Abstract
The MICs of triclosan for 31 clinical isolates of Staphylococcus aureus were 0.016 μg/ml (24 strains), 1 to 2 μg/ml (6 strains), and 0.25 μg/ml (1 strain). All the strains for which triclosan MICs were elevated (>0.016 μg/ml) showed three- to fivefold increases in their levels of enoyl-acyl carrier protein (ACP) reductase (FabI) production. Furthermore, strains for which triclosan MICs were 1 to 2 μg/ml overexpressed FabI with an F204C alteration. Binding studies with radiolabeled NAD+ demonstrated that this change prevents the formation of the stable triclosan-NAD+-FabI complex, and both this alteration and its overexpression contributed to achieving MICs of 1 to 2 μg/ml for these strains. Three novel, potent inhibitors of FabI (50% inhibitory concentrations, ≤64 nM) demonstrated up to 1,000-fold better activity than triclosan against the strains for which triclosan MICs were elevated. None of the compounds tested from this series formed a stable complex with NAD+-FabI. Consequently, although the overexpression of wild-type FabI gave rise to an increase in the MICs, as expected, overexpression of FabI with an F204C alteration did not cause an additional increase in resistance. Therefore, this work identifies the mechanisms of triclosan resistance in S. aureus, and we present three compounds from a novel chemical series of FabI inhibitors which have excellent activities against both triclosan-resistant and -sensitive clinical isolates of S. aureus.
Initially, it was thought that the mode of antibacterial action of triclosan was via a general disruptive effect on bacterial membranes (15). However, it has now been shown that the mode of action of triclosan is via inhibition of enoyl-acyl carrier protein (ACP) reductase (FabI) (11) in organisms such as Staphylococcus aureus and Escherichia coli that rely on this enzyme to perform the ultimate step in the elongation cycle of bacterial fatty acid biosynthesis (3, 4, 5, 11, 16). Resistance to triclosan has been the subject of much discussion in recent years, and laboratory studies with E. coli and S. aureus have shown that mutations in FabI (G93V/S and G23S, respectively) and their overexpression cause decreases in susceptibility to triclosan (3, 4, 5, 11, 16). However, characterization of the triclosan resistance in clinically derived isolates of either organism has not yet been reported.
Triclosan exhibits exquisite activity against S. aureus and is used to control the carriage of methicillin-resistant S. aureus in hospitals (1). However, despite debate on the use of triclosan, few surveys have evaluated the level of resistance to triclosan in this important pathogen. Furthermore, the mechanism of triclosan resistance in clinically derived isolates of S. aureus has not been reported. Consequently, in this work we have identified a set of triclosan-resistant isolates of S. aureus and elucidated the mechanisms that give rise to reduced susceptibility to triclosan. Furthermore, we present some novel inhibitors of FabI which are active against these triclosan-resistant isolates of S. aureus. These compounds are different from the molecules described elsewhere (13), although they are from the same chemical series.
MATERIALS AND METHODS
Bacterial strains.
The 31 clinical isolates of S. aureus included in the antimicrobial activity assays were obtained from the culture collection of GlaxoSmithKline, Upper Providence, Pa. These strains consisted of standard laboratory and reference strains as well as geographically distinct isolates from various clinical sources isolated at different times.
Antimicrobial activity assay.
Whole-cell antimicrobial activity was determined by broth microdilution. The test compounds were dissolved in dimethyl sulfoxide and diluted 1:10 in water to produce a 256-μg/ml stock solution. Fifty microliters of the stock solution was serially diluted into cation-adjusted Mueller-Hinton broth (Becton Dickinson, Cockeysville, Md.) by using a 96-well microtiter plate (Microlab AT Plus 2; Hamilton Co., Reno, Nev.). After the compounds were diluted, a 50-μl aliquot of the test isolate (∼106 CFU/ml) was added to each well of the microtiter plate. The final test concentrations ranged from 0.001 to 128 μg/ml. The inoculated plates were incubated at 35°C in ambient air for 18 to 24 h. The MIC was determined as the lowest concentration of compound that visibly inhibited growth.
Sequencing of FabI from clinical isolates.
