Abstract
Functional crosstalk between G protein-coupled receptors in a homo- or heterodimeric assembly likely involves conformational changes at the dimer interface, but the nature of this interface is not yet established, and the dynamic changes have not yet been identified. We have mapped the homodimer interface in the dopamine D2 receptor over the entire length of the fourth transmembrane segment (TM4) by crosslinking of substituted cysteines. Their susceptibilities to crosslinking are differentially altered by the presence of agonists and inverse agonists. The TM4 dimer interface in the inverse agonist-bound conformation is consistent with the dimer of the inactive form of rhodopsin modeled with constraints from atomic force microscopy. Crosslinking of a different set of cysteines in TM4 was slowed by inverse agonists and accelerated in the presence of agonists; crosslinking of the latter set locks the receptor in an active state. Thus, a conformational change at the TM4 dimer interface is part of the receptor activation mechanism.
Keywords: crosslinking, cysteine, dopamine, oligomer, rhodopsin
G protein-coupled receptors (GPCRs) constitute a large superfamily of receptors that couple binding of a diverse group of ligands to activation of heterotrimeric G proteins (1). Although many GPCRs have been inferred to be dimers or oligomers in the plasma membrane (2-6), the role of GPCR oligomerization remains enigmatic. Functional interactions in heterodimeric receptor complexes have been inferred based on novel pharmacological properties and synergistic or antagonistic effects on signaling attributed to activation of each protomer (7-12). These findings are provocative but do not provide structural insights into the mechanism of this crosstalk.
Another form of crosstalk that requires communication between protomers is transactivation. In the family C heterodimeric GABAB receptor, the N terminus of GB1 (but not GB2) binds GABA (13), whereas cytoplasmic loops of GB2 couple to G protein (14-16). Transactivation has also been reported for a family C metabotropic glutamate receptor and for the family A leutinizing hormone (LH), follicle-stimulating hormone (FSH), and thyroid-stimulating hormone (TSH) receptors (17-20).
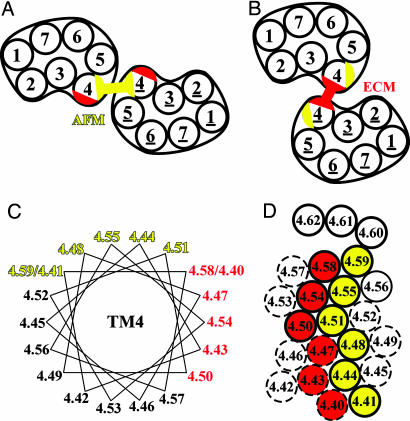
An understanding of the structural basis of crosstalk between receptors in a dimer requires identification of the dimerization interface and its changes, but information about these interfaces is still rather limited. Based on the spatial arrangement of rhodopsin arrays visualized by atomic force microscopy (AFM) of mouse retinal disk membranes in the inactive state, Liang et al. (21) built a molecular model that features a symmetric homodimer interface involving both transmembrane segment (TM) 4 and TM5 (Fig. 1A). In contrast, an 8.4-Å 3D structure was derived from electron cryomicroscopy (ECM) of tilted 2D crystals of squid rhodopsin reconstituted from detergent into lipid (22). This structure, which was proposed to be related to the arrangement of squid rhodopsin in the native membrane, has a symmetric interface involving just TM4 (Fig. 1B) (22). Thus, TM4 forms the major component of the interface in the packing arrangement of both the AFM dimer model of mouse rhodopsin and of the low-resolution ECM squid rhodopsin structure, but there are significant differences. In the AFM model, the interface consists of the face of TM4 closer to TM5 (yellow in Fig. 1) as well as a face of TM5 whereas, in the ECM-packing arrangement, the face of TM4 that is closer to TM3 is at the interface (red in Fig. 1).
Fig. 1.
Models of the rhodopsin dimer interface. Schematic arrangements illustrating the symmetric TM4-TM5 interface (yellow) of mouse rhodopsin in disk membranes in the dark as modeled by Liang et al. (21) based on AFM (A) and the symmetric TM4 interface (red) of squid rhodopsin deduced from ECM of 2D crystalline arrays in reconstituted membrane (22) (B). (C) Helical wheel representation of TM4 showing the indexed positions of the residues. Trp-4.50 (see Methods for indexing) is conserved in 96% of opsins and 99% of amine receptors and allows us to unambiguously align the D2R sequence with the rhodopsin sequence and structure. Residues at the AFM interface are shown in yellow, and those at the ECM interface are shown in red. The positions more extracellular than 4.59 are not colored due to their location in the last distorted turn of helix in bovine rhodopsin and the ambiguity as to the exact end of TM4 in D2R. (D) Helical net of TM4 colored as in C. The extracellular end of TM4 is at the top of the figure. Positions for which the corresponding cysteine mutants in D2R were crosslinked by CuP (see Fig. 3) are shown as solid circles, and those that were not crosslinked are shown as dashed circles.
