Abstract
We have recently reported that OTOF underlies an autosomal recessive form of prelingual sensorineural deafness, DFNB9. The isolated 5-kb cDNA predicted a 1,230 amino acid (aa) C-terminus membrane–anchored cytosolic protein with three C2 domains. This protein belongs to a family of mammalian proteins sharing homology with the Caenorhabditis elegans fer-1. The two other known members of this family, dysferlin and myoferlin, both have six predicted C2 domains. By northern blot analysis, a 7-kb otoferlin mRNA could be detected in the human brain. We isolated the corresponding cDNA, which is expected to encode a 1,977-aa-long form of otoferlin with six C2 domains. A 7-kb cDNA derived from the murine orthologous gene, Otof, was also identified in the inner ear and the brain. The determination of the exon-intron structure of the human and murine genes showed that they are composed of 48 coding exons and extend ∼90 kb and ∼80 kb, respectively. Alternatively spliced transcripts could be detected that predict several long isoforms (six C2 domains) in humans and mice and short isoforms (three C2 domains) only in humans. Primers were designed to explore the first 19 OTOF exons, henceforth permitting exploration of the complete coding sequence of the gene in DFNB9 patients. In a southwestern Indian family affected by DFNB9, a mutation in the acceptor splice site of intron 8 was detected, which demonstrates that the long otoferlin isoforms are required for inner ear function.
Introduction
Deafness is one of the most frequent hereditary disorders in humans. The vast majority of genetic prelingual hearing loss is inherited on the autosomal recessive mode (DFNB forms) (for review, see work by Petit [1996]). To date, 26 DFNB loci have been mapped (see Web site for Nonsyndromic Hearing Impairment Autosomal Recessive Loci) and six genes have been identified (Kelsell et al. 1997; Liu et al. 1997; Weil et al. 1997; Li et al. 1998; Wang et al. 1998; Mustapha et al. 1999; Yasunaga et al. 1999). Using a positional cloning strategy combined to a candidate gene approach, we have identified the OTOF gene, encoding otoferlin, as responsible for DFNB9 (MIM 601071) (Yasunaga et al. 1999). The 5-kb OTOF cDNA from a total fetus polyA+ mRNA preparation, encodes a putative 1,230-aa membrane-anchored cytosolic protein containing three predicted C2 domains (Yasunaga et al. 1999) (GenBank AF107403). A single nonsense mutation has been detected in the four analyzed Lebanese families. This mutation is predicted to truncate the 500 C-terminal amino acids of the protein (Yasunaga et al. 1999). OTOF is a member of a mammalian gene family related to the Caenorhabditis elegans gene fer-1 (Achanzar and Ward 1997). This family also comprises DYSF (Bashir et al. 1998; Liu et al. 1998) and MYOF (Davis et al. 2000), encoding dysferlin (2080 aa) and myoferlin (2061 aa), respectively. Both dysferlin and myoferlin are membrane-anchored cytosolic proteins that contain six predicted C2 domains, four of which are expected to bind Ca2+ (Rizo and Südhof 1998). Subsequent detection of a 7-kb otoferlin mRNA by northern blot analysis prompted us to characterize this transcript which had not been detected in our initial study.
Subjects and Methods
Northern Blot Analysis
Northern blots of adult human and murine tissues (Clontech) were incubated with cDNA probes derived from the 3′-UTR sequences of the otoferlin genes—namely, positions 6722–7103 (GenBank AF183185) and 6615–7005 (AF183184) for the human and murine probes, respectively, in Express Hyb solution (Clontech), according to the manufacturer's instructions. The filters then were exposed to Kodak BioMax X-ray film for 14 d at −80°C. The tissues analyzed were human brain, heart, placenta, lung, liver, pancreas, skeletal muscle, and kidney, and murine brain, heart, spleen, lung, liver, skeletal muscle, kidney, and testis.
RACE-PCR, RT-PCR, and Long-Range PCR
Oligo-dT and random-primed cDNA libraries were constructed from poly(A)+ mRNA of human total fetus, adult brain, heart, kidney, and murine fetal heart, using a Marathon cDNA amplification kit (Clontech). RACE-PCR experiments were performed on these libraries using linker primers and a series of primers selected from the otoferlin cDNA sequence. The PCR products were directly cloned into pGEM-T Easy vector (Promega) and sequenced. To isolate the human cDNA long form (7 kb), a reverse primer (5′-TTCACCTGGGCCCGCAGCATCCT-3′) was designed from the sequence encoding aa 63–70 of the initially reported short form of otoferlin (Yasunaga et al. 1999) (GenBank 107403).
