modified from the original series, viz., a peptide with 6 Lys residues on the N-terminal side of the molecule but with none on the C-terminal side (F25-6K) and a peptide with 4 Phe residues (designated 4F), displayed activities comparable to the those of the parent F25 peptide.
Assay of peptide hemolysis in rabbit and human erythrocytes.
The hemolytic activities of the peptides against rabbit and human erythrocytes were determined at peptide concentrations of 50 and 200 μM (Table 5). Little or no hemolysis was detected, although there were a few exceptions: at concentrations of 200 μM, the peptides 4F, F17-6R, and F25-6K were hemolytic against human erythrocytes and the peptide F17-6R had modest hemolytic activity against rabbit erythrocytes (3%) but displayed significantly higher hemolytic activity against human erythrocytes (26%).
TABLE 5.
Hemolytic activities of antimicrobial peptides in rabbit and human erythrocytes
| Peptide | Rabbit RBC (% lysis)a at:
|
Human RBC (% lysis) at:
|
||
|---|---|---|---|---|
| 200 μM | 50 μM | 200 μM | 50 μM | |
| S25 | 3 | 0 | 0 | 0 |
| A25 | 1 | 0 | ND | ND |
| F25 | 4 | 0 | 1 | 0 |
| F25-6K | ND | ND | 17 | 11 |
| 4F | 34 | 14 | 42 | 17 |
| F21 | 3 | 1 | 0 | 0 |
| F17 | 2 | 0 | 0 | 0 |
| F17-R | 2 | 0 | 0 | 0 |
| F17-6K | 1 | 0 | 0 | 0 |
| All-d F17-6K | 2 | 0 | 0 | 0 |
| F17-6R | 3 | 1 | 26 | 14 |
| KAFW | 1 | 0 | 0 | 0 |
| 3F176K | 2 | 0 | 0 | 0 |
| W17-6K | 2 | 0 | 0 | 0 |
RBC, red blood cells. Percentages are given to the nearest 0.5%. Values are representative of two to three experiments. ND, not determined.
Helical structures of peptides inserted into membranes.
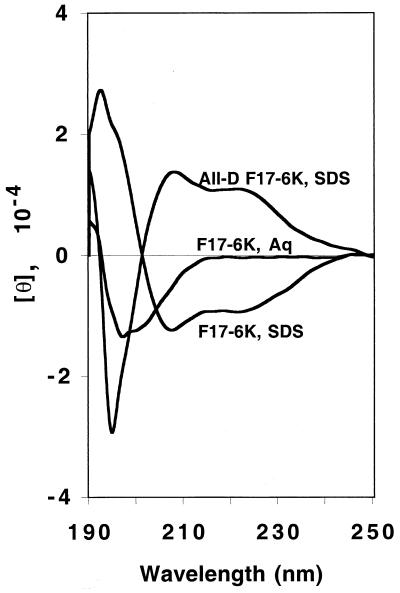
Conformations of the peptides under investigation in the present study, particularly those of the prototypic “25” series (Table 1), have been widely studied in aqueous environments and in the environments inside membranes (18, 19). Those studies have demonstrated that while the peptides take random or mixed random and partially helical structures in aqueous phases as a function of the guest residue X, all peptides with hydrophobicities above the threshold value take α-helical structures as their hydrophobic core regions become inserted into phospholipid micelles (19) and vesicles (18). In fact, peptides suitable for transmembrane insertion have been demonstrated to require residues of significant core hydrophobicities in combination with suitable helix promotion within nonpolar phases (7). These phenomena are illustrated in Fig. 1, which presents CD spectra of one of the most active peptides, F17-6K, in an aqueous phase, where it is largely unstructured, and upon insertion into SDS micelles, where it becomes essentially fully α-helical. The mirror image helical CD spectrum of all-d F17-6K inserted into SDS micelles is presented for comparison.
FIG. 1.
CD spectra of the F17-6K peptide in an aqueous buffer (Aq) containing 20 mM Tris-HCl and 20 mM NaCl, pH 7.4, and in the same buffer with the addition of 20 mM SDS. The spectrum of all-d F17-6K in SDS is presented for comparison. [θ], ellipticity (measured in degrees × square centimeter per decimole). The peptide concentration was 30 to 40 μM, with an estimated uncertainty of ±5%. Spectra are as indicated on the diagram. See the text for a further discussion.