The cell pellet from a 150-μl S. aureus culture was lysed in 50 μl of lysis buffer (10 mM Tris [pH 8], 1 mM EDTA, 0.35 M sucrose, 200 μg of lysostaphin per ml) by incubation at 37°C for 15 min, at 95°C for 5 min, and on ice for 5 min. After centrifugation, equal volumes of the supernatant and water were mixed to yield the chromosomal DNA preparation. By using DNA oligonucleotides complementary to the upstream and downstream DNA sequences of fabI, the S. aureus fabI gene was amplified by PCR (with a mixture of 1 μl of chromosomal DNA, 25 pmol of primers, and 2.5 U of Pfu DNA polymerase for 30 cycles at 55°C). The PCR products were purified and sequenced.
Examination of FabI abundance in various S. aureus strains by Western immunoblotting.
Mid-log-phase cell cultures for various S. aureus strains were collected and normalized to 1 optical density OD unit at 600 nm (∼5 × 108 CFU/ml). Cell samples were lysed and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting by protocols described earlier (18). Polyclonal rabbit antibodies were generated with purified FabI, FtsZ, and SpsB as antigens by a previously described method (16).
Preparation of F204C FabI enzyme.
As described previously (D. J. Payne, W. E. DeWolf, D. R. Gentry, H. Kallender, K. H. Pearce, and S. C. Pearson, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. K131, 1999), FabI was cloned from S. aureus WCUH29 by PCR to create vector pET16FabI. To provide recombinant F204C protein for biochemical studies, the S. aureus FabI protein with the F204C mutation (the F204C FabI protein) was created by mutating wild-type S. aureus FabI in an overexpression vector (pET16FabI) by using the Stratagene (La Jolla, Calif.) QuikChange kit. The overexpressed proteins were purified in a two-step process by using blue Sepharose and anion-exchange chromatography. FabI activity was monitored spectrophotometrically by following the oxidation of NAD(P)H at 340 nm in a Spectramax plate reader (Molecular Devices, Corp. Sunnyvale, Calif.). The standard conditions for the assay were as follows: 100 mM N-(2-acetamido)-2-iminodiacetic acid (ADA; pH 6.5), 4% glycerol, 25 μM crotonoyl-ACP, 50 μM NAD(P)H, and FabI. Assays were carried out in a volume of 150 μl in Costar half-area plates.
Characterization of F204C FabI enzyme.
By using the buffer described above, kinetic characterization of both S. aureus wild-type FabI and the F204C mutant was carried out by using crotonoyl-ACP and both NADH and NADPH as substrates. For Km determinations, the concentrations used for NAD(P)H were 200, 100, 75, 50, 25, 20 15, and 10 μM with crotonoyl-ACP at a fixed concentration of 25 μM. The concentrations used for determination of the crotonoyl-ACP Km were 25, 15, 10, 7.5, 5, 3, 2, and 1.25 μM with NAD(P)H at a fixed concentration of 50 μM. NAD(P)H Kms were determined by using the standard Michaelis-Menten equation. Crotonoyl-ACP exhibits substrate inhibition, and therefore, Km values were determined by using an equation describing substrate inhibition: v = (Vmax · S)/(Km + S + S2)/Ki), where S is the concentration of crotonoyl-ACP. Fifty percent inhibitory concentrations (IC50s) were determined by performing assays with 25 μM crotonoyl-ACP and 50 μM NAD(P)H. Ten different concentration of triclosan between 10 and 0.078 μM were used. All kinetic parameters (kcat, Km, and IC50) were determined with Grafit software (Erithacus Software Ltd.).
Binding of triclosan to wild-type and F204C FabI.
The binding affinity of triclosan to FabI was measured by a filter binding assay (5). Various amounts of enzyme (5, 2.5, 1.25, 0.625, and 0 μg) were incubated with 5 μM [3H]NAD+ and 10 μM triclosan in a final assay volume of 50 μl. After a 15-min incubation at room temperature, 40 μl of each reaction mixture was transferred to an MHAB N45 filter plate (Millipore, Bedford, Mass.) prewetted with 100 μl of 100 mM ADA (pH 6.5). Samples were filtered with a MultiScreen vacuum manifold (Millipore). The plate was washed three times with 100 μl of 100 mM ADA (pH 6.5) to remove free [3H]NAD+. The plate was then dried in an oven at 37°C for 1 h, 100 μl of scintillation cocktail (Microscint 40; Packard) was added to each sample well, and counts were recorded on a Packard Top Count scintillation counter.