Total RNAs were extracted from 2-d-old mouse cochlea and brain by the guanidium isothiocyanate procedure. RT-PCR experiments were performed in various murine and human RNA sources, according to the GeneAmp RNA PCR kit protocol (Perkin Elmer Cetus). To reconstitute the entire murine cDNAs derived from the brain and the cochlea, we used two primer pairs, one from the exon 1 5′-UTR (5′-AGGCGTGTGAGCCACACTCCACCA-3′) and exon 22 (5′-CATAACCTCAGCTTGTCCCGAACA-3′), and the other from the exon 18-19 junction (5′-GGCCCCAGATCACGGACAGGAAC-3′) and exon 48 3′-UTR (5′-GGCCAGTACACCTGATTCACACT-3′). To reconstitute the entire 5′ part of the human brain cDNA long form, primers derived from the 5′-UTR exon 1 (5′-GGAGGAGGCAGCGGCAGAGAAGA-3′) and exon 22 (5′-TTCACCTGGGCCCGCAGCATCCT-3′) were used. Alternative splicings were also detected by RT-PCR (see fig. 4). To show the skipping of exon 6, primers from exon 4 (5′-AATCGGGTAGAGGTGACCGACAC-3′) and exon 10 (5′-CCGAGCCTCAATCACTGTGATGC-3′) were selected in mouse, and primers from exon 5 (5′-GTGGAGGTCCGGTATCAGGCCAC-3′) and exon 8 (5′-ACACCGAGTCGGGATCCAGTCCA-3′) were designed in humans. To reveal the alternative splicing of exon 31, a primer pair from the murine exon 29-30 (5′-CAAGTGGTTTGAAGTGGACCTCCC-3′) and exon 32-33 (5′-GCCACATCCACCTTGACCACAGC-3′) junctions was used, both in mice and in humans, as the human and murine sequences were identical. Finally, to show the alternative splicing of exon 47, primers from exon 44 (5′-GGAGTCTATGTTCTCCTGGGATGAGAC-3′) and exon 48 3′-UTR (5′-GTCTTGCTCAAGGCTGGCAGGCG-3′) and from exon 45 (5′-GACAGCCAAGCAGTGCACCATGG-3′) and exon 48 3′-UTR (5′-AGGCAGGCTCGGCCCAAGGCATG-3′) were chosen in mice and in humans, respectively. Long-range PCR was performed using Expand Long Template PCR Systems. PCR primers with a predicted melting temperature (Tm) of 72°C were designed, and a two-step PCR procedure (35 cycles of 92–94°C for 15 sec and 68°C for 4–30 min) was adopted.
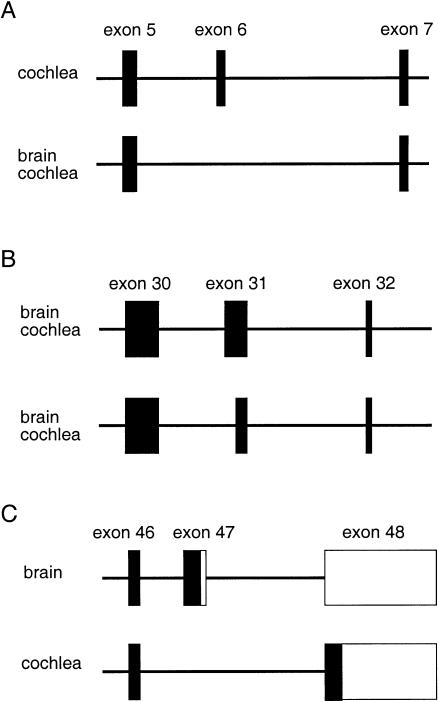
Figure 4.
Alternative splicings of the Otof exons 6, 31, and 47 in the murine brain and cochlea. The genomic regions encompassing exons 5–7 (A), exons 30–32 (B) and exons 46–48 (C) are schematized. The exonic sequences found in the different murine transcripts are boxed. The coding regions are indicated by blackened boxes, the untranslated regions by unblackened boxes.
Patients
The consanguineous family J2 originates from southwestern India (Maharashtra state). All members were subjected to clinical investigation, including a general examination and audiometric tests. Audiometry was performed with a pure-tone, portable audiometer. Affected individuals demonstrated severe-to-profound prelingual sensorineural hearing impairment.