We previously demonstrated that the dopamine D2 receptor (D2R) can be oxidatively crosslinked via Cys-1684.58 at the extracellular end of TM4 (23), at a position consistent with the ECM interface but not with the AFM interface (Fig. 1C). [Residues are identified by using an indexing method (24) that facilitates comparison with other family A GPCRs; see Methods for details.] To address this apparent discrepancy, as well as other evidence that TM4 may contribute to the dimer interface of several GPCRs (25-28), we now have mapped the entire contribution of TM4 to the homodimer interface. We show crosslinking of a subset of substituted cysteines along the entire length of TM4 in the D2R. The susceptibilities to crosslinking were differentially altered by the presence of agonists and inverse agonists. Symmetric crosslinking of residues in TM4, which would not be proximal in the AFM model of inactive receptor, activated the unliganded receptor. These results demonstrate that a change of the dimer interface is part of the receptor activation mechanism.
Methods
Numbering of Residues, Site-Directed Mutagenesis, and Transfection. Residues are numbered both according to their positions in the human D2short receptor sequence and also relative to the most conserved residue in the TM in which they are located (24). The most conserved residue is assigned the position index “50,” e.g., in TM4, Trp-1604.50, and therefore Val-1594.49 and Val-1614.51. Mutations were generated and confirmed as described (23). Each mutation was generated in an N-terminally FLAG-tagged background D2R construct in which Cys-1183.36, Cys-1684.58, Cys-3706.61, and Cys-3726.63 were mutated to Ser (23) and stably transfected pools of HEK293 cells were created as described (23). We previously demonstrated that [3H]N-methylspiperone bound to this background construct (FLAG-D2R-C1684.58S) with a KD of 74 pM and a Bmax of 5.7 pmol/mg protein; sulpiride had a KI of 1.4 nM and dopamine a KI of 2.0 μM (23). These values are very similar to those observed in the background construct used for our previous studies, in which only Cys-1183.36 was mutated to Ser (29); in this background, the TM4 cysteine mutants had near-normal binding affinities for the antagonist N-methylspiperone (29).
Crosslinking, Drug Treatment, and Immunoblotting. The crosslinkers Cu2+(phenanthroline)2 (CuP) and HgCl2 were applied to intact adherent cells stably expressing the indicated cysteine mutants as described (23, 30). The concentrations indicated are those of Cu2+ or Hg2+. When indicated, the inverse agonist sulpiride was incubated with the cells at 37°C for 60 min before crosslinking and maintained in the buffer during crosslinking. Dopamine was added for 15 s and then removed immediately before addition of CuP; similar results were obtained when dopamine was added with the CuP. The crosslinking reaction was stopped by addition of 10 mM N-ethylmaleimide (NEM), the cells were harvested and extracted, 20 μg of protein was loaded per sample, and SDS/PAGE and immunoblotting using the M1 anti-FLAG monoclonal antibody (1 μg/ml, Sigma) were performed as described (23).
GTPγS Binding and cAMP Accumulation Assays. Intact adherent cells stably expressing the appropriate cysteine mutants were treated with vehicle or with CuP at the indicated concentrations, and, after washing, membranes were prepared and GTPγS binding was performed as described (31). D2R activation was also assayed by measuring the cAMP level resulting from the inhibition of adenylyl cyclase by Gi; the adenylyl cyclase was first activated by 100 μM forskolin as described (32). cAMP was extracted from cells and assayed by using a competitive binding assay as described (32).
Results
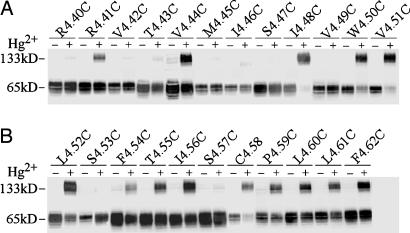
We first used HgCl2 as a crosslinking reagent to increase the likelihood that we would crosslink cysteines within the membrane. In other studies of membrane proteins, CuP was unable to crosslink cysteines deep in the transmembrane domain, whereas HgCl2 efficiently crosslinked a group of these residues that were inferred to be at an oligomeric interface (30, 33).
A background construct D2R in which Cys-1684.58 was mutated to serine (23) was not crosslinked by 20 μM HgCl2 (data not shown). In contrast, 14 of 23 substituted cysteine mutants in TM4 in the same background construct were crosslinked to the extent of 30-80% of total receptor (Fig. 2). The higher order band observed on the immunoblots after crosslinking was shown previously to be a homodimer of D2R by coimmunoprecipitation of differentially tagged D2Rs (23). Analysis of the shift of the bands with deglycosylation was also consistent with the band being a D2R homodimer and not D2R crosslinked to another protein (data not shown). The crosslinked residues spanned six helical turns over the entire predicted length of TM4. Closer to the cytoplasmic end of TM4, crosslinking was observed over a narrow stripe of cysteines substituted for Arg-1514.41, Val-1544.44, and Ile-1584.48, consistent with the proximity of these positions in the AFM model (Fig. 1D). However, the experimentally observed face of crosslinking broadened substantially starting with W1604.50C, in contrast to the predictions of the AFM model (Fig. 1D).
Fig. 2.
