Abstract
X-linked nonspecific mental retardation (MRX) has a frequency of 0.15% in the male population and is caused by defects in several different genes on the human X chromosome. Genotype-phenotype correlations in male patients with a partial nullisomy of the X chromosome have suggested that at least one locus involved in MRX is on Xp22.3. Previous deletion mapping has shown that this gene resides between markers DXS1060 and DXS1139, a region encompassing ∼1.5 Mb of DNA. Analyzing the DNA of 15 males with Xp deletions, we were able to narrow this MRX critical interval to ∼15 kb of DNA. Only one gene, VCX-A (variably charged, X chromosome mRNA on CRI-S232A), was shown to reside in this interval. Because of a variable number of tandem 30-bp repeats in the VCX-A gene, the size of the predicted protein is 186–226 amino acids. VCX-A belongs to a gene family containing at least four nearly identical paralogues on Xp22.3 (VCX-A, -B, -B1, and -C) and two on Yq11.2 (VCY-D, VCY-E), suggesting that the X and Y copies were created by duplication events. We have found that VCX-A is retained in all patients with normal intelligence and is deleted in all patients with mental retardation. There is no correlation between the presence or absence of VCX-B1, -B, and VCX-C and mental status in our patients. These results suggest that VCX-A is sufficient to maintain normal mental development.
Introduction
X-linked nonspecific mental retardation (MRX [MIM 309530) accounts for ∼25% of mental retardation in males (Turner and Opitz 1980), which equals ∼0.15% of the male population (Gedeon et al. 1996; Chelly 1999; Gécz and Mulley 2000). Despite this high frequency, little is known about the biological basis of this clinical entity. Patients with MRX have learning disabilities as a single clinical feature and show no other consistent abnormalities. During the past few years, positional-cloning approaches and subsequent mutation analysis have identified seven different genes involved in the phenotype of MRX. These genes include that for oligophrenin (OPHN1), PAK3, GDI1, RPS6KA3, IL1RAPL, and that for tetraspanin (TM4SF2), all of which have been shown to participate in various stages of intracellular signaling, and the gene FMR2, the function of which remains unknown (reviewed in Gécz and Mulley 2000). RPS6KA3, on Xp22.13, has been shown to be involved in both nonspecific and syndromal (Coffin-Lowry syndrome) mental retardation (Trivier et al. 1996; Merienne et al. 1999).
Genotype-phenotype correlations in male patients with a partial nullisomy for the X chromosome have suggested at least one further MRX locus, distal to the RPS6KA3 gene. Initial deletion mapping assigned this putative MRX gene to an interval of ∼3 Mb on Xp22.3 (Ballabio et al. 1989). This assignment was narrowed to a region between DXS1145 and GS1 (Herrell et al. 1995), between DXS1060 and DXS1139 (Muroya et al. 1996), and between DXS1060 and GS1 (Weissortel et al. 1998), with a common overlap of ∼1.5 Mb.
Six disease genes have been mapped to Xp22.3; these include the genes for short stature (SS), X-linked recessive chondrodysplasia punctata (CDPX), MRX, X-linked ichthyosis (XLI), Kallmann syndrome (KAL), and ocular albinism type I (OA1) (Ballabio and Andria 1992). Of these six genes, only the gene for SS resides within the pseudoautosomal region 1 and is identical on both the X and the Y chromosomes; the other five genes have been assigned to the X-specific region and are inherited as X-linked recessive traits. Extensive positional-cloning studies have been carried out during the past 12 years, resulting in the successful cloning of SHOX (for SS), ARSE (for CDPX), STS (for XLI), KAL (for KAL), and OA1 (for OA1); however, the gene for MRX has remained elusive.
We have studied 15 male individuals with rearrangements and specific clinical features of the contiguous gene–deletion syndrome on Xp22.3 (Ballabio et al. 1989). Comparison between the deletion breakpoints and the clinical phenotype revealed a critical MRX region of only 15 kb, in which the small full-length gene VCX-A (variably charged, X chromosome mRNA on CRI-S232A) was isolated.
Material and Methods
Deletion Mapping
Genomic DNA was extracted from peripheral blood leukocytes (cases 1, 6-11, 13, and 511), lymphoblast cell lines (cases 2–5 and 12), or fibroblast cell lines (case 88) and was examined by PCR analysis. Primers newly established in this study are shown in table 1. The other primer sequences and conditions have been described elsewhere (Schaefer et al. 1993; Herrell et al. 1995; Muroya et al. 1996). PCR analysis was carried out at least three times for each locus. For detailed mapping of the members of the VCX family, specific primers termed “MAP EII” were used for PCR, and the resulting fragments were cleaved as described in the “Reverse Transcription–PCR (RT-PCR)” subsection below.
Table 1.