Genotyping and Linkage Analysis
Informed consent was obtained from all individuals (Consultative Committee for People Protection in Biomedical Research in France and IRB approval nr OH93-DC-0116 in the USA). Genotyping was carried out using a set of primer pairs amplifying three polymorphic markers from the DFNB9 chromosomal region (D2S174, D2S2350, and D2S2223) (Yasunaga et al. 1999). A multipoint LOD score of 2.4 was obtained for family J2.
Mutation Detection
Each of the 48 OTOF coding exons was PCR amplified on 50 ng of genomic DNA, using the primers described in table 1 for exons 1–19, and in table 2 (Yasunaga et al. 1999) for exons 20–48. The sequences of both DNA strands were determined using the same primers as for PCR amplification.
Table 1.
Primers for PCR Amplification of OTOF Exons 1–19
|
Primer (5′→3′) |
|||
| Exon | Forward | Reverse | Product Size (bp) |
| 1 | AGGCAGCGGCAGAGAAGA | CTCCCAGCCCTGTCCTAT | 270 |
| 2 | CACGAGGTCCCATGTTGCA | CGGCCAGTGCCTGGGATT | 295 |
| 3 | TCCCTGGGGAGCACTGG | AGGTTGGGAGTGTAGGTCC | 334 |
| 4 | CCAAGCAGTCACAGCCCTT | ACCTCGCCATGCATGAGAG | 302 |
| 5 | TCCAGTGAGGCAAGGGTGT | CTTGGATGTCTCTCCAGAAG | 434 |
| 6 | TCTGCAGACCTAGGCTTGC | CGACAGCCCACTCCTGAG | 334 |
| 7 | TTCTCTCATGTTTGGCTCTTC | GAGGGCCACGCATCACTG | 304 |
| 8 | TAACTCTCAGCTTTCTGGATG | TACCCAAATTCCAATCATGGC | 303 |
| 9 | GTGCTTGAGTGTTTAAAGACC | AGTATAGTGGATAATGCACATC | 265 |
| 10 | CAGATGAGAGGCAGTGGTC | GCTCCTTGACTTCCAGCCA | 385 |
| 11 | AGGGAGGGGCCCAACACT | CTCTTTATCATGGGTCTAGAC | 239 |
| 12 | TCCCACTTCACCACAAAGCT | ACAGTCGCCAGACTGTGGT | 301 |
| 13 | GACAGCATTTGGTTCTGCCA | GTGGCAGGTGCTCTCAGC | 351 |
| 14 | GTGCCAGGACCCAGGAGT | TGATTTCCAGCCTTGTCTTAC | 387 |
| 15 | TCCCACGCCCTCACCTGT | TGAAGAGAGGGCATCTCACA | 302 |
| 16 | TCAGCACCCAGGAGCTGG | CCTGGGACCCAGGTGACT | 408 |
| 17 | CGGCCTGTCTGTGAGACG | GAGCCTCACACTTACCACC | 290 |
| 18 | AAGGCAGCACCGGATTGGA | TCGCTCCAGGTAGGGCAG | 371 |
| 19 | TCCTCCACTCCACCAATGC | CCTCTGACAGCGCCGTCT | 300 |
Table 2.