Crosslinking of D2R TM4 cysteine mutants with mercuric chloride. Stably expressed cysteine mutants in TM4 from R1504.40C to F1724.62C [denoted by using the indexing system described in Methods (24)] were crosslinked by 20 μM HgCl2 at 25°C for 10 min, stopped with N-ethylmaleimide and analyzed by immunoblotting. All experiments were repeated at least three times, and a representative experiment is shown.
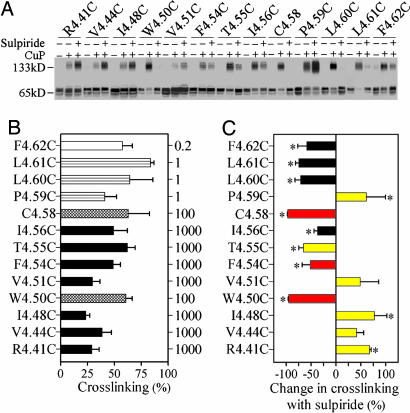
Every cysteine mutant that was crosslinked by HgCl2, with the exception of L1624.52C, was also oxidatively crosslinked by 1 mM CuP (Fig. 3A and data not shown). For mutants that were >85% crosslinked by 1 mM CuP, we crosslinked in the presence of 10-fold lower CuP concentrations until the fraction of crosslinked receptor was <85%. The lower the concentrations reached using this protocol (see Fig. 3B), the higher are the susceptibilities of the cysteines to crosslinking. The susceptibilities of the substituted cysteines to crosslinking were greatest at the extracellular end of TM4 but did not differentiate the AFM and ECM faces.
Fig. 3.
Effects of sulpiride on crosslinking of D2R TM4 cysteine mutants with copper phenanthroline. (A) Cysteine mutants were crosslinked by copper sulfate and 1,10-phenanthroline in a 1:2 molar ratio at the Cu2+ concentrations indicated in B at 25°C for 10 min in the absence or presence of 10 μM sulpiride. All experiments were repeated at least three times, and a representative experiment is shown. (B) Cysteine mutants were crosslinked by CuP at the concentrations in μM shown on the right axis at 25°C for 10 min, and the fraction of crosslinking is shown as mean ± SEM (n = 3-8). (C) The sulpiride-induced change in crosslinking fraction for each mutant is shown as mean ± SEM (n = 3-5). Crosslinking in the presence or absence of sulpiride was compared by one-way ANOVA and Bonferroni post hoc test with statistical significance at the P < 0.05 level indicated by *. Bars are shown in yellow or red for those positions predicted to be at the dimer interface in the AFM or ECM models, respectively (see Fig. 1).
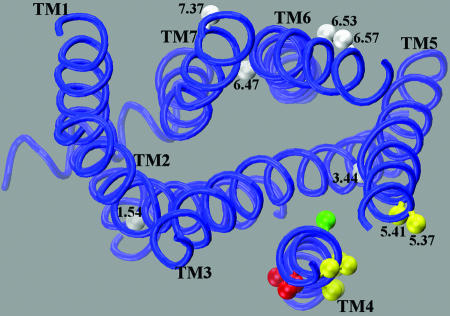
To validate further the central importance of TM4 in forming the homodimer interface and to support the specificity of the crosslinking, we carried out additional crosslinking experiments in TM6 and TM7, helices proposed to play a role in dimerization of D2R (34). We substituted cysteines for 6.53, 6.57, and 7.37, positions within the extracellular portions of TM6 and TM7 that are predicted to face outward in the rhodopsin monomer (Fig. 4), but not to face a symmetrical interface in any of our dimer or dimer array models (see below). We observed no crosslinking of these mutants by 1 mM CuP (data not shown). In addition, 1.54, 3.44, and 6.47, the endogenous cysteines present in the background construct, are not crosslinked by CuP or by HgCl2. In contrast to the absence of crosslinking at these positions in TM1, TM3, TM6, and TM7, we did observe crosslinking at positions 5.37 and 5.41, as well as other positions in TM5 (W.G. and J.A.J., unpublished results). These findings argue against random collision of D2R in the membrane as the basis for crosslinking and support a central role for TM4 and TM5 in forming the homodimer interface.
Fig. 4.
Homology model of the D2R based on the structure of bovine rhodopsin (52). The Cβ positions that were crosslinked by CuP are shown in yellow or red depending on their predicted location at the AFM or ECM interfaces (see Fig. 1). Position 4.56, which is predicted to face inward is shown in green. For simplicity, we omit the segment 4.60-4.62, which is the most extracellular part of TM4 and may not be in an α-helical conformation. Endogenous cysteines that did not crosslink, including 1.54, 3.44, and 6.47, are shown in gray, as are the positions of substituted cysteines 6.53, 6.57, and 7.37 in TM6 and TM7 that did not crosslink despite their predicted positions facing outward in the rhodopsin monomer.
Because the pattern of crosslinking in TM4 seemed incompatible with a single conformation, we reasoned that the receptor fluctuates between functional states and that the TM4 interface differs in these states. Therefore, we tried to trap the receptor in one of these states with sulpiride, a D2R inverse agonist (35). The striking result was that the susceptibilities to crosslinking were decreased at eight positions but were increased at five other positions (Fig. 3C).