Construction of isogenic strains to evaluate effects of FabI overexpression and F204C on FabI inhibitors.
The wild-type fabI gene from S. aureus WCUH29 was amplified by PCR and cloned into vector pYH4 (J. Huang et al., unpublished data, 2002) yielding plasmid pI-wt. pYH4 is an E. coli-S. aureus shuttle expression vector that allows overexpression of S. aureus genes in its native host. Unlike pET16FabI, which can replicate only in E. coli, pYH4-FabI can replicate in both E. coli and S. aureus hosts. This plasmid was then used as a template to generate plasmid pI-F204C by site-directed mutagenesis with a QuikChange kit from Stratagene. These plasmids were separately introduced into S. aureus RN4220. The plasmid DNA was then purified from RN4220 and used to transform WCUH29. Transformation of S. aureus was carried out by electroporation (9).
Preparation of novel FabI inhibitors.
The three novel FabI inhibitors were designed and synthesized as described previously (D. J. Payne et al., Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1688, 2001; W. H. Miller et al., Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1689, 2001; M. A. Seefeld et al., Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1690, 2001). These compounds were different from those described by Payne et al. (13) but originated from the same chemical series. The inhibitory potency of each compound for FabI was determined by previously described methods (13).
Tools for determination of mode of antibacterial action.
By using a methodology described previously (16), an S. aureus strain overexpressing FabI was used to confirm that the mode of antibacterial action of the novel FabI inhibitors was via FabI.
Computational modeling of F204C FabI.
A model of F204C FabI was created from the X-ray crystal structure of triclosan complexed to wild-type E. coli FabI (14). F204 was converted to a cysteine with the MOE software package (Chemical Computing Group, Montreal, Quebec, Canada), and the resulting structure was minimized.
RESULTS
Characterization of triclosan resistance in clinical isolates of S. aureus.
The MICs of triclosan for 31 random clinical isolates of S. aureus were 0.016 μg/ml (24 strains), 1 to 2 μg/ml (6 strains), and 0.25 μg/ml (1 strain).
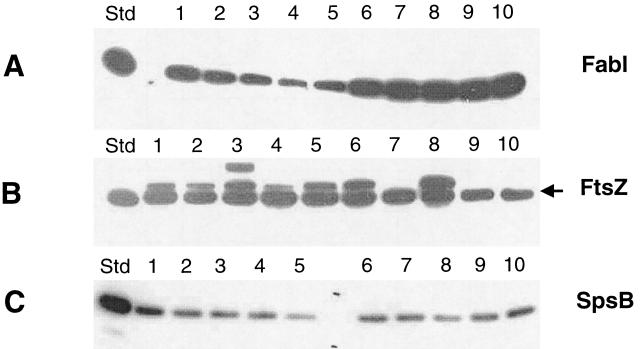
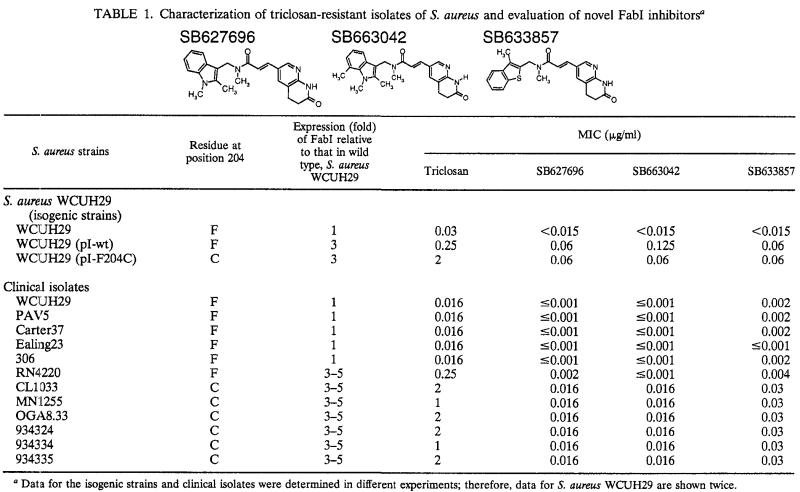
It has previously been shown that overexpression of FabI in S. aureus results in reduced susceptibility to triclosan (5, 16). Therefore, the abundance of FabI and two other proteins (FtsZ, a cell division protein, and SpsB, leader peptidase) were examined by Western immunoblotting for five of the triclosan-sensitive strains and seven of the strains for which triclosan MICs were elevated. The results are shown in Fig. 1 and Table 1. The abundance of FabI in the seven strains for which triclosan MICs were elevated was three to five times higher than that observed in the five sensitive strains (Table 1), while the abundance of FtsZ and SpsB remained the same. Therefore, higher levels of expression of FabI in these clinical isolates correlated with the observed decrease in sensitivity to triclosan.