Primer Sequences and PCR Conditions Established in This Study
|
Primer(5′→3′) |
||||
| Locus | Forward | Reverse | Annealing Temperature(°C) | Size(bp) |
| DXS6837 | attcatcatatatacatcag | agtcatactttaatcttgc | 62 | 88 |
| BT47 | agccagcgcagcatctatgaa | taagtttacttgcccaattcc | 58 | 123 |
| K7-2 | tgtcgtgtcgtgtgtgttttacca | aaaaaaatggcccagagca | 58 | 393 |
| K2H | ctgcagggatctgaggattgt | ctggcaccttatgttagaagt | 58 | 186 |
| K6a | gtcctgctctaatcagtttctaagtgct | tgcagaaatcaaggaggaagtgggag | 58 | 186 |
| K5 | ggttgcagaactctgatttagagcatctac | caggagctctcatcaagaaagtggg | 58 | 300 |
| DXS6834 | ccaatcagagagctcag | atctgctcatgcttctc | 57 | 148 |
| VCX EIb | tgatggtgcgttgtgacgtataagg | cgggccactggggctcggctg | 60 | 281 |
| VCX EIIb | aaaccagagcccttctgtgatctcc | cttagtcgctgctctcggag | 61 | Variablec |
| MAP EII | tgtagacggccagctactcc | atcttcagaatggcaagcac | 61 | 106 |
| Reverse transcriptase | gatgagtccaaagccgagag | atcttcagaatggcaagcac | 61 | Variable |
Positive in all individuals tested (including cases 1–4, which had large deletions of Xp22.3), indicating that this locus is repetitive.
“VCX EI” and “VCX EII” indicate specific primers for exon I and exon II of the VCX family, respectively.
Described in the text.
Physical Mapping
Cosmids and plasmid artificial chromosomes (PACs) were derived from the Imperial Cancer Research Fund (ICRF) X chromosome–specific library (ICRFc104), the Lawrence Livermore Laboratory Biology and Biotechnology Research Program X chromosome–specific library (LLNc110), and the Roswell Park Cancer Institute (RPCI [see the “BACPAC Resources” Web site of the Children's Hospital Oakland Research Institute]) human PAC library (Ioannou et al. 1994). YAC clones were derived from either the Fondation Jean Dausset CEPH Human Mega-YAC library or the X chromosome YAC collection maintained at the Max-Planck Institut for Molecular Genetics (Berlin) and bacterial artificial chromosome (BAC) clones of the BAC (I) library constructed by Genome Systems. Clones were identified by PCR, by means of sequence-tagged sites mapping to this region. To verify overlaps, end probes from clones were used in cases in which overlaps could not be proved by use of known primers.
FISH Analysis
Biotinylated DNA was hybridized to metaphase chromosomes from stimulated lymphocytes of the patients, under conditions that have been described elsewhere (Lichter and Cremer 1992). The hybridized probe was detected via avidin-conjugated FITC.
PCR Amplification
All PCRs were performed in 50-μl volumes containing 100 pg–200 ng of template, 20 pmol of each primer, 200 μM of each dNTP (MBI), 1.5 mM MgCl2, 75 mM Tris/HCl pH 9.0, 20 mM (NH4)2SO4, 0.01% (w/v) Tween-20, and 2 U of Goldstar DNA polymerase (Eurogentec). Thermal cycling was carried out in a thermocycler GeneE (Techne). PCRs with newly established primers were carried out under the following conditions: 35 cycles of 93°C for 2 min, 93°C for 20 s, annealing temperature for 20 s, and 72°C for 40 s; and 72°C for 7 min.
Exon Amplification
The cosmids were partially digested with Sau3A. Gel-purified fractions in the size range of 4–10 kb were cloned into the BamHI-digested pSPL3B vector and, with minor modification, were used for the exon-amplification experiments, as described elsewhere (Church et al. 1994; Burn et al. 1995). The second PCR amplification was performed with UDG-tailed SA4 and SD2 primers, and the product was cloned in the vector pAMP system (Gibco-BRL), according to the manufacturer's recommendations.
Northern Hybridization
Human adult and fetal multiple-tissue northern-blot filters (Clontech) were hybridized in ExpressHyb hybridization solution (Clontech) at 65°C and were washed at a final stringency of 0.1 × SSC, 0.1% SDS. The probe used for northern hybridization was a PCR product amplified with primers VCX EI (forward) and VCX EII (reverse).
ReverseTranscription–PCR (RT-PCR)
Human poly(A)+ RNA of liver, heart, kidney, brain, muscle, stomach, and placenta was purchased from Invitrogen, and amygdala, thalamus, hippocampus, and caudate nucleus was purchased from Clontech. Total RNA was isolated from fetal heart, kidney, brain, muscle, and testis by TRIZOL reagent (Gibco-BRL), according to the manufacturer's recommendations. First-strand cDNA synthesis was performed with the Superscript first-strand cDNA synthesis kit (Gibco-BRL), starting with either 20–50 ng of poly(A)+ RNA or 10 μg of total RNA and using oligo-(dT) adaptor primer 5′-GGCCACGCGTCGACTAGTAC(dT)20N-3′. After first-strand cDNA synthesis, the reaction mix was diluted 1/10. For further PCR experiments, 5 μl of the dilutions were used. A “Marathon adaptor-ligated ds cDNA library” was constructed from poly(A)+ RNA, with the Marathon cDNA amplification kit (Clontech) used according to the manufacturer's recommendations. RT-PCR was carried out by performing 40 cycles with primer pairs shown in table 1. PCR products were digested with BsmAI (New England Biolabs), RsaI, or AluI (Roche Diagnostics).
Cloning and Sequencing of PCR Products
PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen). The clones were sequenced with Cy5-labeled vector primers M13, universal and reverse, by the cycle sequencing method described by the manufacturer (ThermoSequenase kit; Amersham), and were analyzed on an ALF-express automated sequencer (Pharmacia).