These data are consistent with a sulpiride-stabilized conformation in which some cysteine pairs are closer together and others are more distant than in the unliganded receptor. Notably, in the presence of sulpiride positions that are brought together symmetrically in the AFM model (yellow in Fig. 1), 4.41, 4.44, 4.48, 4.51, and 4.59 showed increased crosslinking, whereas the residues that were not apposed in the AFM model (red in Fig. 1), 4.50, 4.54, 4.58, showed decreased crosslinking (Fig. 3C). At the several positions tested, including 4.48, 4.50, 4.54, and 4.58, the inverse agonists N-methylspiperone and butaclamol (35) had effects similar to sulpiride (data not shown).
Although 4.55 is expected to be on the AFM face indicated in yellow, sulpiride decreased crosslinking at this position. It is likely, however, that 4.55 is protected through a local steric effect of ligand binding, rather than through a global rearrangement, because the impact of crosslinking 4.55 on receptor function (see below) was consistent with it being on the AFM face. Crosslinking at positions 4.60-4.62 was also decreased by sulpiride, but we cannot interpret this because the secondary structure of 4.60-4.62, which is at the extracellular end of TM4 or at the beginning of the second extracellular loop, is not known (29). Inhibition by sulpiride of crosslinking at position 4.56, predicted to face inward (Fig. 1), is likely due to steric hindrance, consistent with the previously inferred role of this residue in ligand binding (36).
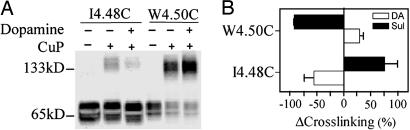
We also studied the effect of the agonist dopamine on the susceptibilities to crosslinking of a selected pair of mutants. We chose positions 4.48 and 4.50 because these loci exhibit opposite effects with sulpiride and are deep within the transmembrane domain where a change in crosslinking is likely due to movement of the helix. These residues, predicted to be on nearly opposite faces of TM4, were differentially affected by sulpiride, which decreased crosslinking of W1604.50C and increased crosslinking of I1584.48C (Fig. 3 A and C). Conversely, dopamine had the opposite effects on the crosslinking of the two cysteine mutants, consistent with the expectation that agonist and inverse agonist stabilize different receptor conformations (Fig. 5 A and B). Moreover, quinpirole, another D2R agonist, had the same effect as dopamine on the crosslinking of these two mutants (data not shown).
Fig. 5.
Effects of dopamine on crosslinking of selected D2R TM4 cysteine mutants with copper phenanthroline. (A) I1584.48C and W1604.50C were crosslinked by the concentrations of CuP indicated in Fig. 3B in the absence or presence of 10 μM dopamine and detected by immunoblotting. (B) The sulpiride- and dopamine-induced changes in crosslinking fraction for each mutant are shown as mean ± SEM (n = 3-5) for dopamine (open bars) or sulpiride (filled bars). DA, dopamine.
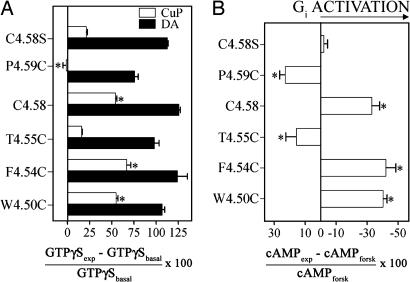
Taken together, the different effects of sulpiride and dopamine on the susceptibilities to crosslinking suggest that Trp-1604.50, Phe-1644.54, and Cys-1684.58 are closer to their counter-parts in the dimer interface in the active state, and are more distant in the inactive state. In these dynamics, activation would still take place after crosslinking of Cys-1684.58, as we observed previously (23). As a key inference, we hypothesized that disulfide trapping of the appropriate residues on this dynamic dimer interface might stabilize the active state even in the absence of agonist. The predicted observation would be that crosslinking of these residues would activate unliganded D2R, measurable as an increase in GTPγS binding. Consistent with our prediction, crosslinking by CuP of W1604.50C, F1644.54C, and Cys-1684.58 increased GTPγS binding significantly more than CuP treatment of C1684.58S, which was not crosslinked (Fig. 6A). In the absence of CuP treatment, GTPγS binding to each of these mutants was increased by dopamine (Fig. 6A), and the extent of activation of these mutants by 100 μM CuP ranged from 43-54% of full activation by dopamine (Fig. 6A). In contrast, when compared with C1684.58S, CuP treatment of P1694.59C significantly decreased GTPγS binding, and crosslinking of T1654.55C had no significant effect on GTPγS binding (Fig. 6A), consistent with their predicted positions near the interface in the inactive AFM model (Fig. 1). At 1,000 μM CuP, the increase in GTPγS binding for W1604.50C, F1644.54C, Cys-1684.58, and C1684.58S was 93 ± 14%, 91 ± 18%, 52 ± 4%, and 6 ± 2%, respectively (n = 3-4), of full activation by dopamine.