FIG. 1.
Abundance of FabI in various S. aureus strains determined by Western immunoblotting of the same set of S. aureus cell samples with antibodies against FabI (A), FtsZ (B), and SpsB (C). Ten nanograms of purified protein was used as a standard (Std) in each blot. Lanes: 1, WCUH29; 2, PAV5; 3, Carter37; 4, Ealing23; 5, 306; 6, RN4220; 7, CL1033; 8, MN1255; 9, OGA8.33; 10, 934324 (data for strains 934334 and 934335 are not shown).
TABLE 1.
Characterization of triclosan-resistant isolates of S. aureus and evaluation of novel FabI inhibitorsa
Data for the isogenic strains and clinical isolates were determined in different experiments; therefore, data for S. aureus WCUH29 are shown twice.
The FabI genes from five of the sensitive strains and the seven strains for which triclosan MICs were elevated were sequenced. The sequences of the FabI genes from each of the sensitive strains and the strain for which the MIC was 0.25 μg/ml were identical. However, the DNA sequence of the FabI produced by each of the strains for which triclosan MICs were 1 to 2 μg/ml all predicted a cysteine at amino acid residue 204, which is a phenylalanine in FabI from the sensitive organisms.
Biochemical characterization of F204C.
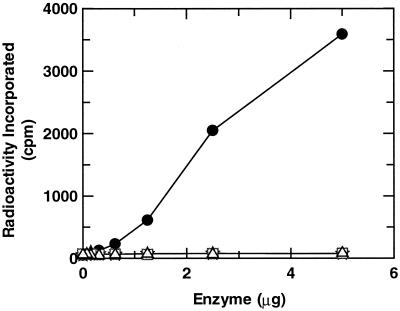
In order to examine the effects of the amino acid residue at position 204 on inhibition by triclosan, the mutant FabI (F204C FabI) was purified and found to possess kinetic parameters similar to those of wild-type FabI (Table 2). The IC50s of triclosan for the mutant enzyme were only twofold higher. Progress curves for the native S. aureus FabI indicated that inhibition by triclosan was time dependent, as described previously (5), but this was not observed with the mutant enzyme. Consequently, the measured IC50s do not truly reflect the relative affinities of triclosan for the two enzymes. Previously, binding studies with radiolabeled NAD+ demonstrated that triclosan inhibits E. coli FabI via the formation of a stable triclosan-NAD+-FabI complex (17), and this complex is thought to be responsible for the excellent activity of triclosan against S. aureus (5). We demonstrated that a similar complex is formed between triclosan-NAD+ and wild-type S. aureus FabI, but similar studies performed with the mutant FabI demonstrated no evidence for the formation of this stable complex (Fig. 2). Consequently, F204C prevents the formation of this complex and thus reduces the inhibitory effect of triclosan on the target. This suggests that this substitution contributes to the elevated MICs of triclosan observed for the clinical isolates harboring it.
TABLE 2.
Kinetic characterization of F204C FabI
| FabI |
Km (μM)
|
kcat (min−1)
|
Triclosan IC50 (μM)
|
||||
|---|---|---|---|---|---|---|---|
| NADH | NADPH | Crotonoyl-ACP (NADPH) | NADH | NADPH | NADPH | NADH | |
| Wild type | 124 | 32 | 9 | 12 | 28 | 0.35 | 0.54 |
| Mutant (F204C) | 158 | 26 | 12 | 22 | 25 | 0.74 | 1 |
FIG. 2.
