Abstract
The antibiotic growth promoter avilamycin inhibits protein synthesis by binding to bacterial ribosomes. Here the binding site is further characterized on Escherichia coli ribosomes. The drug interacts with domain V of 23S rRNA, giving a chemical footprint at nucleotides A2482 and A2534. Selection of avilamycin-resistant Halobacterium halobium cells revealed mutations in helix 89 of 23S rRNA. Furthermore, mutations in helices 89 and 91, which have previously been shown to confer resistance to evernimicin, give cross-resistance to avilamycin. These data place the binding site of avilamycin on 23S rRNA close to the elbow of A-site tRNA. It is inferred that avilamycin interacts with the ribosomes at the ribosomal A-site interfering with initiation factor IF2 and tRNA binding in a manner similar to evernimicin.
Molecular models resulting from the recent identification of X-ray crystal structures of ribosomal subunits and rRNA components of the binding site of antibiotics (6, 9, 15, 22) are notable new tools for drug design. New efficient “wonder drugs” are thus to be expected from this development. Unfortunately, based on experience, we know that resistance will evidently appear at some point. Knowledge about resistance mechanisms might help decide whether it is worth developing a drug for commercial use and how to minimize the occurrence and spread of resistance. Elucidation of mechanisms of resistance to current antibiotics is also essential if we are to control this increasing and widespread problem. Identification of the binding site of a drug can guide both the elucidation of its inhibitory mechanisms and the identification of resistance mechanisms.
The antibiotic avilamycin is used as a growth promoter and thus is fed in large amounts to animals. Bacterial resistance to avilamycin increases in proportion to consumption of the drug and, conversely, decreases again when the drug is withdrawn (3). The organism producing avilamycin accommodates two rRNA methyltransferases and an ATP binding transporter conferring avilamycin resistance (20). The only drug that so far shows cross-resistance to avilamycin is the structurally similar antibiotic evernimicin (1), which has been considered for clinical use. Consistent with this, an rRNA methyltransferase has been isolated from an Enterococcus faecium strain exhibiting resistance to avilamycin and evernimicin (13). Recent reports on evernimicin have revealed details of its binding to 23S rRNA and have shown that resistance to evernimicin can be generated by mutations in both 23S rRNA and L16 genes (4, 5, 7, 14). Also, resistance to avilamycin has been associated with alterations in the ribosomal protein L16 (2).
The interaction of avilamycin with rRNA has been investigated in two ways in this study. First, chemical footprinting was used to define the binding site of avilamycin on Escherichia coli ribosomes. Second, Halobacterium halobium, which contains only one rRNA operon, was used to select for avilamycin-resistant mutants with changes in the 23S rRNA. We found resistance mutations in the same region where mutations causing resistance to evernimicin have previously been found, and we assayed those mutants for cross-resistance. From these and other results, we draw conclusions about the avilamycin binding site, its inhibitory mechanism, and a resistance mechanism.
MATERIALS AND METHODS
Growth of cells and isolation of ribosomes and 50S ribosomal subunits.
E. coli MRE 600 cells were grown to an optical density at 550 nm of 0.5 to 0.6 in Luria broth (LB). The cells were harvested, lysed, and ribosomal subunits isolated as described previously (17).
Footprinting of avilamycin on E. coli ribosomal subunits.
The final experiments were performed as follows. Ribosomes (5 pmol) were incubated with avilamycin in modification buffer (50 mM HEPES-KOH [pH 8.0], 10 mM MgCl 2, 100 mM KCl, 5 mM dithiothretiol) for 30 min at 37°C. Modification procedures were described previously (8). Aliquots (50 μl) of the antibiotic-ribosome complexes were modified with either 2 μl of dimethyl sulfate (DMS) (1:6 dilution in 96% ethanol) for 10 min at 37°C or 50 μl of CMCT [1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate; 42 mg/ml in modification buffer] for 20 min at 37°C. DMS probes the accessibility of the N-1 position of adenosine and N-3 of cytosine residues, and CMCT probes the N-3 of uridine and N-1 of guanosine residues. The DMS reactions were stopped by adding 25 μl of stop buffer (1 M Tris-HCl [pH 7.75], 1 M 2-mercaptoethanol, 1 mM EDTA) followed by precipitation. The CMCT reactions were stopped by precipitating the ribosomes. After centrifugation, the ribosomes were resuspended in 0.25 M sodium acetate and extracted with phenol-chloroform. The rRNA was precipitated and resuspended in water.