DISCUSSION
Cationic hydrophobic peptides with nonamphipathic core sequences that promote membrane insertion have been observed in the present work to be active against S. epidermidis, C. xerosis, and E. coli. Activity coincides with the threshold hydrophobicity required for insertion of the core hydrophobic segment, as peptides with hydrophobicities on the Liu-Deber scale below that of the peptide in which X is Ala (Table 1) were inactive. Upon shortening of the F25 peptide to one with 17 total residues and with 11 amino acids in the hydrophobic core, we initially observed reduced antimicrobial activity (Tables 3 and 4). However, activity could be increased by grouping the Lys or Arg residues at the N terminus of the molecule, which, in effect, converts the molecules into true amphiphilic structures. The latter findings suggest that the cation-free C-terminal region may interact more effectively with the bacterial membrane.
Interestingly, the grouping of cationic residues at the N terminus—which does not alter the hydrophobicity of the peptide core segment—induced significant effects on peptide retention times in HPLC experiments. For example, the retention time of F25 on the Vydac C4 column (see Materials and Methods) was 31.4 min, while the retention time of F25-6K was 35.9 min (data not shown). Similarly, F17 was eluted at 23.1 min while F17-6K was eluted at 26.0 min. Increased retention time is nominally associated with increased peptide hydrophobicity as perceived by the HPLC column, leading one to suppose initially that peptides with Lys or Arg residues grouped at the N terminus may be more antimicrobially active than their counterparts with Lys or Arg residues on both the N and C termini. However, MIC data in Tables 3 and 4 do not support this notion, as F25-6K is generally less active than F25 but F17-6K is much more active than F17. Hemolysis values also vary in a noncorrelated manner. These findings suggest that the activity of each peptide is determined by a subtle confluence of factors which are likely to include some (or all) of the following: length, hydrophobicity, sequence, cationic residue positioning, and conformation adopted in the membrane.
In syntheses designed to identify the essential features of activity for this category of peptides, we prepared the peptides F17-6K, all-d F17-6K, KAFW, and W17-6K (Table 2). These materials all displayed similar antimicrobial activities, e.g., MICs of these peptides were 8 to 16 μM (ca. 15 to 30 μg/ml) for P. aeruginosa ATCC 27853, and showed no hemolytic activity. Although the hydrophobic core of each peptide is 11 residues long—too short to span a typical lipid bilayer—these peptides show higher antimicrobial activity than the original F25 counterpart, even when the 25-residue peptide has all Lys residues placed at the N terminus of the molecule (F25-6K). A fully d-amino acid analog of the peptide F17-6K showed an antimicrobial activity equal to or often twofold higher than that of the l-peptide, which is in agreement with results reported by other groups with respect to d-residue incorporation (3, 23) and which indicates that antimicrobial activity by this category of peptides is not stereospecific. In addition, while the 25-residue peptides F25-6K and 4F showed some hemolytic activity, the 17-residue peptides (with Lys) had no hemolytic activity at concentrations up to 200 μM.
In the consideration of possible mechanisms of action for the materials described herein, hydrophobicity, amphipathicity, and net positive charge have been implicated as important features of antimicrobial peptides (14, 23). A model that explains the membrane-penetrating activity of most naturally occurring α-helical antimicrobial peptides is the carpet model (24, 27). In this model, the peptides bind to the phospholipid head groups at the surface of the membrane but do not become inserted into the hydrophobic core of the membrane. When a threshold concentration (distinct from the threshold hydrophobicity discussed in the present work) of the peptide is reached, displacement of lipids and alteration of the membrane structure follow, resulting in generalized membrane disruption. For peptides of the design described above, we note that the core regions are not amphipathic, as are those of most naturally occurring cationic antimicrobial peptides. Nevertheless, similar to what occurs in the carpet model, the first step of interaction of these peptides with the bacterial membrane is likely to be one in which the lysines grip the membrane via the preponderance of anionic lipids prevalent in bacteria. It is tempting to speculate that in a subsequent (or concurrent) step, the hydrophobic peptide core—not constrained by amphipathicity to the membrane surface—may dip partially into the lipid bilayer, ostensibly producing a disruptive effect on lipid packing.
Although peptides become inactive as their average segmental hydrophobicities fall below the threshold value (such as in those in the 25-residue series in which X is Ser) (19), sequences with increased cationic character (those in which X is Lys, Arg, or His) convert the peptides into amphipathic molecules that likely have the capacity to embed effectively into the bacterial membrane surface and possibly to function in a manner akin to that described in the carpet model.