Binding of 3H-NAD+ to the S. aureus FabI wild-type and F204C mutant enzymes in the presence and absence of triclosan. ✫, wild-type FabI; •, wild-type FabI plus triclosan; □, mutant F204C FabI; ▵, mutant F204C FabI plus triclosan.
Analysis of relative contributions of FabI overexpression and F204C to resistance to triclosan (isogenic strains).
The relative contribution of FabI overexpression and F204C to the overall resistance observed in the strains for which triclosan MICs are elevated was then investigated. Expression plasmids for wild-type and F204C FabI were constructed and introduced into triclosan-sensitive strain WCUH29. The expression of the fabI gene was under the control of a regulated promoter, Pxy/tet (18). After induction with the inducer, the abundance of FabI was threefold higher than that in the parent strain, S. aureus WCUH29, and FabI was expressed at levels similar to those observed in the triclosan-resistant strains (data not shown). It was observed that overexpression of wild-type FabI in S. aureus WCUH29 gave rise to an increase in the MIC of triclosan from 0.016 to 0.25 μg/ml. The same was observed with S. aureus clinical isolate RN4220, which overexpresses only the wild-type FabI (Table 1). However, similar levels of overexpression of the mutant FabI in S. aureus WCUH29 caused the MIC to increase to 1 to 2 μg/ml (Table 1).
Characterization of novel FabI inhibitors against triclosan-resistant strains of S. aureus.
SB633857, SB627696, and SB663042 were all potent inhibitors of the S. aureus FabI (IC50s, 52, 64, and <3 nM, respectively). These three novel inhibitors of FabI were tested against the 31 clinical strains of S. aureus (Table 1). The compounds demonstrated between >16- and 8-fold greater activity than triclosan against the triclosan-sensitive strains and up to 1,000-fold better activity than triclosan against the strains for which triclosan MICs were elevated. Primary FabI assay data for compounds from this series of inhibitors did not demonstrate progressive time-dependent inhibition, and in the binding studies with radiolabeled NAD+, no stable complex with FabI and NAD+ was observed with any of the compounds tested from this series (data not shown). Examination of the activities of the novel FabI inhibitors against the isogenic strains revealed that the overexpression of either wild-type or mutant FabI gave rise to similar increases in MICs, suggesting that F204C has little effect on the antibacterial activities of the novel FabI inhibitors.
DISCUSSION
Our study with a relatively small collection of S. aureus isolates has identified seven strains for which triclosan MICs were elevated, and our data suggest that clinical isolates of S. aureus use two mechanisms for protection against triclosan. These data demonstrate not only that a mutation in FabI is required for triclosan resistance but also that this altered FabI needs to be overexpressed at three- to fivefold higher levels than the level of expression in triclosan-sensitive strains. Intermediate resistance (MICs, 0.25 μg/ml), as observed with strain RN4220, was achieved via overexpression of the wild-type FabI protein only. Binding studies with radiolabeled NAD+ demonstrated that F204C negates the ability of triclosan to form a stable NAD+-FabI-triclosan complex. Therefore, this substitution contributed to the decrease in sensitivity of the host strain to triclosan. Isogenic strains overexpressing the wild-type and mutant FabI clearly confirmed that overexpression of the wild-type FabI caused the MIC of triclosan to increase by eightfold, but overexpression of F204C was required to achieve the >50-fold increase in MIC which gave rise to an MIC of 2 μg/ml for the triclosan-resistant isolates of S. aureus.
The mechanism for higher levels of expression of FabI in the triclosan-resistant strains is unclear at this point. Sequencing of the promoter region of the fabI gene in all the strains revealed some variations between triclosan-sensitive and -resistant strains. However, these variations did not appear to have significant effects on the promoter activities when the activities were tested with a reporter gene (data not shown).
If the collection of strains tested in this study is representative of the species, overexpression of wild-type FabI (giving rise to an MIC of 0.25 μg/ml) does not appear to be advantageous in S. aureus, since it was identified in only one of the strains that we tested. Overexpression of the F204C FabI, giving rise to triclosan MICs of 1 to 2 μg/ml, appeared to be the preferred resistance mechanism.