Genomic Sequencing
Mechanically sheared fragments of cosmid ICRFc104-A05122 and EcoRI/HindIII-digested fragments of cosmid LLNc110-45O15 were subcloned in pUC18 vectors. For fragments >3 kb, internally deleted fragments were generated. Remaining gaps were closed by primer walking. The sequencing was done via cycle sequencing with the AmpliTaq S core kit (Applied Biosystems) and standard forward and reverse primers labeled with either FITC or CY5. An MJ Research (Waltham) PT-200 cycler was used to perform 25 cycles (97°C for 15 s, 55°C for 30 s, and 68°C for 30 s). Reactions were loaded onto the 72-clone porous-membrane combs, were applied to 60-cm-long polyacrylamide gels (4.5% Hydrolink Long Ranger gel solution; FMC), and were analyzed by the ARAKIS sequencing system with array detectors (Erfle et al. 1997). This system allows simultaneous online sequencing on both strands (“doublex” sequencing), with the two sequencing products obtained in a single sequencing reaction, each labeled with a different fluorescent dye (Wiemann et al. 1995). Sequencing data were assembled and edited, by the software packages (i.e., LANE TRACKER and GENE SKIPPER) developed by EMBL.
Results
Defining and Cloning the MRX Critical Region
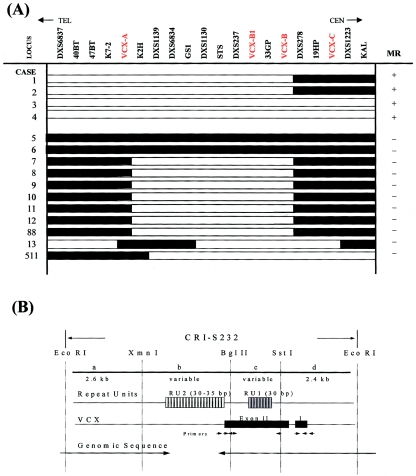
Fifteen males with terminal and interstitial deletions of Xp22.3 were analyzed in our study (table 2). Their mental status was evaluated either with the aid of WISC-R or WAIS or on the basis of developmental history. Cases 1–4 had obvious mental retardation, with developmental quotient (DQ) or intellectual quotient (IQ) in the 46–75 range, in addition to other specific clinical features consistent with the contiguous gene–deletion syndrome on Xp22.3. Patients 5–13, 88, and 511 had normal intelligence (defined here as IQ >91) yet one or more other clinical features (table 2). For the deletion analysis, individuals with IQ 75–90 were excluded, as being “borderline” cases (Muroya et al. 1996). To define the breakpoint regions on the rearranged X chromosomes, PCR was performed with primers specific for 25 different loci of the chromosome X–specific region on Xp22.3. As depicted in figure 1, only patient 13 had a complex deletion; the other patients had simple terminal or interstitial deletions. To define the MRX critical interval, individual breakpoints were correlated with the mental status of the respective cases. All patients with mental retardation lack a DNA interval including markers DXS1139 and STS, whereas all individuals with normal intelligence retain the marker(s) DXS6837 and/or DXS1139, suggesting that there is an MRX critical region between DXS6837 and DXS1139.
Table 2.
Cytogenetic and Clinical Findings in 15 Patients with Deletions on Xp22.3
| Case | Karyotype | Clinical Feature(s)a | Intellectual Assessment | Reference |
| 1 | 46,XY | MR, XLI | DQ = 60 | Authors' unpublished data |
| 2 | 46,Y,der(X)t(X;Y)(p22.3;q12) | SS, CDPX, MR, XLI | No single words (at age 2 years) | Agematsu et al. (1988) |
| 3 | 46,Y,der(X)t(X;Y)(p22;q11) | SS, CDPX, MR, XLI, KAL, (OA1) | DQ = 65–75 | Nishimura et al. (1991) |
| 4 | 46,XY | (SS), MR, XLI, KAL, OA1 | IQ = 46 | Muroya et al. (1996) |
| 5 | 46,Y,der(X)t(X;Y)(p22.3;q11) | SS | Average (based on school records) | Muroya et al. (1996) |
| 6 | 46,Y,r(X)(p22.3q28) | SS, (CDPX) | Average (based on school records) | Ogata et al. (1990) |
| 7 | 46,XY | XLI | Average (based on school records) | Authors' unpublished data |
| 8 | 46,XY | XLI | Average (based on school records) | Authors' unpublished data |
| 9 | 46,XY | XLI | Average (based on school records) | Authors' unpublished data |
| 10 | 46,XY | XLI | Average (based on school records) | Authors' unpublished data |
| 11 | 46,XY | XLI | DQ = 110 | Authors' unpublished data |
| 12 | 46,XY | XLI, epilepsy | DQ = 103 | Authors' unpublished data |
| 88 | 46,XY | XLI | Normal development | Schaefer et al. (1993) |
| 13 | 46,XY | SS, CDPX, XLI | High (based on school records) | Schaefer et al. (1993) |
| 511 | 46,XY | (SS), XLI, KAL | Normal development | Unpublished |
Parentheses indicate very mild or borderline phenotypes. Clinical reports of all individuals except cases 1, 7–12, and 511 have been published previously.
Figure 1.
Deletion map and mental status of 15 patients. In the “MR” column, a plus sign (+) denotes mental retardation, and a minus sign (−) denotes normal intelligence. Sequence-tagged-site loci are shown in black, gene loci are shown in red. The RPS6KA3 gene is thought to reside proximal to DXS85 and is not depicted on this map. The blackened portions of the horizontal bars represent positive loci confirmed by molecular studies, and the unblackened portions represent absent loci. The physical locus order is based on published reports (Schaefer et al. 1993; van Slegtenhorst et al. 1994; Ferrero et al. 1995; Herrell et al. 1995), with one exception—the locus order of DXS6834 and DXS1139 is based on the physical map established in the present study. The underlined loci indicate the boundaries of the critical region of the gene for MRX.