The chemical modifications were monitored by primer extension analysis on 23S rRNA using aurian myeloblastosis virus reverse transcriptase (Life Sciences) and 5′-32P-labeled deoxyoligonucleotides (18). The cDNA products of the primer extension reactions were separated on 6% polyacrylamide sequencing gels. The positions of the stops in cDNA synthesis were visualized on a gel autoradiogram and identified by reference to dideoxy sequencing reactions on 23S RNA that were run in parallel.
Isolation of avilamycin-resistant mutants of H. halobium.
H. halobium strain R1 was grown in liquid high-salt SW-25 medium at 37°C as described previously (12). Avilamycin-resistant mutants were obtained in two ways. Approximately 10 7 cells were spread on SW-25 agar plates containing 66 μg of avilamycin per ml and incubated at 37°C for 2 weeks. Also, essentially as described previously (16), approximately 107 cells were inoculated into 100 ml of SW-25 medium containing avilamycin at 1.3 μg/ml, and every second day the culture was diluted into fresh medium containing avilamycin at 1.3 μg/ml. During dilution, 107 cells were also spread on SW-25 agar plates containing 13.2 μg of avilamycin per ml, and the plates were sealed and incubated at 37°C for 2 weeks. Individual colonies were picked for isolation of chromosomal DNA, and mutations were mapped by sequencing. A segment of 23S rDNA was PCR amplified by using a forward primer, GGCCCGGTGAACTGTACG (positions 2004 to 2021), and a reverse primer, GTTCCTCTCGTACTATACG (positions 2649 to 2667), and sequenced.
Testing of avilamycin resistance.
Avilamycin sensitivity of the wild-type and mutant cells was determined by diluting an exponentially growing culture 1,000-fold in duplicate into 2 ml of SW-25 medium with avilamycin. Growth was measured after 3 days of incubation at 37°C, and the relative growth of the mutants and wild type in the absence and presence of avilamycin was determined.
Peptidyltransferase assays.
The effect of avilamycin on peptidyltransferase activity was measured as described previously (17) by incubating ribosomes with N-acetyl-[14C]Phe-tRNAPhe, poly(U), and puromycin and counting the released radioactivity after extraction. The peptidyltransferase activity was analyzed in the presence of avilamycin at concentrations ranging from 0 to 660 μg/ml.
RESULTS AND DISCUSSION
Footprinting of avilamycin on E. coli ribosomes.
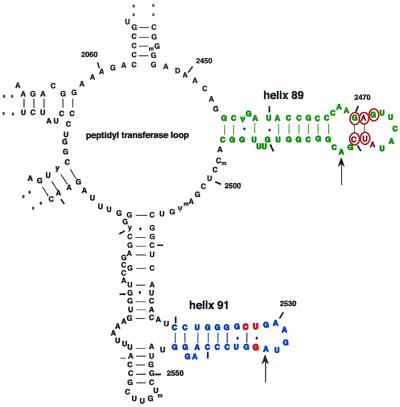
The drug binding site was studied by chemical probing of accessible nucleotides. After binding of avilamycin to ribosomes, the complexes and free ribosomes were treated with DMS and CMCT and alterations in the chemical reactivity of specific nucleotides in domain V of 23S rRNA induced by the binding of the drug to the ribosome were identified by primer extension with reverse transcriptase. The avilamycin footprints were investigated over a range of concentrations to identify the lowest concentration giving maximal protection effects. Positions A2482 and A2534 were affected, as shown in the autoradiogram in Fig. 1. These positions are marked on the secondary-structure model of 23S rRNA in Fig. 2. No other footprint was observed in domain V of 23S rRNA. In contrast to most other classes of antibiotics interacting in domain V of 23S rRNA, the footprint of avilamycin is not close to the central ring structure constituting part of the peptidyltransferase area. Only the structurally similar evernimycin has so far been observed to footprint in this region at A2468, A2469, A2476, A2478, and A2482 in helix 89 and at A2534 in helix 91 (7). Evernimicin thus affects more nucleotides and the protection seems to be stronger, indicating that it binds more tightly to the ribosomes than does avilamycin. Evernimicin has additional substituents relative to avilamycin, which probably enhance binding.
FIG. 1.