OTOF Exon/Intron Boundaries
|
Splice Site (5′→3′) |
||||
| Exon Numbera | Size (bp) | Acceptor | Donor | Intron Size(kb) |
| 1 | 206 | ACTTTCCGAG/gtagggaagccgctgtctca | 11 | |
| 2 | 59 | tatttcctctttccccacag/GGCAATCCTT | CTTTGATGAG/gtaggaggcccaagactccc | 8.5 |
| 3 | 89 | agttgtccctctctccccag/ACATTTCGGT | TCAGCAACAA/gtaagtgtgggatggggcag | 7 |
| 4 | 100 | gccccctctgccatccccag/GCTCATCGGG | TATCATCAAG/gtgggtatgccccacggagt | 2.5 |
| 5 | 182 | caccgcctccctcccttcag/ACCAGCCTGT | GCTTCCGGAG/gtaacaggcaggacccctct | 6 |
| 6 | 45 | catttcttccccctactcag/CAAAGGCAGA | ACGAGCACAA/gtaggtgtcccaggcgaggg | 9 |
| 7 | 74 | tctggtggcccccctcacag/AGCCGGGAGG | CAAAGACCAG/gtaggacaggtgccgcgtga | 1.2 |
| 8 | 127 | cagtctttcatcttccccag/ATGAACCGGC | CCAACAAGCG/gtgagtggacgggagccctg | .492 |
| 9 | 55 | tggtacattctcttcctcag/ATCTAAGCCA | GGATTACCAG/gtaggtgggcacctggaggc | 7 |
| 10 | 132 | cccccttctcctgcctgcag/GTCAGCATCA | TTACAACGAG/gtcagtggccctgtggggaa | 6 |
| 11 | 63 | ctggatccttccccctccag/TACTTCGTCT | CAAGATTTCG/gtgagtggggagcagcccct | .383 |
| 12 | 82 | gctgtggttctgcctgaaag/GTGATTCACT | TCGCAGCCAG/gtgagtacctgcgcatggcc | 5 |
| 13 | 160 | cccacccctcatcattgcag/AGCACCAGTT | ACATTGAGGG/gtgaggcccagctaccccag | .8 |
| 14 | 187 | ctggctgtatgtgctggcag/GAACTTGCTG | TGGCCAGAAG/gtactggggtatgaggtaca | .9 |
| 15 | 187 | tgtgctcccccttccaccag/GGCAAGACTT | GGAGACAAAG/gtcagcagcgggagacccgg | 1.397 |
| 16 | 224 | ttttgttcccactgccacag/GCTTCCTGCC | CATCTCGGAG/gtgagaccccaggcactgtg | .474 |
| 17 | 109 | tttgaactgcctccccacag/AGCTGTGCAG | GTCACCATAG/gtgagtgctgggcccacggg | .548 |
| 18 | 181 | ctgggcccattctgctccag/GCAACTATGG | TCACCGACAG/gtgggcccagcctcccatcc | .081 |
| 19 | 121 | cccgcctctgcccaccccag/GAACTACTTC | CGACAAGCTG/gtcagggccaggccaggggc | 1.512 |
| 5′UTRsf1b | 199 | AGATCATGAG/gtccctcactctcctgatca | ||
| 20 | 101 | cccacactcctgattcacag/GAAGAAGGCC | GTGGCTGCTG/gtgagaagggaggggcggca | .142 |
| 21 | 91 | ccggctcgccctacccccag/CCGCTTCCTC | GAGGGAGCTG/gtgaggacgcaactggacgg | .127 |
| 22 | 117 | tgacaccccctccttcgcag/GAAAACATGG | GGCGGACGAG/gtgcggcccaaggggtcggg | .128 |
| 23 | 153 | ggccaccccccaacccccag/CCCCAGCACA | CTTCCTTAAG/gtgctggagggggcaggatg | .549 |
| 24 | 190 | gaccccatgcccacccccag/CTGCCAGGGA | GTCTACACCA/gtgagtgaggacccctcact | .089 |
| 25 | 125 | caggctgcccttccccacag/AGAAGCAGGC | GTGCACAGAG/gtgagggcctgggaggaggg | .42 |
| 26 | 135 | ctccccatcccatgctgcag/GTGCTGAATG | GGATTCCATG/gtatgggtgggctctggtgc | .684 |
| 27 | 162 | tgggttgtctgttgcgtcag/GGCAAAGCTG | GCTGCTGCAG/gtgggactgcagggaagagg | .402 |
| 28 | 120 | ctgtgacccccattccccag/ATTGGACCAG | CCGAGTGGAG/gtgcgagggctctgtgtggc | .426 |
| 29 | 162 | caccctccaacctctcccag/GTGCTGTTCT | GTTTGAAGTG/gtgagtgcaggccctggcgg | .111 |
| 30 | 163 | tcctgtcgccaacaccccag/GACCTCCCAG | AACACCACGG/gtatggccacatccaccctg | .482 (.542) |
| 31c | 131 | ccatctatccatctgtccag/TCAGGCTTCT | GCTGGACGCG/gtaaggcgggtggggtcagc | |
| ctcctcctctcactccacag/GGGAGGTTGT | 1.369 | |||
| 32 | 30 | tctccttggttcctctgcag/ACTTCTGAAG | GGTGGATGTG/gtgagtgtgggggccatgca | .399 |
| 33 | 129 | tggacctggccaacctgcag/GCTGAGGAGG | CATGAAGGAG/gtgagacctcgggtagggtg | 2.119 |
| 34 | 67 | ttcatctcatcttttggcag/CAACTTCGAC | AATACCGAGG/gtgagccctggaacctggaa | .905 |
| 35 | 137 | ccttttctgcttctggccag/GCCTGAAGGG | TGAGCTTAAG/gtgagagcctaggagcagac | .131 |
| 36 | 135 | aacagcctggtctgtctcag/GTATACCCCA | ACGCTTCAAG/gtcaggccaggagcacgggc | .247 |
| 37 | 138 | cccacatttgtcttgcccag/GGCTCCCTCT | TGTGGTCCGG/gtgagactcccgtcgctttc | .637 |
| 38 | 128 | agcccttctccccattccag/GCCACGGACC | TCTTTGGGAA/gtaaggcttcttggtgccct | .106 |
| 39 | 171 | tcctaactccaccccaccag/GTCCTTTGAC | CCTACTCCAC/gtatgtggggcatggggcag | .642 |