Previous laboratory studies identified G93V/S or G23S FabI mutations as giving rise to triclosan resistance in E. coli and S. aureus, respectively (3, 4, 5, 11, 16). However, in this study we show for the first time that the F204C change appears to be the substitution found in triclosan-resistant clinical strains of S. aureus. It will be interesting to evaluate what resistance mechanism(s) that the novel FabI inhibitors would select in S. aureus.
The novel FabI inhibitors described in this study were intrinsically more active than triclosan against S. aureus. Moreover, they also exhibited exceptional activity against the triclosan-resistant strains of S. aureus. Overexpression of FabI decreased the susceptibilities to these compounds. Although F204C prevented triclosan from forming a stable NAD+-FabI-triclosan tertiary complex, there is no evidence to show that the novel FabI inhibitors form such a complex, and their binding orientation is likely to be different from that of triclosan. Therefore, it was not surprising that the F204C change had no effect on the exceptional antibacterial activities exhibited by these novel inhibitors. This was demonstrated by the isogenic strains, in which overexpression of mutant FabI did not give rise to further increases in MICs compared to those for the strain overexpressing wild-type FabI.
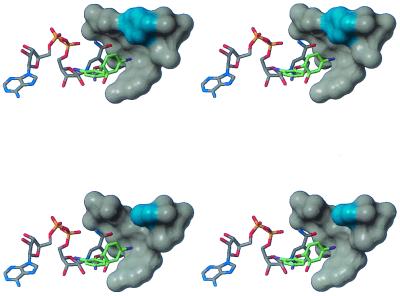
Examination of the X-ray crystal structure of triclosan bound to wild-type E. coli FabI (14) suggested a plausible structural explanation for the loss in inhibitory potency of triclosan against S. aureus F204C FabI. As bound in the crystal structure, one of the chlorine atoms of triclosan binds in a hydrophobic pocket near the nicotinamide ring of the cofactor and near F203, which corresponds to F204 of the S. aureus sequence. F203 of E. coli (shown in cyan in the upper panel of Fig. 3) forms part of the interaction surface between the protein and this chlorine atom, while computational mutation of F203 to cysteine (shown in cyan in the lower panel of Fig. 3) results in the loss of this interaction. The twofold loss of potency of triclosan against the mutant FabI was consistent with estimates of the energetic cost of removing a single hydrophobic group (6, 7, 10). The novel FabI inhibitors are not expected to bind in this particular region of the protein; the expected difference in binding provided a plausible explanation of why F204C had no significant effect on the potencies of these compounds.
FIG. 3.
Structural consequences of F204C mutation: X-ray crystal structure of triclosan bound to wild-type E. coli FabI (upper figure) and model of triclosan bound to F203C E. coli FabI (lower figure). The side-chain atoms of F203 and C203 are highlighted in cyan. Stereographic images were created with the program MolMol (8).
This work has further illustrated that for triclosan to exhibit potent antibacterial activity, a stable tertiary complex with FabI is required. This perhaps suggests that such a complex is a prerequisite for a FabI inhibitor to exhibit good antibacterial activity. However, no evidence for such a complex was observed with the novel FabI lead compounds; therefore, it was demonstrated with these compounds that potent antibacterial activity can also be achieved by sufficiently active reversible inhibitors of FabI. In addition, the novel FabI lead compounds possessed greater antibacterial activity than triclosan, suggesting that optimization of reversible inhibitors of enoyl-ACP reductase may be a more successful strategy for designing anti-infective agents against this target.
In conclusion, we have identified the mechanisms of triclosan resistance in isolates of S. aureus. In addition, we present three compounds from a novel chemical series of FabI inhibitors which demonstrate excellent potencies against both triclosan-resistant and -sensitive isolates of S. aureus. This work further illustrates the potential of fatty acid biosynthesis as a source of novel antibacterial strategies (12).