This critical region was subsequently isolated in YACs, PACs, and cosmid clones (fig. 2A). PCR analysis of newly derived sequence-tagged-site markers revealed that cases 11 and 13, both with normal intelligence, share only a small region between markers 40BT and DXS1139. The entire gene for MRX is predicted to be located in this interval, which is covered by two overlapping cosmids, LLNc110-45O15 and ICRFc104-A05122. To confirm the deletion map, FISH analysis of metaphase chromosomes from patients 11 and 13 was performed. Cosmids LLNc110-45O15 and ICRFc104-A05122 were hybridized to metaphases from patient 11 and produced signals of normal intensity, whereas ICRFc104-E10129 produced no signal. In patient 13, YAC332B12, YAC924P6, RPCI1-118G17, LLNc110-21L7, LLNc110-21J18, and ICRFc104-H09154 produced no signal, LLNc110-45O15 produced a weak signal, and ICRFc104-A05122 and ICRFc104-E10129 were fully preserved. Because of chimerism, YAC 903F3 was not used as a FISH probe.
Figure 2.
Physical map and detailed deletion map of patients 11 and 13. A, Physical maps outlining the relationship between chromosomal markers, the clone contig, and DNA deletions in patients 11 and 13. Solid lines represent YAC clones (Ferrero et al. 1995), double lines cosmid clones, and the dotted line a PAC clone. The names of PACs and cosmid clones are as follows: 118G17, RPCI1-118G17; 21L7, LLNc110-21L7; 21J18, LLNc110-21J18; H09154, ICRFc104-H09154; 45O15, LLNc110-45O15; A05122, ICRFc104-A05122; and E10129, ICRFc104-E10129. Black dots represent positive markers in the respective clones. Cosmids 45O15 and A05122 overlap. “11” and “13” denote cases 11 and 13, respectively. The blackened portions of the horizontal bars represent loci that are present, unblackened portions represent absent loci, and gray-shaded portions represent the breakpoint region. Underlined loci indicate the borders of the MRX critical region. B, Diagram presenting the MRX critical interval (as defined by individuals 11 and 13) flanked by markers 40BT and DXS1139 (shown in boldface), which reside on two overlapping cosmids. Deletion intervals in cases 11 and 13 are indicated. Black dots represent sequence-tagged sites established on the basis of the genomic sequence data, and gray rectangles represent trapped putative exons. Genomic sequencing was carried out between markers 40BT and K6 (28 kb in total). The position of the trapped putative exons was established on the basis of the genomic sequence.
Exon Trapping, Genomic Sequencing, and Isolation of the Candidate Gene for MRX
By means of exon trapping on two cosmid clones (LLNc110-45O15 and ICRFc104-A05122), eight putative exons were isolated. At the same time, >28 kb of these cosmid clones were sequenced. The genomic region between 40BT and K6 was subjected to sequence analysis with the NIX analysis software package. Two of the previously trapped exons, KE14 and KO4, also were identified by the exon-prediction programs; KO4 and KE14 likely were trapped because of cryptic splice sites. On the basis of the genomic sequence data (DDBJ/EMBL/GenBank accession number AJ243947), PCR primers for K7-2, K2H, K6, and K5 were established, and they were tested on patients 11 and 13. The MRX critical region was finally narrowed to an interval of ⩽15 kb between K7-2 and K2H (fig. 2B). Both trapped exon clones, KE14 and KO4, reside within this minimal critical region.
RT-PCR experiments carried out on RNA from adult testis subsequently showed that KO4 and KE14 are part of a single gene encoding a predicted protein of 186 amino acids. The protein was enriched in serine, proline, and glutamine residues and did not present any known motifs. Exon I contains the putative start codon at position 147, exon II the first in-frame stop codon at nucleotide 704 and a polyadenylation signal at position 824. The VCX-A gene has a genomic size of 1,019 bp. Exon II contains a 30-bp repeat unit termed “RU1,” which is present, in variable numbers, in individuals with normal intelligence (see fig. 3B). Analyzing 50 unrelated white male patients, we found that 39 (78%) of them have 10 repeat units, 9 (18%) have 8 repeat units, 1 (2%) has 11 repeat units, and 1 (2%) has 12 repeat units.
Figure 3.
A, Refined deletion map indicating the region between markers DXS6837 and KAL. The position of VCX-A, -B1, -B, and -C is shown in red. For the VCX-C locus, marker DXS1134 was tested. Cases are as depicted in figure 1. Blackened portions of the horizontal bars represent loci present in the respective cases, and unblackened portions represent absent loci. B, Sequence organization of the CRI-S232 element, modified from that reported by Li et al. (1992). All instances of CRI-S232 share a very similar structure. The size of CRI-S232 is variable between individuals, covers ∼7 kb of DNA, and is not drawn to scale. CRI-S232 is divided into four regions by EcoRI, XmnI, Bgl II, and SstI. Regions b and c contain different repeat units. The VCX cDNA is 1,019 bp long, divided into two exons, and not drawn to scale. Repeat unit RU1 is contained within the translated portion of exon II. VCX is located in regions b–d. The arrowheads below the VCX exons indicate primers used to amplify the exons. The long arrows below the arrowheads represent the DNA interval that was sequenced in the present study, except for the unstable RU2 repeat unit of <2 kb. The GenBank accession number for the genomic sequence is AJ243947 (GenBank Overview).
