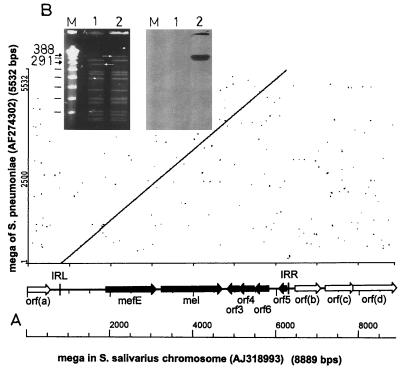
Macrolide resistance in pneumococci and oral streptococci is a common phenomenon (8). The macrolide resistance determinants erm(B) and mef(A) are widespread (4, 7, 9). We characterized the erythromycin resistance determinant of a Streptococcus salivarius strain isolated from a healthy person with no exposure to antibiotics 6 months prior to isolation. It was identified as mef(A) conferring an erythromycin MIC of 6 μg/ml (6). mef(A) was part of a macrolide efflux genetic assembly (mega) recently identified in Streptococcus pneumoniae (GenBank accession no. AF274302 [3]) in the United States. The nucleotide sequence of the mega from S. salivarius Sp6 (GenBank accession no. AJ318993) was 99.8% identical to that of the pneumococcal element (Fig. 1A). Deletions resulted in a 5,511-bp element in S. salivarius compared to 5,532 bp in S. pneumoniae. orf1 is identical to mef(A) from S. pneumoniae (U83667 [11]); orf2 encodes an ABC transporter identical to mel in the mega of S. pneumoniae. The open reading frames orf1 to -5 of the S. salivarius mega are homologous to orf4 to -8 from Tn1207.1, a mef(A)-carrying defective transposon of S. pneumoniae (AF227520 [10]). orf6 is not present in Tn1207.1. Upstream of mef(A) of the S. salivarius mega is a 16-bp gap that eliminates a direct repeat in the mega of S. pneumoniae (AF274302). This gap is also present in Tn1207.1 and in megas in Italian pneumococcal strains (2). Contrary to Orf4 in the mega of S. pneumoniae, Orf4 in the S. salivarius sequence is 19 amino acids larger due to a single base pair deletion but is the same size as Orf7 in Tn1207.1. Another deletion in Orf5 of the mega in S. salivarius causes a shortened open reading frame.
FIG. 1.
(A) Comparison of the mega of S. salivarius (GenBank accession no. AJ3188993) and S. pneumoniae (GenBank accession no. AF274302) in a dot plot analysis. IR, inverted repeats described by Gay and Stephens (3). (B) Identification of the S. pneumoniae R800 chromosomal DNA fragment containing the S. salivarius mega by digestion with SmaI, pulsed-field gel electrophoresis, and Southern hybridization. Lanes M, Lambda Ladder PFG Marker (in kilobases) (Biolabs); lanes 1, S. pneumoniae R800; and lanes 2, R800-transformant RSp6-II. Arrows indicate the fragment into which mega and adjacent sequences integrated.
The mega in S. salivarius was located on the chromosome. Downstream of the S. salivarius mega, three open reading frames with homologies to a regulator protein (orfb), an ABC transporter (orfc), and a transmembrane protein (orfd) were found.
The mega from S. salivarius Sp6 was transformed in two independent experiments with low but significant frequency (two transformants per 107 recipient cells and 100 ng of S. salivarius DNA) into erythromycin-susceptible S. pneumoniae strain R800 by in vitro transformation using accepted protocols and control DNA (1, 5). No spontaneous mutation of the recipient cells was obtained. The MIC for the transformants was comparable to that for the donor strains. SmaI-digested genomic DNA of the recipient strain and of transformants of S. pneumoniae R800 was analyzed by pulsed-field gel electrophoresis. The mega integrated into a 291-kbp fragment to yield a 388-kbp fragment detected by Southern hybridization (Fig. 1B).
The mega was detected by PCR with specific primers for nucleotides 1 to 2723 and 2291 to 5510 in oral streptococci from six healthy persons, including S. salivarius (10 strains), Streptococcus mitis (six strains), Streptococcus parasanguis (five strains), and Streptococcus cristatus (three strains).
This is the first report of the pneumococcal mega in S. salivarius and of a successful transformation of S. pneumoniae with S. salivarius DNA. The mechanism of worldwide dissemination of the mega in oral streptococci remains to be established.
REFERENCES
- 1.Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75-83. [DOI] [PubMed] [Google Scholar]
- 2.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gay, K., and D. S. Stephens. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 4.Ioannidou, S., P. T. Tassios, A. Kotsovili-Tseleni, M. Foustoukou, N. J. Legakis, and A. Vatopoulos. 2001. Antibiotic resistance rates and macrolide resistance phenotypes of viridans group streptococci from the oropharynx of healthy Greek children. Int. J. Antimicrob. Agents 17:195-201. [DOI] [PubMed] [Google Scholar]
- 5.Martin, B., M. Prudhomme, G. Alloing, C. Granadel, and J. P. Claverys. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 38:867-878. [DOI] [PubMed] [Google Scholar]
- 6.NCCLS. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, p. M100-S107, 4th ed., vol. 17. NCCLS, Wayne, Pa.
- 7.Perez-Trallero, E., D. Vicente, M. Montes, J. M. Marimon, and L. Pineiro. 2001. High proportion of pharyngeal carriers of commensal streptococci resistant to erythromycin in Spanish adults. J. Antimicrob. Chemother. 48:225-229. [DOI] [PubMed] [Google Scholar]
- 8.Roberts, M. C. 1998. Antibiotic resistance mechanisms in bacteria of oral and upper respiratory origin. Int. J. Antimicrob. Agents 9:255-267. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Avial, I., C. Rodriguez-Avial, E. Culebras, A. Benitez, and J. J. Picazo. 2001. Distribution of mef(A) and erm(B) genes in macrolide-resistant blood isolates of viridans group streptococci. J. Antimicrob. Chemother. 47:727-728. [DOI] [PubMed] [Google Scholar]
- 10.Santagati, M., F. Iannelli, M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux genemef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]