Fig. 6.
Activation of unliganded D2R results from crosslinking selected TM4 cysteine mutants. (A) GTPγS binding to membranes prepared after treating cells stably expressing the appropriate TM4 cysteine mutants with 100 μM CuP (open bars) or by untreated membranes exposed to 10 μM dopamine for 15 min at 30°C (filled bars). The dopamine- or CuP-induced change in GTPγS binding (GTPγSexp) compared with untreated membranes exposed to vehicle alone (GTPγSbasal) is shown as mean ± SEM (n = 3-5). The effects were analyzed by one-way ANOVA and Dunnett's post hoc test, and statistical significance, comparing the effect of CuP treatment on each cysteine mutant with that on uncrosslinked C1684.58S treated with the same concentration of CuP, at the P < 0.05 level is indicated by *. (B) The change in [cAMP] in the presence of forskolin (100 μM) after treatment with CuP (100 μM) in cells stably expressing the appropriate cysteine mutants is shown as the mean ± SEM (n = 3-6). One-way ANOVA and Dunnett's post hoc test were performed as in A. The x axis is plotted so that activation of the receptor, which causes inhibition of adenylyl cylase through Gi, is shown to the right of the y intercept. In C1684.58S, the maximal change of cAMP induced by dopamine was -74% (23). DA, dopamine.
Because D2R inhibits adenylyl cyclase through activation of Gi, we also determined cAMP levels in the mutants before and after crosslinking. The overall effects were nearly identical to the GTPγS-binding data, with enhanced activity of unliganded receptor, manifested as greater inhibition of forskolin-stimulated cAMP levels, after crosslinking of W1604.50C, F1644.54C, and Cys-1684.58, and less inhibition after crosslinking of T1654.55C and P1694.59C (Fig. 6B). Treatment with CuP of C1684.58S (Fig. 6B) or of untransfected cells (data not shown) had no significant effect on cAMP levels, consistent with the lack of an effect in the absence of crosslinking.
Discussion
Our results demonstrate an extensive and dynamic involvement of TM4 in forming a symmetrical dimer interface in D2R and show that conformational changes at this interface are an important component of receptor activation that was previously unappreciated. GPCR activation has been associated with dynamic changes in a monomer, resulting principally from a conformational change in TM6 and an associated opening of a binding cleft for G protein between TM6 and TM3 (37). Our data suggest that, in addition to these conformational changes, a rearrangement of the dimer interface is a critical component of activation. The data supporting this inference are (i) that the pattern of crosslinking in the presence of sulpiride and other inverse agonists is consistent with the TM4-TM4 proximities in the AFM model of the inactive GPCR, (ii) that, at the positions probed, the agonists dopamine and quinpirole produce an effect on crosslinking opposite to that of sulpiride, and (iii) that crosslinking of W1604.50C, F1644.54C, and Cys-1684.58 (mutants in which crosslinking is inhibited by sulpiride) leads to activation of unliganded receptor. Crosslinking of any one of these three residues presumably maintains the dimer interface in the active conformation. Our data also suggest that the unliganded receptor moves readily between the active and inactive conformations, a conclusion consistent with the significant constitutive activity of the unliganded D2R (35, 38). Crosslinking presumably traps the active conformation.
The pattern of crosslinking in the presence of sulpiride provides support for the orientation of TM4 in the AFM model generated by Liang et al. (21) for the inactive state (Fig. 1 A). Further support for the AFM model comes from experiments in which we have observed efficient crosslinking of cysteines substituted in the extracellular end of TM5 of the D2R at positions predicted by Liang et al. (21) to be in close proximity (Figs. 1 A and 4). In contrast, crosslinking of TM5 would be much slower in the ECM model (Fig. 1B).
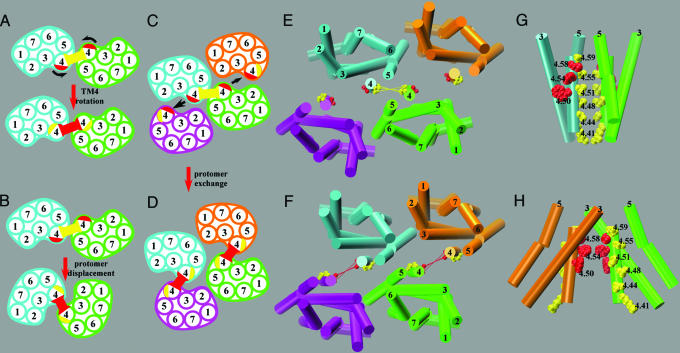
Three structural mechanisms that are not mutually exclusive might account for our data. In the first mechanism, TM4 might rotate upon activation (Fig. 7A). The β-ionone ring of retinal was photocrosslinked to Ala-1694.58 in the lumirhodopsin, metarhodopsin I, and metarhodopsin II intermediate states of rhodopsin (39). Because 4.58 (red in Figs. 1 and 7) faces lipid in the rhodopsin crystal structure, such a crosslink requires a significant conformational change upon activation. A conformational change in metarhodopsin I has been supported by spectroscopy studies (40) but not by electron crystallography studies (41).