Gel autoradiogram of the avilamycin footprint in domain V of 23S rRNA after binding to ribosomes, showing DMS modifications detected by reverse transcriptase of rRNA. U, C, G, and A, dideoxy sequencing lanes; No mod., chemically unmodified rRNA; No drug, chemically modified ribosomes in the absence of avilamycin. The triangle denotes modification in the presence of 1.3, 6.6, 13.2, 66, 132, and 264 μg of avilamycin per ml. Positions with altered reactivities are indicated by arrows. Reverse transcriptase stops one nucleotide before the corresponding nucleotide in the sequencing tracks.
FIG. 2.
Secondary-structure model of the central loop region of domain V of E. coli 23S rRNA. Small numbers are nucleotide positions in 23S rRNA, and large numbers are helix numbers. The avilamycin footprints are indicated by arrows. The corresponding positions of mutations in H. halobium selected with avilamycin (this study; indicated by circles) or evernimicin (7; indicated by red letters and circles) are marked. The helix 89 region is in green; the helix 91 region is in blue. (Adapted from Gutell Lab., comparative RNA web site http://www.rna.icmb.utexas.edu/.)
Selection of 23S rRNA mutations in H. halobium conferring avilamycin resistance.
Antibiotic sensitivity testing demonstrated that avilamycin efficiently inhibits the growth of H. halobium, and because this organism possesses a single-copy rRNA operon, it is convenient for investigating whether avilamycin resistance can be caused by mutations in rRNA. Spontaneous mutants were obtained by plating H. halobium cells on avilamycin-containing agar plates as described in Materials and Methods. Five different mutant clones were obtained. Sequencing of part of the 23S rRNA gene showed the following mutations: G2470 to U, A2471 to G, G2472 to U, U2479 to C, and C2480 to U (the positions are in E. coli numbering). The mutations are positioned close to each other in the secondary structure in helix 89, as illustrated in Fig. 2. The growth of the mutants is either not or only slightly impaired compared to that of the wild type, demonstrating that none of the mutations interfered severely with the ribosome function.
The mutations obtained are considered solely responsible for the avilamycin resistance. First, it is well established that single mutations in rRNA can cause antibiotic resistance. Second, single mutations in the same region were found in a similar study to cause resistance to evernimicin, and two of these mutations were transferred to wild-type H. halobium and conferred resistance (7). Third, an rRNA methyltransferase exhibiting resistance to avilamycin and evernimicin methylates position G2470 (13), also indicating a close interaction between the antibiotic and this region of rRNA.
Avilamycin resistance conferred by different 23S rRNA mutations.
The level of avilamycin resistance was determined in mutants from both this study and the evernimicin study by Belova et al. (7) to determine whether all mutations conferring evernimicin resistance would also cause avilamycin resistance. This study was performed with the strains in liquid SW-25 medium (19) as described in Materials and Methods. All mutants exhibited resistance to avilamycin, and in general, the changes in helix 89 conferred higher resistance than did the changes in helix 91 (Table 1). This pattern is very similar to that observed with evernimicin (7). Selection of the same or nearby mutations and the generation of cross-resistance with the two drugs underline the concern about using distinct but chemically similar drugs for growth promotion and therapeutic use.
TABLE 1.
Sensitivity of mutants to avilamycina
| Strainb | IC50 (μg/ml)c |
|---|---|
| Wild type | <0.3 |
| U2470 | >66 |
| G2471 | >66 |
| C2471 | >66 |
| U2472 | 6.6 |
| C2478 | >66 |
| C2479 | >66 |
| U2480 | >66 |
| A2527 | >66 |
| C2528 | 6.6 |
| A2535 | 6.6 |
Growth was tested at the following concentrations of avilamycin: 0, 0.3, 1.3, 6.6, 13.2, 33, and 66 μg/ml.
The underlined mutations were obtained in this study by selection with avilamycin. Except for U2470 and C2472, all mutants were obtained by Belova et al. (7) by selection with evernimicin.
IC50, minimal concentrations of avilamycin where growth (measured by absorbance) was 50% or less of growth in the absence of antibiotic.
Mutations have also been obtained in the same region at 23S rRNA positions A2469C, C2480U, G2535A, and G2536C in evernimicin-resistant Streptococcus pneumoniae (4), although this organism has four rRNA operons. This emphasizes that this kind of resistance is not confined to organisms with one or two operons.