| 40 | 161 | ctgtgtgtcccctcccacag/ACATGGCTAC | GACGAGAACG/gtaatggggcactggatgca | .766 |
| 41 | 143 | tgagccgccggcacccacag/GTCAGAGGAA | CATCGAGCAG/gtgacacttgcatggccaag | .392 |
| 42 | 89 | gccacaaccctcttcctcag/GGCCGCCTGG | AGCCCAAGAA/gtgagcggcctggggcccag | 1.287 |
| 43 | 99 | ccttctctccctgggcccag/GTACGAGCTG | TCGTGAGGGG/gtgggtgagcagtccctgca | .145 |
| 44 | 242 | tggagatgtggcgcccacag/GTGGCTGAAG | GACTTCCTGG/gtgcagagcagggcagggat | .664 |
| 45 | 179 | tgacactgaggttgccacag/GGGCCATCGA | TGAGCTCACG/gtgcgcaccccttcctgctc | .104 |
| 46 | 101 | gacctgtctgtcctctgcag/GGCAAGGTGG | AGAAACCCAA/gtgagtgccctgccggccca | .441 |
| 47 | 198 | ctctacccttcatactccag/CCGGCCCGAC | GGCCTCCTGG/gtctgatatttcttacttct | 1.695 |
| 48 | 1018 | tcctctcttccacttcccag/CCGGCCCGAC | ||
Exons 20–47 were referred to as exons 1–28 in Yasunaga et al. (1999).
“5′UTRsf1” refers to an additional exon which corresponds to the 5′-untranslated region of the transcript encoding the otoferlin short form 1 and encodes the predicted first 17 aa of the short form 2 (see fig. 3).
Exon 31 contains an alternative acceptor splice site at nucleotide position 61 (see fig. 4B).
DNA Sequencing
PCR products (500 ng) were treated with exonuclease I and shrimp alkaline phosphatase (Amersham Life Science), and subsequently sequenced using a DYEnamic ET-terminator cycle sequencing premix kit (Amersham Pharmacia Biotech) on an ABI 377 DNA sequencer. For the direct sequencing of BAC DNA, we used a BigDye-terminator RR mix (PE Applied Biosystems).
DNA and Protein Sequence–Analysis Tools
Sequence-comparison analysis was carried out using BLAST and FASTA. The search for protein motifs was carried out by SMART and Pfam. PSORT II was used to predict the subcellular localization of the proteins. The secondary structure of the protein was predicted by the Pôle Bio-informatique Lyonnais server. Putative enzyme posttranslational modification sites were predicted using the Prosearch 2.0 program.
Results and Discussion
Characterization of Human and Murine cDNAs Encoding Long Forms of Otoferlin
The previously characterized otoferlin 5-kb cDNA was derived from total human fetus RNA (Yasunaga et al. 1999). In our initial study of the otoferlin mRNA, we had failed to detect a signal by Northern blot analysis of poly(A)+ RNA from various adult human tissues. However, upon longer exposure of the blots, we were able to detect a 5-kb transcript in the heart, placenta, liver, pancreas, skeletal muscle, kidney, and brain, as well as an additional 7-kb band in the brain (data not shown). In order to characterize the longer brain transcript, 5′-RACE-PCR experiments were performed with primers derived from the 5′ coding sequence of the short (5-kb) otoferlin cDNA (see Subjects and Methods). A 5′-RACE-PCR product extending the previous cDNA sequence was obtained from brain mRNA, but not from heart, kidney, or total fetus mRNA. Successive rounds of extension of the brain product permitted the reconstitution of a 7,156-bp poly(A)+ cDNA sequence (GenBank AF183185). The putative translation-initiation site was identified at position 128 (Kozak consensus sequence ACCAGCatgGCC [Kozak 1996]) and is preceded by an in-frame stop codon 51 bp upstream. The initiation codon is followed by a 5,991-bp ORF and a 1,038-bp 3′-UTR with a polyadenylation signal (AATAAA) at position 7146. The deduced aa sequence predicts a 1,997-aa (227 kDa) protein (fig. 1), showing 31% and 33% identity (55% and 43% similarity) with the human dysferlin and myoferlin sequences, respectively, and 23% identity (49% similarity) with the Caenorhabditis elegans fer-1.