Fig. 7.
Illustration of potential dimer rearrangements and molecular model representations of the proposed dimer interfaces. (A) Rotation of TM4 that could bring the red interface closer together in the active state. (B) A protomer displacement in which the same two protomers move so that they now interact at the red interface. (C and D) A protomer exchange in which crosslinking is between different protomer pairs in the inactive state (C) and in the active state (D). (E and G) Extracellular and side views, respectively, of a symmetric TM4-TM5 interface (yellow) as proposed by Liang et al. (21) incorporated into a tetrameric arrangement. (F and H) Extracellular and side views of a symmetric TM4 interface (red) deduced from the squid rhodopsin 2D electron density map (22) incorporated into an alternative tetrameric arrangement. These two models are proposed to correspond to the inactive and active states, respectively. The backbone of the transmembrane segments is rendered as cylinders. E-H were prepared by using VMD (53) for a homology model of the D2R based on the structure of bovine rhodopsin (52). For simplicity, we omit the segment 4.60-4.62, which is the most extracellular part of TM4 and may not be in an α-helical conformation.
In the second mechanism, a rearrangement of the dimer interface would take place by a displacement of the protomers (Fig. 7B). In this case, a large movement and reorganization would be necessary for receptor activation.
In the third mechanism, protomers in an extended array change partners. An extended array of dimers has been suggested for rhodopsin from AFM analysis of native mouse retinal membranes (21, 42). In the higher order oligomeric assembly, a relatively small conformational change upon binding of an agonist or an inverse agonist may be sufficient to reorganize the oligomeric packing, thereby changing the protomers that are crosslinked. Thus, in the inactive state, crosslinking occurs at the interface consistent with the AFM model (Fig. 7C) whereas, in the active state, crosslinking occurs via an interface consistent with the ECM model (Fig. 7D). Molecular modeling of the two interfaces in an oligomeric assembly shows that both the “inactive” and “active” oligomeric arrangements shown in (Fig. 7 C and D) are feasible and that the symmetric pairs of residue positions are consistent with our data (Fig. 7 E-H). That inverse agonist binding promotes one arrangement and agonist binding promotes the other arrangement indicate that the structure of the interface is coupled to the structure of the binding site. Further predictions from these models need to be tested experimentally by using constructs with more than one cysteine (e.g., in both TM4 and TM5) to establish whether the crosslinking we observed occurs at more than a single symmetric interface.
Although our results are consistent with the proposal of protomer exchange, they are not consistent with an alteration in the oligomeric status of D2R upon ligand binding. Thus, although sulpiride decreases crosslinking of particular cysteines at the TM4 dimer interface, other cysteines are crosslinked more efficiently, indicating that dimerization is preserved in the presence of sulpiride.
Ligand binding to one protomer in a GPCR dimer seems to be sufficient to cause an activation-like conformational change in the unoccupied protomer in family A chemokine and leukotriene receptor dimers (43-45), consistent with transfer of information between the protomers through a conformational change at the dimer interface similar to what we have identified here. Similarly, negative binding cooperativity between protomers in a chemokine receptor heterodimer (46) requires crosstalk at the interface. In addition, transactivation of GPCRs (14-20) requires that binding to one protomer be transduced to the second protomer, and this conformational change is also likely mediated through the dimer interface.
The proposal that the signaling unit may be a GPCR dimer complexed to a single heterotrimeric G protein (47, 48) opens the possibility that both cis- and transactivation are manifestations of the normal signaling process in which G protein interacts with two GPCR protomers at a conformationally sensitive interface. Based on x-ray crystallography studies of the soluble N-terminal binding sites of metabotropic glutamate receptors, agonist binding is thought to alter the relative orientation of the dimer formed by the two extracellular binding domains (17, 49, 50). Moreover, in metabotropic glutamate receptors, agonist binding has been shown to lead to a change in the distance between the cytoplasmic loops of the protomers (51). Such a movement must be associated with an altered relationship between the transmembrane domains of the two protomers. We have now shown in a family A receptor the existence of such a change in the TM dimer interface and its integral role in receptor activation.
Acknowledgments
We thank J. C. Fowler and R. B. Mailman for help in setting up the GTPγS-binding assays. This work was supported by National Institutes of Health Grants MH57324 and MH54137 (to J.A.J.) and DA12923 and DA00060 (to H.W.), and by the National Alliance for Research on Schizophrenia and Depression Vicente Investigator Award and the Lieber Center for Schizophrenia Research.
Conflict of interest statement: No conflicts declared.
Abbreviations: TM, transmembrane segment; CuP, copper phenanthroline; AFM, atomic force microscopy; ECM, electron cryomicroscopy; GPCR, G protein-coupled receptor; D2R, dopamine D2 receptor.