The VCX Gene Family: Structural Features and Expression Profile
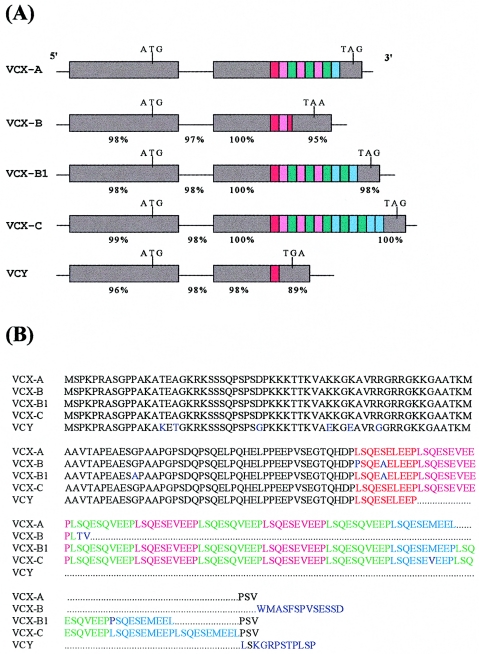
The genomic sequence revealed that the entire gene is located on the sex chromosome–specific low-copy repetitive element CRI-S232A (Li et al. 1992). The BLASTN 2.0 homology-search program detected high homology between this gene and VCY, previously known as putative DNA- or RNA-binding protein BPY1. VCY (BPY1) was isolated by cDNA selection on cosmids randomly selected from the LLN0YC03 “M” Y-chromosome cosmid library (Lahn and Page 1997). Since, on the nucleotide level, this gene is 97% identical to VCY, we termed it “VCX-A” (variably charged X chromosome mRNA on CRI-S232A). In contrast to VCY, which has only one 30-bp repeat unit in exon II, VCX-A was observed to have 8 repeat units in the cosmid sequence (fig. 4).
Figure 4.
A, DNA sequence comparison of different members of the VCX and VCY family. Gray-shaded portions of the horizontal bars represent exons, and the colored portions within exon II represent the 30-bp repeat units. In VCX-A, the number of repeat units is 8–12, on the basis of data on 50 control males investigated in the present study. In three individuals, 10, 12, and 14 repeat units were detected in VCX-C. The percentages indicate the degree of similarity between VCX-A and other VCX-family members. B, Amino acid comparison of members of the VCX family. Red, pink, green, and light blue represent the specific 10-amino-acid (30-bp) repeats schematically outlined in figure 5A. Differing amino acids are shown in dark blue.
CRI-S232 (DXS278) is known to have at least four copies on the human X chromosome and to have two on the Y chromosome (Li et al. 1992). Each CRI-S232 unit contains a 5-kb unique sequence in addition to two elements, RU1 and RU2, composed of a variable number of tandem repeats (VNTR) (fig. 3B). To isolate the paralogues of VCX-A and to compare the different members with one another, we investigated clones containing other CRI-S232 sequences (proximal to VCX-A). VCX-B and -C were isolated from physically mapped YAC NB2E11 and BAC GS-478I2 clones, respectively, by PCR primers (i.e., VCX EI [forward] and VCX EII [reverse]) established from VCX-A (Ferrero et al. 1995). The sequence of VCX-B1 was derived from a BAC clone, GSHB-214D18, from The Genome Database. One copy of VCY—VCY-D—is located on CRI-S232D, which is contained within BAC clones NH0292P09 and NH0264A13. The other copy on the Y chromosome—VCY-E—was found in CRI-S232E, which is contained within BAC clone NH0053K10 published by The Genome Database. Family members VCX-A, -B1, -B, and -C all reside within an interval of ∼2.5 Mb on Xp22.3 (with one member distal and three members proximal to the STS gene); VCY-D and VCY-E reside within an interval of ∼50 kb on Yq11.2 (Ballabio et al. 1990; Bardoni et al. 1991; Herrell et al. 1995; Nelson et al. 1995; Lahn and Page 1997).
Genomic analysis revealed that all family members have a very similar exon/intron organization (fig. 4). In addition, all members of the VCX-gene family are highly homologous to each other, except for the number of repeat units in exon II (fig. 4). A VNTR is detected within all X-chromosome copies (VCX-A, -B, -B1, and -C), whereas VCY-D and -E consistently have one repeat unit (fig. 4). In a recent paper (Lahn and Page 2000), VCX-A has been found to correspond to VCX-8r, VCX-B to VCX-2r, and VCX-B1 to VCX-10r; VCX-C has not been described prior to the present study.
To assess the pattern of expression, northern-blot analysis and RT-PCR were carried out. When VCX-A cDNA was used as a probe, a strong signal of ∼1.0 kb was detected in adult testis; no signals were observed in the other tissues tested (fig. 5). Subsequent RT-PCR experiments in various fetal and adult tissues (fetal tissues—muscle, heart, kidney, brain, and testis; adult tissues—placenta, stomach, liver, muscle, heart, kidney, brain, thalamus, hippocampus, and caudate nucleus) did not reveal any expression at standard PCR conditions (data not shown), suggesting that there is apparently exclusive expression in adult testis tissue.
Figure 5.

Expression study by northern analysis. The 1.0-kb signal in testis was revealed after overnight exposure. The hybridization probe does not distinguish among the various members of the VCX/-Y family.