Figure 1.
Deduced amino acid sequence of human otoferlin brain long isoform. The predicted C-terminal transmembrane domain is indicated in boldface; the six C2 domains are underlined; the five aspartyl residues (D) that presumably bind Ca2+ in each of the last four C2 domains (Sutton et al. 1995; Rizo and Südhof 1998) are in boldface.
By RT-PCR experiments, an expression of the murine orthologous gene, Otof, had been detected in various tissues—including the brain and the inner ear, both in the cochlea (the auditory organ) and in the vestibule (the balance organ) (Yasunaga et al. 1999). By northern blot analysis, only a 7-kb transcript was detected in the murine tissues analyzed (see Subjects and Methods, and data not shown). We thus hypothesized that this transcript encodes a long form of otoferlin. The 75A2MH otoferlin cDNA clone (Yasunaga et al. 1999), which was expected to encode the C-terminal 205 aa of the protein, had been identified from subtractive murine cochlear cDNA libraries (Cohen-Salmon et al. 1997; Verpy et al. 1999). To extend this cDNA sequence, 5′- and 3′-RACE-PCR experiments were performed on mouse fetal head mRNA (see Subjects and Methods). The entire cDNA sequences were eventually reconstituted by RT-PCR experiments on cochlear and brain total RNA. This resulted in 6,872-bp (cochlea) and 7,090-bp (brain) poly(A)+ cDNA sequences (GenBank AF183183 and AF183184). The putative translation-initiation site was identified in an appropriate Kozak consensus site (ACCAGCatgGCC, (Kozak 1996) at position 105, preceded by an in-frame stop codon located 66 bp upstream. The initiation codon is followed by a 5,976-bp (cochlea) or 5,991-bp (brain) ORF, and a 792-bp (cochlea) or 995-bp (brain) 3′-UTR with a polyadenylation signal (AATAAA) at position 6856 (cochlea) or 7074 (brain). These cDNAs encode predicted 1992 aa (cochlea) and 1997 aa (brain) proteins, showing 95% of sequence identity (98% similarity) with the human otoferlin. RACE-PCR experiments failed to detect short cDNA forms in all the murine tissues tested, namely the cochlea, vestibule, brain, and testis.
Analysis of the deduced sequences of the human and murine otoferlins showed the presence of a C-terminal transmembrane (TM) domain (see fig. 2) and the absence of a leader peptide or subcellular targeting signal. Six fragments with homology to C2 domains (C2A-F) were recognized (figs. 1 and 2) using SMART and Pfam programs. C2 domains are composed of two four-stranded β sheets and have highly similar structures (Sutton et al. 1995; Rizo and Südhof 1998). The extensive analysis of the C2A domain of rat synaptotagmin-1 (Syt-1 C2A) has permitted the recognition of five aspartyl residues which bind Ca2+; two are located in the loop between β strands 2 and 3, and three are located in the loop between β strands 6 and 7. The C2A domain of otoferlin is incomplete (the β1 strand is missing as in dysferlin) and both C2A and C2B domains lack several of the five aforementioned aspartyl residues. The amino acid sequences of the C2A and C2B domains show 16% and 30% identity (28% and 46% similarity) with that of Syt-1 C2A, respectively. The C2C domain is a full C2 domain, with the five aspartyl residues predicted to bind Ca2+ being conserved. This domain shows 29% identity (40% similarity) with Syt-1 C2A. The subsequent three C2 domains are also complete domains predicted to bind Ca2+ (Yasunaga et al. 1999). Therefore, as in myoferlin and dysferlin, only the last four C2 domains of otoferlin are predicted to bind Ca2+. The C2 domain-containing proteins are known to interact with phospholipids and proteins. In addition, synaptotagmins, rabphilin 3A, munc 13, DOC2, and RIM, all of which possess C2 domains, are involved in the docking of the synaptic vesicles to the plasma membrane and/or their fusion (Rizo and Südhof 1998). Along the same line, even though the function of fer-1 in C. elegans is not yet entirely elucidated, this protein seems to be required for the final step of the fusion between vesicles and the plasma membrane (Achanzar and Ward 1997). On the basis of these data and the presence of the otoferlin transcript in the sensory hair cells of the inner ear (Yasunaga et al. 1999), we hypothesized that otoferlin is involved in the Ca2+-triggered synaptic vesicle-plasma membrane fusion. No other domain of homology with known proteins could be identified in the otoferlin deduced sequences. Finally, by use of the Prosearch 2.0 program, several putative phosphorylation sites for cAMP-dependent protein kinase (5–7), casein kinase II (42–43), tyrosine kinase (4) and protein kinase C (29–31), and 9–11 putative N-glycosylation sites were predicted in the murine and human sequences.