References
- 1.Gether, U. (2000) Endocr. Rev. 21, 90-113. [DOI] [PubMed] [Google Scholar]
- 2.Terrillon, S. & Bouvier, M. (2004) EMBO Rep. 5, 30-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milligan, G. (2004) Mol. Pharmacol. 66, 1-7. [DOI] [PubMed] [Google Scholar]
- 4.Angers, S., Salahpour, A. & Bouvier, M. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 409-435. [DOI] [PubMed] [Google Scholar]
- 5.Park, P. S., Filipek, S., Wells, J. W. & Palczewski, K. (2004) Biochemistry 43, 15643-15656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pin, J. P., Galvez, T. & Prezeau, L. (2003) Pharmacol. Ther. 98, 325-354. [DOI] [PubMed] [Google Scholar]
- 7.Jordan, B. A. & Devi, L. A. (1999) Nature 399, 697-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes, I., Jordan, B. A., Gupta, A., Trapaidze, N., Nagy, V. & Devi, L. A. (2000) J. Neurosci. 20, RC110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.So, C. H., Varghese, G., Curley, K. J., Kong, M. M., Alijaniaram, M., Ji, X., Nguyen, T., O'Dowd B, F. & George, S. R. (2005) Mol. Pharmacol. 68, 568-578. [DOI] [PubMed] [Google Scholar]
- 10.Lee, S. P., So, C. H., Rashid, A. J., Varghese, G., Cheng, R., Lanca, A. J., O'Dowd, B. F. & George, S. R. (2004) J. Biol. Chem. 279, 35671-35678. [DOI] [PubMed] [Google Scholar]
- 11.Yoshioka, K., Saitoh, O. & Nakata, H. (2001) Proc. Natl. Acad. Sci. USA 98, 7617-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filizola, M. & Weinstein, H. (2005) Curr. Opin. Drug Discov. Devel. 8, 577-584. [PubMed] [Google Scholar]
- 13.Galvez, T., Duthey, B., Kniazeff, J., Blahos, J., Rovelli, G., Bettler, B., Prezeau, L. & Pin, J. P. (2001) EMBO J. 20, 2152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robbins, M. J., Calver, A. R., Filippov, A. K., Hirst, W. D., Russell, R. B., Wood, M. D., Nasir, S., Couve, A., Brown, D. A., Moss, S. J. & Pangalos, M. N. (2001) J. Neurosci. 21, 8043-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duthey, B., Caudron, S., Perroy, J., Bettler, B., Fagni, L., Pin, J. P. & Prezeau, L. (2002) J. Biol. Chem. 277, 3236-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havlickova, M., Prezeau, L., Duthey, B., Bettler, B., Pin, J. P. & Blahos, J. (2002) Mol. Pharmacol. 62, 343-350. [DOI] [PubMed] [Google Scholar]
- 17.Kniazeff, J., Bessis, A. S., Maurel, D., Ansanay, H., Prezeau, L. & Pin, J. P. (2004) Nat. Struct. Mol. Biol. 11, 706-713. [DOI] [PubMed] [Google Scholar]
- 18.Ji, I., Lee, C., Song, Y., Conn, P. M. & Ji, T. H. (2002) Mol. Endocrinol. 16, 1299-1308. [DOI] [PubMed] [Google Scholar]
- 19.Ji, I., Lee, C., Jeoung, M., Koo, Y., Sievert, G. A. & Ji, T. H. (2004) Mol. Endocrinol. 18, 968-978. [DOI] [PubMed] [Google Scholar]
- 20.Urizar, E., Montanelli, L., Loy, T., Bonomi, M., Swillens, S., Gales, C., Bouvier, M., Smits, G., Vassart, G. & Costagliola, S. (2005) EMBO J. 24, 1954-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang, Y., Fotiadis, D., Filipek, S., Saperstein, D. A., Palczewski, K. & Engel, A. (2003) J. Biol. Chem. 278, 21655-21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies, A., Gowen, B. E., Krebs, A. M., Schertler, G. F. & Saibil, H. R. (2001) J. Mol. Biol. 314, 455-463. [DOI] [PubMed] [Google Scholar]
- 23.Guo, W., Shi, L. & Javitch, J. A. (2003) J. Biol. Chem. 278, 4385-4388. [DOI] [PubMed] [Google Scholar]
- 24.Ballesteros, J. & Weinstein, H. (1995) Methods Neurosci. 25, 366-428. [Google Scholar]
- 25.Lee, S. P., O'Dowd, B. F., Rajaram, R. D., Nguyen, T. & George, S. R. (2003) Biochemistry 42, 11023-11031. [DOI] [PubMed] [Google Scholar]
- 26.Klco, J. M., Lassere, T. B. & Baranski, T. J. (2003) J. Biol. Chem. 278, 35345-35353. [DOI] [PubMed] [Google Scholar]