Discussion
We have defined a small MRX critical region by deletion mapping of patients with chromosomal rearrangements on Xp22.3 and with clinical features of a contiguous gene–deletion syndrome. This was made possible by comparison of small overlaps in two patients with deletion who had normal intelligence and who therefore were expected to harbor an intact gene for cognitive function. Genomic sequence analysis as well as exon trapping revealed VCX-A as the only gene residing in the defined critical interval (fig. 3A). The proximal border of this interval (K2H) was established on the basis of data from several patients (cases 7–12 and 88), as well as from most of the reported patients with XLI (Ballabio et al. 1989; Shapiro et al. 1989; Schnur et al. 1990; Yen et al. 1990), whereas the distal border (K7-2) is based on PCR and FISH analysis of only one patient (case 13), a postgraduate student with high intelligence. We were asking whether this patient has further DNA distal to the critical region, yet the results of FISH analysis with Xp22.3-specific YACs, PACs, and cosmid clones argue against this.
We have shown that VCX-A is preserved in all individuals with normal intelligence but is deleted in patients with Xp22.3 rearrangements and mental retardation (fig. 3A). These results suggest that one intact copy of VCX-A is sufficient to maintain normal mental development. There is no correlation between the presence or absence of VCX-B, -B1, and -C and the mental status of our patients. The finding that repeat elements in CRI-S232A and -B contain VCX-A and VCX-B also reveals that steroid sulfatase deficiency results from unequal crossover between the VCX-A gene (which remains preserved) and the VCX-B gene (which is interrupted), which flank the steroid sulfatase gene.
To further determine the role of VCX-A in cognitive function and to explain the normal IQ in patients 13 and 511, who had deletion of VCX-B1, -B, and -C, expression profiles of the individual genes were analyzed by RT-PCR. Expression at detectable levels was found only in testicular tissue in all VCX members investigated and was not detected in any of the postnatal brain structures analyzed. The apparent absence of VCX-A expression is not easy to reconcile with a putative cognitive function, since most genes involved in mental retardation have been shown to be expressed in postnatal brain structures—for example, in the cerebral cortex and hippocampus; however, this may suggest that VCX-A has a highly distinctive spatial and temporal expression pattern, with elevated expression occurring only during specific stages of neuronal development. In support of this notion, among X-Y homologous genes the Y-linked genes are usually expressed only in testis, whereas the X-linked genes are shown to be expressed in a range of tissues (Lahn and Page 1997).
To demonstrate a possible involvement of VCX-A in the physiology of the CNS, mutation analysis was carried out in five index patients with MRX (MRX21, MRX24, MRX36, MRX37, and MRX49) whose families showed linkage to Xp22, to the region including the VCX-A, -B1, and -B genes (Kozak et al. 1993; Martinez et al. 1995; Bar-David et al. 1996; Claes et al. 1996, 1997). Unfortunately, the genetic intervals of all five pedigrees are quite large. The smallest genetically defined region was for MRX49, spanning ∼15 Mb of DNA, and the largest was for MRX21, spanning ∼38 Mb of DNA. Sequence analysis of all five index patients failed to detect any mutation in the VCX genes.
The fascinating question of what differentiates us from our closest mammalian relatives provokes thoughts regarding the quality (regulation) and quantity (number) of genes with a putative involvement in cognitive function. Our finding that orthologues of VCX-A exist in primates and in old- and new-world monkeys but not in prosimians, marsupials, and other mammals (data not shown) suggests that VCX originated ∼40 million years ago. This occurred just after the separation of the prosimian and the higher-primate lineages. VCX was then amplified rapidly during the evolution of the hominoids. In addition to the amplification of the entire locus, the 30-bp gene internal repeat unit has been amplified up to 14 times. The discovery of a multiple 30-bp (10-amino-acid) repeat unit predicted to be highly negatively charged also raises the question of whether the expansion of these repeats above a certain threshold may lead to disease. So far, all known expansion diseases are considered to be neurological in a broad sense, involving mostly triplets or 12-bp expansions (Mandel 1997). Further investigations of the VCX-gene family, with regard to neurological diseases and sex-chromosome evolution, are warranted.