Figure 2.
Schematic representation of the human otoferlin (long and short forms), myoferlin, dysferlin, and nematode fer-1. The six C2 domains of otoferlin (long form) are designated C2A (aa 1–97), C2B (aa 254–352), C2C (aa 417–528), C2D (aa 960–1067), C2E (aa 1493–1592), and C2F (aa 1733–1863). Asterisks (*) indicate the C2 domains expected to bind Ca2+.
Exon-Intron Structure of the Human and Murine Otoferlin Genes
To determine the exon/intron boundaries and to estimate the size of the introns, long-range PCR amplifications were performed on genomic DNA using primers selected within the cDNA sequences, and the PCR products were directly sequenced. Direct sequencing on BAC (bacterial artificial chromosome) DNA was also performed (on human BAC clone 93m21 and murine BAC clone 233j15). From the comparative analysis of the genomic sequence and the sequence of the various cDNA isoforms that were identified (see below), we were able to conclude that the human OTOF and murine Otof are both composed of 48 coding exons. The first and last exons also encode the 5′-UTR and 3′-UTR, respectively. OTOF and Otof extend over 90 kb and 80 kb, respectively (fig. 3). The exon-intron junctions of the human gene are listed in table 2. All the splice sites follow the GT/AG rule (Shapiro and Senapathy 1987). In addition, specific primers flanking each OTOF exon from exon 1 to exon 19 were designed (table 1), henceforth allowing sequence analysis of the 48 exons of the gene (see also Yasunaga et al. 1999) for a mutation search in DFNB9-affected individuals (see below).
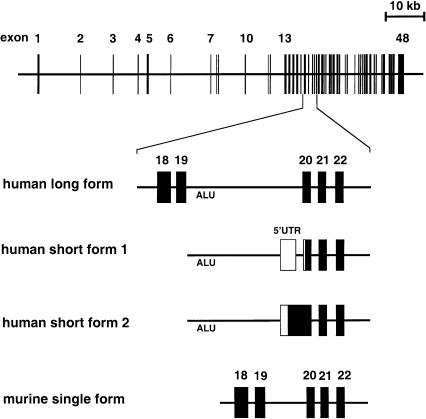
Figure 3.
Schematic representation of the OTOF exon-intron structure (top) and detail of the genomic region encompassing exons 18–22 in humans and in mice (bottom). The exonic sequences found in the three different human transcripts (long form and short forms 1 and 2) and in the single murine long transcript are boxed. The predicted coding sequences are indicated by blackened boxes, the untranslated regions by unblackened boxes.
Multiple Otoferlin Isoforms Predicted from Alternatively Spliced Transcripts
In the human brain, in addition to the otoferlin short cDNA (5 kb) initially reported in the fetus (Yasunaga et al. 1999) (GenBank AF107403) and referred to as human short form 1 in fig. 3, we isolated another, slightly longer cDNA (GenBank AF183186) which differs by its 5′ region (human short form 2 in fig. 3). In this form, the initiation codon identified within a weak Kozak consensus (TTCCTCatgATG) is preceded by an in-frame stop codon 57 bp upstream. The corresponding predicted protein of 1,307 aa (149 kDa), possesses 57 additional aa in the N-terminal region, which does not present any similarity with known proteins. From the analysis of the genomic sequence, we could conclude that this form results from use of both the 3′ part of exon 5′-UTR sf1 (for 5′ untranslated region of short form 1) and the following optional intron (i.e., used as an intron in short form 1), to encode the predicted 57 additional aa of the protein (fig. 3) (table 2). The absence of a detected short form (i.e., encoded by exons 20–48) of the otoferlin transcript in the mouse deserves further comment. No open reading frame was found in the murine genomic sequence corresponding to the human 5′-UTR sf1 additional exon and following intron, which is evidence against the existence of a murine equivalent of the human brain otoferlin short form 2 (see fig. 3). Moreover, even though the human putative translation initiation site of exon 20 is conserved in the mouse, no appropriate donor splice-site consensus sequence could be predicted within murine intron 19, strongly suggesting that a type 1 short form of otoferlin (fig. 3) does not exist either.