It was gratifying to note the general lack of hemolysis of human and rabbit erythrocytes by most peptides studied here, even at peptide concentrations up to 200 μM. Such selectivity of cationic nonamphipathic antimicrobial peptides for bacterial membranes may be explicable, in part, by the differences in the compositions of eukaryotic and prokaryotic membranes (21, 37). The outer leaflet of mammalian cells is predominantly composed of the zwitterionic phospholipids phosphatidylcholine and sphingomyelin (32), along with cholesterol (30), while bacterial membranes contain mainly anionic phospholipids and no cholesterol (6). In addition, the outer surfaces of gram-negative bacteria contain lipopolysaccharides, while those of gram-positive bacteria contain teichoic acid, which in both cases adds to the negative charge of the bacterial surface (6). The cationic nature of native antimicrobial peptides clearly contributes to their preferential recognition by the negatively charged outer surfaces of bacterial membranes (24, 27). Whether the peptides described above bind erythrocytes too weakly to incur damage—and/or are stymied by steric clash with cholesterol—remains a question for further investigation.
Acknowledgments
This work was supported, in part, by grants to C.M.D. from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Translation Initiatives research grant program of the Hospital for Sick Children. M.S. held a fellowship from the Sweden-American Foundation. L.-P.L. held a Research Training Award from the Hospital for Sick Children.
References
- 1.Amsterdam, D. 1996. Susceptibility testing of antimicrobials in liquid media, p. 52-111. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, Md.
- 2.Bechinger, B. 1999. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta 1462:157-183. [DOI] [PubMed] [Google Scholar]
- 3.Blondelle, S. E., E. Pérez-Payá, and R. A. Houghten. 1996. Synthetic combinatorial libraries: novel discovery strategy for identification of antimicrobial agents. Antimicrob. Agents Chemother. 40:1067-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boman, H. G. 1995. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 13:61-92. [DOI] [PubMed] [Google Scholar]
- 5.Bonomo, R. A. 2000. Multiple antibiotic-resistant bacteria in long-term-care facilities: an emerging problem in the practice of infectious diseases. Clin. Infect. Dis. 31: 1414-1422. [DOI] [PubMed] [Google Scholar]
- 6.Brock, T. D. 1974. Biology of microorganisms, 2nd ed. Prentice-Hall, Inc., Englewood Cliffs, N.J.
- 7.Deber, C. M., C. Wang, L. P. Liu, A. S. Prior, S. Agrawal, B. L. Muskat, and A. J. Cuticchia. 2001. TM Finder: a prediction program for transmembrane protein segments using a combination of hydrophobicity and nonpolar phase helicity scales. Protein Sci. 10:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock, R. E. 1997. Peptide antibiotics. Lancet 349:418-422. [DOI] [PubMed] [Google Scholar]
- 9.Hancock, R. E. 1999. Host defence (cationic) peptides: what is their future clinical potential? Drugs 57:469-473. [DOI] [PubMed] [Google Scholar]
- 10.Hancock, R. E., T. Falla, and M. Brown. 1995. Cationic bactericidal peptides. Adv. Microb. Physiol. 37:135-175. [DOI] [PubMed] [Google Scholar]
- 11.Hancock, R. E., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, R. E., and M. G. Scott. 2000. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. USA 97:8856-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock, R. E. W. 2001. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 1: 156-164. [DOI] [PubMed] [Google Scholar]
- 14.Hong, J., Z. Oren, and Y. Shai. 1999. Structure and organization of hemolytic and nonhemolytic diastereomers of antimicrobial peptides in membranes. Biochemistry 38:16963-16973. [DOI] [PubMed] [Google Scholar]
- 15.Hwang, P. M., and H. J. Vogel. 1998. Structure-function relationships of antimicrobial peptides. Biochem. Cell Biol. 76:235-246. [DOI] [PubMed] [Google Scholar]