Acknowledgments
For help and discussion, we thank our colleagues from the unit, as well as N. Matsuo, S. Lindsay, and U. Wirkner; for providing the blood samples from patients with MRX and from family members, we thank the following clinicians: N. Sakura, T. Matsumoto, K. Agematsu, Y. Tanabe, M. Yoshino, Y. Tanaka, H. Ohashi, T. Nagai, F. Martinez, D. Abeliovich, G. Neri, J. Chelly, and J. P. Fryns. We would also like to express our gratitude to all patients and families who participated in this study. This work was supported by a Deutscher Akademischer Auslandsdienst fellowship to M.F. and by the Deutsche Forschungsgemeinschaft.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Biology and Biotechnology Research Program (Lawrence Livermore Laboratory), http://www-bio.llnl.gov (for X chromosome–specific library [accession number LLNc110])
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST (for sequence alignments)
- Children's Hospital Oakland Research Institute, http://www.chori.org/bacpac/ (for human PAC library)
- DDBJ, DNA Databank of Japan, http://www.ddbj.nig.ac.jp (for VCX-A, -B, -C, and -B1 [DDBJ/EMBL/GenBank accession numbers AF167078–AF167081 and AJ243947]; VCY [DDBJ/EMBL/GenBank accession number gi4759303]; and genomic sequence [DDBJ/EMBL/GenBank accession number AJ243947])
- EMBL Outstation, European Bioinformatics Institute, http://www.ebi.ac.uk/ (for LANE TRACKER and GENE SKIPPER)
- Fondation Jean Dausset CEPH, http://www.cephb.fr (for YAC clones)
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html
- Genome Database, The, http://www.gdb.org (for BAC clones GSHB-214D18 [accession number RC005296], NH0053K10 [accession number RC010720], and NH0264A13 [accession number RC018677])
- Imperial Cancer Research Fund, http://www.icnet.uk (for X chromosome–specific library [accession number ICRFc104])
- Max-Planck Institut, http://www.mpimg-berlin-dahlem.mpg.de (for X chromosome YACs)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for MRX [MIM 309530])
References
- Agematsu K, Koike K, Morosawa H, Nakahori Y, Nakagome Y, Akabane (1988) Chondrodysplasia punctata with X;Y translocation. Hum Genet 80:105–107 [DOI] [PubMed] [Google Scholar]
- Ballabio A, Andria G (1992) Deletions and translocations involving the distal short arm of the human X-chromosome: review and hypotheses. Hum Mol Genet 1:221–227 [DOI] [PubMed] [Google Scholar]
- Ballabio A, Bardoni B, Carrozzo R, Andria G, Bick D, Campbell L, Hamel B, Ferguson-Smith MA, Gimelli G, Fraccaro M, Maraschio P, Zuffardi O, Guioli S, Camerin G (1989) Contiguous gene syndromes due to deletions in the distal short arm of the human X chromosome. Proc Natl Acad Sci USA 86:10001–10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio A, Bardoni B, Guioli S, Basler E, Camerino G (1990) Two families of low-copy-number repeats are interspersed on Xp22.3: implications for the high frequency of deletions in this region. Genomics 8:263–270 [DOI] [PubMed] [Google Scholar]
- Bar-David S, Lerer I, Sarfaty CK, Kohan ZG, Meiner V, Zlotogora J, Abeliovich D (1996) Localization of two X-linked mental retardation (XLMR) genes to Xp: MRX37 gene at Xp22.31-p22.32 and a putative MRX gene on Xp22.11-p22.2. Am J Med Genet 64:83–88 [DOI] [PubMed] [Google Scholar]
- Bardoni B, Zuffardi O, Guioli S, Ballabio A, Simi P, Cavalli P, Grimoldi MG, Fraccaro M, Camerino G (1991) A deletion map of the human Yq11 region: implications for the evolution of the Y chromosome and tentative mapping of a locus involved in spermatogenesis. Genomics 11:443–451 [DOI] [PubMed] [Google Scholar]
- Burn TC, Connors TD, Klinger KW, Landes ML (1995) Increased exon-trapping efficiency through modification to the pSPL3 splicing vector. Gene 161:183–187 [DOI] [PubMed] [Google Scholar]
- Chelly J (1999) Breakthroughs in molecular and cellular mechanisms underlying X-linked mental retardation. Hum Mol Genet 8:1833–1838 [DOI] [PubMed] [Google Scholar]
- Church DM, Stotler CJ, Rutter JL, Murrell JR, Trofatter JA, Buckler AJ (1994) Isolation of genes from complex sources of mammalian genomic DNA using exon amplification. Nat Genet 6:98–105 [DOI] [PubMed] [Google Scholar]
- Claes S, Gu XX, Leguis E, Lorenzetti E, Marynen P, Fryns JP, Cassiman JJ, Raeymaekers P (1996) Linkage analysis in three families with nonspecific X-linked mental retardation. Am J Med Genet 64: 137–146 [DOI] [PubMed] [Google Scholar]
- Claes S, Vogels A, Holvoet M, Devriendt K, Raeymaekers P, Cassiman JJ, Fryns JP (1997) Regional localization of two genes for nonspecific X-linked mental retardation to Xp22.3-p22.2 (MRX49) and Xp11.3-p11.21 (MRX50). Am J Med Genet 73:474–479 [PubMed] [Google Scholar]
- Erfle H, Ventzki R, Voss H, Rechmann S, Benes V, Stegemann J, Ansorge W (1997) Simultaneous loading of 200 sample lanes for DNA sequencing on vertical and horizontal, standard and ultrathin gels. Nucleic Acids Res 25:2229–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero GB, Franco B, Roth EJ, Firulli BA, Borsani G, Delmas Mata J, Weissenbach J, Halley G, Schlessinger D, Chinault AC, Zoghbi HY, Nelson DL, Ballabio A (1995) An integrated physical and genetic map of a 35 Mb region on chromosome Xp22.3-Xp21.3. Hum Mol Genet 4:1821–1827 [DOI] [PubMed] [Google Scholar]