REFERENCES
- 1.Bamber, A. I., and T. J. Neal. 1999. An assessment of triclosan susceptibility in methicillin-resistant and methicillin sensitive Staphylococcus aureus. J. Hosp. Infect. 41:107-109. [DOI] [PubMed] [Google Scholar]
- 2.Cleland, W. W. 1979. Statistical analysis of enzyme kinetic data. Methods Enzymol. 63:103-138. [DOI] [PubMed] [Google Scholar]
- 3.Heath, R. J., Y.-T. Yu, M. A. Shapiro, E. Olson, and C. O. Rock. 1998. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 273:30316-30320. [DOI] [PubMed] [Google Scholar]
- 4.Heath, R. J., J. R. Rubin, D. R. Holland, E. Zhang, M. E. Snow, and C. O. Rock. 1999. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J. Biol. Chem. 274:11110-11114. [DOI] [PubMed] [Google Scholar]
- 5.Heath, R. J., U. T. Yu, M. A. Shapiro, E. Olson, and C. O. Rock. 2000. Inhibition of the Staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J. Biol. Chem. 275:4654-4659. [DOI] [PubMed] [Google Scholar]
- 6.Holder, J. B., A. F. Bennett, J. Chen, D. S. Spencer, M. P. Byrne, and W. E. Stites. 2001. Energetics of side chain packing in staphylococcal nuclease assessed by exchange of valines, isoleucines, and leucines. Biochemistry 40:13998-14003. [DOI] [PubMed] [Google Scholar]
- 7.Kellis, J. T., Jr., K. Nyberg, D. Sali, and A. R. Fersht. 1988. Contribution of hydrophobic interactions to protein stability. Nature (London) 333:784-786. [DOI] [PubMed] [Google Scholar]
- 8.Koradi, R., M. Billeter, and K. Wüttrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics 14:51-55. [DOI] [PubMed] [Google Scholar]
- 9.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:373-376. [Google Scholar]
- 10.Kuntz, I. D., K. Chen, K. A. Sharp, and P. A. Kollman. 1999. The maximal affinity of ligands. Proc. Natl. Acad. Sci. USA 96:9997-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Triclosan targets lipid synthesis. Nature (London) 394:531-532. [DOI] [PubMed] [Google Scholar]
- 12.Payne, D. J., P. V. Warren, D. J. Holmes, Y. Ji, and J. T. Lonsdale. 2001. Bacterial fatty acid biosynthesis: a genomics driven target for antibacterial drug discovery. Drug Discovery Today 6:537-544. [DOI] [PubMed] [Google Scholar]
- 13.Payne, D. J., et al. 2002. Discovery of a novel and potent class of FabI-directed antibacterial agents. Antimicrob. Agents Chemother. 46:3118-3124. [DOI] [PMC free article] [PubMed]
- 14.Qiu, X., C. A. Janson, R. I. Court, M. G. Smyth, D. J. Payne, and S. S. Abdel-Meguid. 1999. Molecular basis for triclosan activity involves a flipping loop in the active site. Protein Sci. 8:2529-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regos, J., and H. R. Hitz. 1974. Investigations on the mode of action of triclosan, a broad spectrum antimicrobial agent. Zentbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. Reihe A 226:390-401. [PubMed] [Google Scholar]
- 16.Slater-Radosti, C., G. Van Aller, R. Greenwood, R. O. Nicholas, P. Keller, W. E. DeWolf, F. Fan, D. J. Payne, and D. D. Jaworski. 2001. Biochemical and genetic characterisation of the action of triclosan on Staphylococcus aureus. J. Antimicrob. Chemother. 48:1-6. [DOI] [PubMed] [Google Scholar]
- 17.Ward, W. H. J., G. A., Holdgate, S. Rowsell, E. G. McLean, R. A. Paupit, E. Clayton, W. W. Nichols, J. G. Colls, C. A. Minshull, D. A. Jude, A. Mistry, D. Timms, R. Camble, N. J. Hales, C. J. Britton, and I. W. F. Taylor. 1999. Kinetic and structural characteristics of the inhibition of enoyl(acyl carrier protein) reductase by triclosan. Biochemistry 38:12514-12525. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, L., F. Fan, L. M. Palmer, M. A. Lonetto, C. Petit, L. L. Voelker, A. St. John, B. Bankosky, M. Rosenberg, and D. McDevitt. 2000. Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene 255:297-305. [DOI] [PubMed] [Google Scholar]