- 16.Kuroiwa, T., M. Sakaguchi, K. Mihara, and T. Omura. 1991. Systematic analysis of stop-transfer sequence for microsomal membrane. J. Biol. Chem. 266:9251-9255. [PubMed] [Google Scholar]
- 17.La Rocca, P., Y. Shai, and M. S. Sansom. 1999. Peptide-bilayer interactions: simulations of dermaseptin B, an antimicrobial peptide. Biophys. Chem. 76:145-159. [DOI] [PubMed] [Google Scholar]
- 18.Liu, L. P., and C. M. Deber. 1997. Anionic phospholipids modulate peptide insertion into membranes. Biochemistry 36:5476-5482. [DOI] [PubMed] [Google Scholar]
- 19.Liu, L. P., and C. M. Deber. 1998. Guidelines for membrane protein engineering derived from de novo designed model peptides. Biopolymers 47:41-62. [DOI] [PubMed] [Google Scholar]
- 20.Liu, L. P., S. C. Li, N. K. Goto, and C. M. Deber. 1996. Threshold hydrophobicity dictates helical conformations of peptides in membrane environments. Biopolymers 39:465-470. [DOI] [PubMed] [Google Scholar]
- 21.Matsuzaki, K. 1999. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta 1462:1-10. [DOI] [PubMed] [Google Scholar]
- 22.Melnyk, R. A., A. W. Partridge, and C. M. Deber. 2001. Retention of native-like oligomerization states in transmembrane segment peptides: application to the Escherichia coli aspartate receptor. Biochemistry 40:11106-11113. [DOI] [PubMed] [Google Scholar]
- 23.Oren, Z., J. Hong, and Y. Shai. 1997. A repertoire of novel antibacterial diastereomeric peptides with selective cytolytic activity. J. Biol. Chem. 272:14643-14649. [DOI] [PubMed] [Google Scholar]
- 24.Oren, Z., and Y. Shai. 1998. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers 47:451-463. [DOI] [PubMed] [Google Scholar]
- 25.Richmond, M. H., D. C. Clark, and S. Wotton. 1976. Indirect method for assessing the penetration of beta-lactamase-nonsusceptible penicillins and cephalosporins in Escherichia coli strains. Antimicrob. Agents Chemother. 10:215-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson, C. 2000. The twin-arginine translocation system: a novel means of transporting folded proteins in chloroplasts and bacteria. Biol. Chem. 381:89-93. [DOI] [PubMed] [Google Scholar]
- 27.Shai, Y. 1999. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1462:55-70. [DOI] [PubMed] [Google Scholar]
- 28.Tossi, A., L. Sandri, and A. Giangaspero. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4-30. [DOI] [PubMed] [Google Scholar]
- 29.Tossi, A., C. Tarantino, and D. Romeo. 1997. Design of synthetic antimicrobial peptides based on sequence analogy and amphipathicity. Eur. J. Biochem. 250:549-558. [DOI] [PubMed] [Google Scholar]
- 30.Turner, J. D., and G. Rouser. 1970. Precise quantitative determination of human blood lipids by thin-layer and triethylaminoethylcellulose column chromatography. I. Erythrocyte lipids. Anal. Biochem. 38:423-436. [DOI] [PubMed] [Google Scholar]
- 31.van't Hof, W., E. C. Veerman, E. J. Helmerhorst, and A. V. Amerongen. 2001. Antimicrobial peptides: properties and applicability. Biol. Chem. 382:597-619. [DOI] [PubMed] [Google Scholar]
- 32.Verkleij, A. J., R. F. Zwaal, B. Roelofsen, P. Comfurius, D. Kastelijn, and L. L. van Deenen. 1973. The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochim. Biophys. Acta 323:178-193. [DOI] [PubMed] [Google Scholar]
- 33.Williams, J. D. 2001. Antibiotic resistance in hospital pathogens—acquisition or spread? Int. J. Antimicrob. Agents 18:295-298. [DOI] [PubMed] [Google Scholar]
- 34.Wong, A. H., R. P. Wenzel, and M. B. Edmond. 2000. Epidemiology of bacteriuria caused by vancomycin-resistant enterococci—a retrospective study. Am. J. Infect. Control 28:277-281. [DOI] [PubMed] [Google Scholar]
- 35.Wu, M., and R. E. Hancock. 1999. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J. Biol. Chem. 274:29-35. [DOI] [PubMed] [Google Scholar]
- 36.Zähner, H., and H.-P. Fiedler. 1995. The need for new antibiotics: possible ways forward, p. 67-85. In P. A. Hunter, G. K. Darby, and N. J. Russell (ed.), Fifty years of antimicrobials: past perspectives and future trends. Cambridge University Press, Cambridge, England.
- 37.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]