- 27.Hernanz-Falcon, P., Rodriguez-Frade, J. M., Serrano, A., Juan, D., del Sol, A., Soriano, S. F., Roncal, F., Gomez, L., Valencia, A., Martinez, A. C. & Mellado, M. (2004) Nat. Immunol. 5, 216-223. [DOI] [PubMed] [Google Scholar]
- 28.Filizola, M. & Weinstein, H. (2002) Biopolymers 66, 317-325. [DOI] [PubMed] [Google Scholar]
- 29.Javitch, J. A., Shi, L., Simpson, M. M., Chen, J., Chiappa, V., Visiers, I., Weinstein, H. & Ballesteros, J. A. (2000) Biochemistry 39, 12190-12199. [DOI] [PubMed] [Google Scholar]
- 30.Hastrup, H., Sen, N. & Javitch, J. A. (2003) J. Biol. Chem. 278, 45045-45048. [DOI] [PubMed] [Google Scholar]
- 31.Shapiro, D. A., Renock, S., Arrington, E., Chiodo, L. A., Liu, L. X., Sibley, D. R., Roth, B. L. & Mailman, R. (2003) Neuropsychopharmacology 28, 1400-1411. [DOI] [PubMed] [Google Scholar]
- 32.Liapakis, G., Ballesteros, J. A., Papachristou, S., Chan, W. C., Chen, X. & Javitch, J. A. (2000) J. Biol. Chem. 275, 37779-37788. [DOI] [PubMed] [Google Scholar]
- 33.Soskine, M., Steiner-Mordoch, S. & Schuldiner, S. (2002) Proc. Natl. Acad. Sci. USA 99, 12043-12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng, G. Y., O'Dowd, B. F., Lee, S. P., Chung, H. T., Brann, M. R., Seeman, P. & George, S. R. (1996) Biochem. Biophys. Res. Commun. 227, 200-204. [DOI] [PubMed] [Google Scholar]
- 35.Hall, D. A. & Strange, P. G. (1997) Br. J. Pharmacol. 121, 731-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi, L. & Javitch, J. A. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 437-467. [DOI] [PubMed] [Google Scholar]
- 37.Farrens, D. L., Altenbach, C., Yang, K., Hubbell, W. L. & Khorana, H. G. (1996) Science 274, 768-770. [DOI] [PubMed] [Google Scholar]
- 38.Wiens, B. L., Nelson, C. S. & Neve, K. A. (1998) Mol. Pharmacol. 54, 435-444. [DOI] [PubMed] [Google Scholar]
- 39.Borhan, B., Souto, M. L., Imai, H., Shichida, Y. & Nakanishi, K. (2000) Science 288, 2209-2212. [DOI] [PubMed] [Google Scholar]
- 40.Furutani, Y., Kandori, H. & Shichida, Y. (2003) Biochemistry 42, 8494-8500. [DOI] [PubMed] [Google Scholar]
- 41.Ruprecht, J. J., Mielke, T., Vogel, R., Villa, C. & Schertler, G. F. (2004) EMBO J. 23, 3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fotiadis, D., Liang, Y., Filipek, S., Saperstein, D. A., Engel, A. & Palczewski, K. (2003) Nature 421, 127-128. [DOI] [PubMed] [Google Scholar]
- 43.Percherancier, Y., Berchiche, Y., Slight, I., Volkmer-Engert, R., Tamamura, H., Fujii, N., Bouvier, M. & Heveker, N. (2005) J. Biol. Chem. 280, 9895-9903. [DOI] [PubMed] [Google Scholar]
- 44.Mesnier, D. & Baneres, J. L. (2004) J. Biol. Chem. 279, 49664-49670. [DOI] [PubMed] [Google Scholar]
- 45.Ayoub, M. A., Levoye, A., Delagrange, P. & Jockers, R. (2004) Mol. Pharmacol. 66, 312-321. [DOI] [PubMed] [Google Scholar]
- 46.El-Asmar, L., Springael, J. Y., Ballet, S., Andrieu, E. U., Vassart, G. & Parmentier, M. (2005) Mol. Pharmacol. 67, 460-469. [DOI] [PubMed] [Google Scholar]
- 47.Baneres, J. L. & Parello, J. (2003) J. Mol. Biol. 329, 815-829. [DOI] [PubMed] [Google Scholar]
- 48.Filipek, S., Krzysko, K. A., Fotiadis, D., Liang, Y., Saperstein, D. A., Engel, A. & Palczewski, K. (2004) Photochem. Photobiol. Sci. 3, 628-638. [DOI] [PubMed] [Google Scholar]
- 49.Kunishima, N., Shimada, Y., Tsuji, Y., Sato, T., Yamamoto, M., Kumasaka, T., Nakanishi, S., Jingami, H. & Morikawa, K. (2000) Nature 407, 971-977. [DOI] [PubMed] [Google Scholar]
- 50.Tsuchiya, D., Kunishima, N., Kamiya, N., Jingami, H. & Morikawa, K. (2002) Proc. Natl. Acad. Sci. USA 99, 2660-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tateyama, M., Abe, H., Nakata, H., Saito, O. & Kubo, Y. (2004) Nat. Struct. Mol. Biol. 11, 637-642. [DOI] [PubMed] [Google Scholar]
- 52.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289, 739-745. [DOI] [PubMed] [Google Scholar]
- 53.Humphrey, W., Dalke, A. & Schulten, K. (1996) J. Mol. Graphics 14, 33-38. [DOI] [PubMed] [Google Scholar]