Inhibitory mechanism of avilamycin.
The avilamycin footprint at A2482 and the avilamycin mutations indicate direct binding of the drug to the 23S rRNA helix 89 region. The X-ray crystal structure of the the Thermus thermophilus 70S ribosome with tRNAs (22) identifies the loop region of helix 89 of 23S rRNA as being very close to L16, as illustrated in Fig. 3. This is in good agreement with the observation that mutations around position 52 in L16 in different gram-positive organisms also confer avilamycin resistance (2, 5, 14).
FIG. 3.
The T. thermophilus ribosome with tRNAs (PDB accession no. 1GIX and 1GIY [22]) emphasizing the putative binding site of avilamycin. (A) The rRNA and protein backbone are shown in grey, and tRNA in the A-, P-, and E-site is shown in red, purple and orange. Helix 89, where mutations confer avilamycin resistance, is indicated in green, and helix 91 is indicated in blue. (B) Helix 89 (green) and helix 90 (blue) region with the nearby L16 (yellow) and tRNA in the A-site (red). The positions of the mutations selected by avilamycin are indicated by red balls, and the avilamycin footprints are indicated by black balls. The grey background tubing is the backbone of domain V of 23S rRNA. The molecular visualization was done using VMD (10).
An early study of the effect of avilamycin on ribosome function showed that it inhibited poly(U)-directed poly(Phe) protein synthesis and inhibited the binding of Phe-tRNA to ribosomes (21). To supplement these data, we added avilamycin to a peptidyltransferase assay where puromycin reacts with Phe-tRNA in the P-site of the ribosome. Puromycin mimics the acceptor tRNA in the A-site and forms an amide bond with the Phe on tRNA in the P-site. Avilamycin did not inhibit this peptidyltransferase reaction; it is therefore unlikely to be a general inhibitor of peptidyltransferase (data not shown). This is similar to results obtained with evernimicin, which also did not inhibit peptidyltransferase (7). Helix 89 lies parallel to the acceptor arm of tRNA in the A-site (22), and the mutations causing resistance are very nearby, as illustrated in Fig. 3.
Since avilamycin inhibits tRNA binding by no more than 50% (21) and since it interacts with rRNA proximal to the elbow of tRNA in the A-site, it is conceivable that avilamycin overlaps with the A-site of the ribosomes at some distance from the peptidyltransferase center. Recently, initiation factor IF2 has been shown to footprint at A2476 and A2478 and at residues around position 2660 (11) and is therefore also close to or in the A-site. Evernimicin inhibits the stimulatory activity of IF2 on the formation of initiation complexes (7), and it is likely that avilamycin acts in a similar way. All data are thus consistent with an avilamicin binding site at helix 89 in 23S RNA, probably contacting L16 and overlapping the binding site for IF2 and A-tRNA. This also explains why avilamycin affects both poly(U)-directed Phe synthesis and the formation of initiation complexes (21).
Concluding remarks.
Avilamycin binds to a region of the ribosome constituting helices 89 and 91 of 23S rRNA and probably ribosomal protein L16. It appears to inhibit protein synthesis by sterically blocking the correct positioning of IF2 and tRNA in the A-site. Mutations in helices 89 and 91 of 23S rRNA confer resistance to avilamycin. The similarity of the binding and resistance mechanisms of avilamycin and evernimicin is obvious, thus emphasizing the concerns about growth promoters, which might enhance resistance and thereby interfere with the clinical success of potentially useful drugs.
C2479>66 U2480>66 A2527>66 C25286.6 A25356.6
Acknowledgments
S. Mankin kindly provided evernimicin-resistent H. halobium strains. S. Douthwaite is thanked for critical reading of the manuscript, L. B. Johansson is thanked for excellent technical assistance, and F. M. Aarestrup is thanked for providing avilamycin.
The research was supported by the Danish Medical Research Council.