- Gécz J, Mulley J (2000) Genes for cognitive function: developments on the X. Genome Res 10:157–163 [DOI] [PubMed] [Google Scholar]
- Gedeon AK, Donnelly AJ, Mulley JC, Kerr B, Turner G (1996) How many X-linked genes for non-specific mental retardation (MRX) are there? Am J Med Genet 64:158–162 [DOI] [PubMed] [Google Scholar]
- Herrell S, Novo FJ, Charlton R, Affara NA (1995) Development and physical analysis of YAC contigs covering 7 Mb of Xp22.3-p22.2. Genomics 25:526–537 [DOI] [PubMed] [Google Scholar]
- Ioannou PA, Amemiya CT, Gernes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ (1994) A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet 6:84–89 [DOI] [PubMed] [Google Scholar]
- Kozak L, Chiurazzi P, Genuardi M, Pomponi MG, Zollino M, Neri G (1993) Mapping of a gene for non-specific X linked mental retardation: evidence for linkage to chromosomal region Xp22.1-Xp22.3. J Med Genet 30:866–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC (1997) Functional coherence of the human Y chromosome. Science 278: 675–680 [DOI] [PubMed] [Google Scholar]
- ——— (2000) A human sex-chromosomal gene family expressed in male germ cells and encoding variably charged proteins. Hum Mol Genet 9:311–319 [DOI] [PubMed] [Google Scholar]
- Li XM, Yen PH, Shapiro LJ (1992) Characterization of a low copy repetitive element S232 involved in the generation of frequent deletions of the distal short arm of the human X chromosome. Nucleic Acids Res 20:1117–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Cremer T (1992) Human cytogenetics: a practical approach. IRL, Oxford University Press, Oxford, New York, Tokyo [Google Scholar]
- Mandel JL (1997) Breaking the rule of three. Nature 386:767–769 [DOI] [PubMed] [Google Scholar]
- Martinez F, Gal A, Palau F, Pietro F (1995) Localization of a gene for X-linked nonspecific mental retardation (MRX24) in Xp22.2-p22.3. Am J Med Genet 55:387–390 [DOI] [PubMed] [Google Scholar]
- Merienne K, Jacquot S, Pannetier S, Zeniou M, Bankier A, Gécz J, Mandel JL, Mulley J, Sassone-Corsi P, Hanauer A (1999) A missense mutation in RPS6KA3 (RSK2) responsible for non-specific mental retardation. Nat Genet 22:13–14 [DOI] [PubMed] [Google Scholar]
- Muroya K, Ogata T, Matsuo N, Nagai T, Franco B, Ballabio A, Rappold GA, Sakura N, Fukushima Y (1996) Mental retardation in a boy with an interstitial deletion at Xp22.3 involving STS, KAL1 and OA1: implication for the MRX locus. Am J Med Genet 64:583–587 [DOI] [PubMed] [Google Scholar]
- Nelson DL, Ballabio A, Cremers F, Monaco AP, Schlessinger D (1995) Report of the Sixth International Workshop on human X Chromosome Mapping 1995. Cytogenet Cell Genet 71:307–3428521720 [Google Scholar]
- Nishimura S, Masuda H, Matsumoto T, Skura T, Ueda K (1991) Two cases of steroid sulfatase deficiency with complex phenotype due to contiguous gene deletions. Am J Med Genet 40:260–263 [DOI] [PubMed] [Google Scholar]
- Ogata T, Mastuo N, Shimizu N (1990) A ring X chromosome, 46,Y,r(X)(p22.33q28), as a cause of extreme short stature in a male. Am J Med Genet 35:241–244 [DOI] [PubMed] [Google Scholar]
- Schaefer L, Ferrero GB, Grillo A, Bassi MT, Roth EJ, Wapenaar MC, van Ommen GJB, Mohandas TK, Rocchi M, Zoghbi HY, Ballabio A (1993) A high resolution deletion map of human chromosome Xp22. Nat Genet 4:272–279 [DOI] [PubMed] [Google Scholar]
- Schnur RE, Knowlton RG, Musarella MA, Muenke M, Nussbaum RL (1990) Partial deletions of a sequence family (“DXS278”) and its physical linkage to steroid sulfatase as detected by pulsed-field gel electrophoresis. Genomics 8:255–262 [DOI] [PubMed] [Google Scholar]
- Shapiro LJ, Yen P, Pomerantz D, Martin E, Rolewic L, Mohandas T (1989) Molecular studies of deletions at the human steroid sulfatase locus. Proc Natl Acad Sci USA 86:8477–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivier E, De Cesare D, Jacquot S, Pannetier S, Zackai E, Young I, Mandel JL, Sassone-Corsi P, Hanauer A (1996) Mutations in the kinase Rsk-2 associated with Coffin-Lowry syndrome. Nature 384:567–570 [DOI] [PubMed] [Google Scholar]
- Turner G, Opitz JM (1980) X-linked mental retardation. Am J Med Genet 7:407–415 [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst MA, Bassi MT, Borsani G, Wapenaar MC, Ferrer GB, de Conciliis L, Rugarli EI, Grillo A, Franco B, Zoghbi HY, Ballabio A (1994) A gene from the Xp22.3 shares homology with voltage-gated chloride channels. Hum Mol Genet 3:547–552 [DOI] [PubMed] [Google Scholar]
- Weissortel R, Strom TM, Doerr HG, Rauch A, Meitinger T (1998) Analysis of an interstitial deletion in a patient with Kallmann syndrome, X-linked ichthyosis and mental retardation. Clin Genet 54:45–51 [DOI] [PubMed] [Google Scholar]
- Wiemann S, Stegemann J, Grothues D, Bosch A, Estivill X, Schwager C, Zimmermann J, Voss H, Ansorge W (1995) Simultaneous on-line DNA sequencing on both strands with two fluorescent dyes. Anal Biochem 224:117–121 [DOI] [PubMed] [Google Scholar]
- Yen PH, Li XM, Tsai SP, Johnson C, Mohandas T, Shapiro LJ (1990) Frequent deletions of the human X chromosome distal short arm result from recombination between low copy repetitive elements. Cell 61:603–610 [DOI] [PubMed] [Google Scholar]