REFERENCES
- 1.Aarestrup, F. M. 1998. Association between decreased susceptibility to a new antibiotic for treatment of human diseases, everninomicin (SCH 27899), and resistance to an antibiotic used for growth promotion in animals, avilamycin. Microb. Drug Resist. 4:137-141. [DOI] [PubMed] [Google Scholar]
- 2.Aarestrup, F. M., and L. B. Jensen. 2000. Presence of variations in ribosomal protein L16 corresponding to susceptibility of enterococci to oligosaccharides (avilamycin and evernimicin). Antimicrob. Agents Chemother. 44:3425-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aarestrup, F. M., A. M. Seyfarth, H. D. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adrian, P. V., C. Mendrick, D. Loebenberg, P. McNicholas, K. J. Shaw, K. P. Klugman, R. S. Hare, and T. A. Black. 2000. Evernimicin (SCH27899) inhibits a novel ribosome target site: analysis of 23S ribosomal DNA mutants. Antimicrob. Agents Chemother. 44:3101-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adrian, P. V., W. Zhao, T. A. Black, K. J. Shaw, R. S. Hare, and K. P. Klugman. 2000. Mutations in ribosomal protein L16 conferring reduced susceptibility to evernimicin (SCH27899): implications for mechanism of action. Antimicrob. Agents Chemother. 44:732-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 7.Belova, L., T. Tenson, L. Xiong, P. M. McNicholas, and A. S. Mankin. 2001. A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit. Proc. Natl. Acad. Sci. USA 98:3726-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehresmann, C., F. Baudin, M. Mougel, P. Romby, J.-P. Ebel, and B. Ehresmann. 1987. Probing the structure of RNA in solution. Nucleic Acids Res. 15:9109-9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679-688. [DOI] [PubMed] [Google Scholar]
- 10.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38. [DOI] [PubMed] [Google Scholar]
- 11.La Teana, A., C. O. Gualerzi, and A. E. Dahlberg. 2001. Initiation factor IF 2 binds to the alpha-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA 7:1173-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lázaro, E., C. Rodriguez-Fonseca, B. Porse, D. Urena, R. A. Garrett, and J. P. Ballesta. 1996. A sparsomycin-resistant mutant of Halobacterium salinarium lacks a modification at nucleotide U2603 in the peptidyl transferase centre of 23 S rRNA. J. Mol. Biol. 261:231-238. [DOI] [PubMed] [Google Scholar]
- 13.Mann, P. A., and e. al. 2001. I. Mann, P. A., L. Xiong, A. S. Mankin, A. S. Chau, C. A. Mendrick, D. J. Najarian, C. A. Cramer, D. Loebenberg, E. Coates, N. J. Murgolo, F. M. Aarestrup, R. V. Goering, T. A. Black, R. S. Hare, and P. M. McNicholas. 2001. EmtA, a rRNA methyltransferase conferring high-level evernimicin resistance. Mol. Microbiol. 41:1349-1356. [DOI] [PubMed] [Google Scholar]
- 14.McNicholas, P. M., P. A. Mann, D. J. Najarian, L. Miesel, R. S. Hare, and T. A. Black. 2001. Effects of mutations in ribosomal protein L16 on susceptibility and accumulation of evernimicin. Antimicrob. Agents Chemother. 45:79-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 16.Porse, B. T., and R. A. Garrett. 1999. Sites of interaction of streptogramin A and B antibiotics in the peptidyl transferase loop of 23 S rRNA and the synergism of their inhibitory mechanisms. J. Mol. Biol. 286:375-387. [DOI] [PubMed] [Google Scholar]
- 17.Poulsen, S. M., C. Kofoed, and B. Vester. 2000. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol. 304:471-481. [DOI] [PubMed] [Google Scholar]
- 18.Stern, S., D. Moazed, and H. F. Noller. 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164:481-489. [DOI] [PubMed] [Google Scholar]
- 19.Tan, G. T., A. DeBlasio, and A. S. Mankin. 1996. Mutations in the peptidyl transferase center of 23 S rRNA reveal the site of action of sparsomycin, a universal inhibitor of translation. J. Mol. Biol. 261:222-230. [DOI] [PubMed] [Google Scholar]
- 20.Weitnauer, G., S. Gaisser, A. Trefzer, S. Stockert, L. Westrich, L. M. Quiros, C. Mendez, J. A. Salas, and A. Bechthold. 2001. An ATP-binding cassette transporter and two rRNA methyltransferases are involved in resistance to avilamycin in the producer organism Streptomyces viridochromogenes Tu57. Antimicrob. Agents Chemother. 45:690-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf, H. 1973. Avilamycin, an inhibitor of the 30 S ribosomal subunits function. FEBS Lett. 36:181-186. [DOI] [PubMed] [Google Scholar]
- 22.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]