Interestingly, several alternatively spliced long forms of otoferlin transcripts were detected in both mice and humans. Firstly, in the murine brain and cochlea, cDNAs were found that resulted from the skipping of exon 6 (fig. 4A). Exon 6 encodes 15 aa located in the N-terminal part of the first inter-C2 domain region (aa 169–184 in the murine cochlear form [GenBank AF183183]). RT-PCR experiments on human brain RNA also revealed a single product that resulted from the skipping of exon 6 (data not shown). Secondly, RT-PCR analysis of the cDNA sequence corresponding to exon 31 in the murine brain and cochlea, revealed two different products resulting from the alternative use of an acceptor splice site within exon 31 (table 2) (fig. 4B). The same results were obtained in the human brain (GenBank AF183186). By contrast, in the human heart, kidney, and total fetus, only the shorter PCR product corresponding to the use of the internal splice site was detected (data not shown). The additional 20 aa encoded by the 5′ part of this exon belong to the fourth inter C2 domain region (aa position 1244–1264 in the murine brain sequence GenBank AF183184, and aa position 1245–1265 in the human brain sequence [GenBank AF183185] [fig. 1]). Finally, RT-PCR experiments performed on the murine brain RNA showed the use of exon 47 (encoding the most C-terminal 60 aa) in the otoferlin transcript (fig. 4C), whereas similar experiments performed on the cochlear RNA revealed the skipping of exon 47, and the use of the 5′ part of exon 48 to encode the C-terminal 60 residues (fig. 4C). The murine exon 47 and the 5′ part of exon 48 are expected to code for highly similar 60 aa peptides (fig. 5). The human and murine peptides encoded by exon 47 differ by only 5 aa and those encoded by exon 48 by 1 aa. As in the murine cochlea, exon 47 was found to be skipped in the otoferlin cDNAs isolated from the human adult heart, kidney, and total fetus (data not shown).
Figure 5.
Comparison between the deduced amino acid sequences encoded by exons 47 and 48 in humans and in mice. The predicted transmembrane domains are underlined. Identical residues are indicated by vertical bars and similar residues by colons.
Here, we present evidence for two classes of otoferlin isoforms: the long ones, with six C2 domains and a C-terminal TM domain, and the short ones, containing only the last three C2 domains and the TM domain. In murine tissues, only transcripts encoding long isoforms could be detected, whereas, in human tissues other than the brain, only short forms could be identified. At present, the basis of this difference is enigmatic. So far, no short forms of either dysferlin or myoferlin have been reported (Bashir et al. 1998; Liu et al. 1998; Davis et al. 2000). However, a 542-aa truncated form of myoferlin comprising the first three C2 domains and devoid of a TM domain has been predicted (Davis et al. 2000). In the brain, both in mice and humans, the long isoforms of otoferlin are predicted to lack part of the first inter C2 domain and to contain a TM domain encoded by exon 47. The murine transcripts may or may not contain a region of the fourth inter C2 domain encoded by the 5′ end of exon 31. In the murine cochlea, the two predicted long isoforms contain the part of the first inter C2 domain encoded by exon 6, a C-terminal end encoded by exon 48 and differ by the presence/absence of the region of the fourth inter C2 domain encoded by the 5′ end of exon 31. The presence of the several C2 domains indicates for multiple molecular interactions of otoferlin. In particular, the use of either exon 47 or exon 48 to encode the last 60 aa of this protein most likely has a functional significance, and may underlie membrane targeting of the protein to different cell compartments and/or interaction with different ligands. In this regard, it is noteworthy that although a single transmembrane domain sequence has so far been reported in myoferlin, the protein has been detected as associated with both the nuclear and the cytoplasmic membranes (Davis et al. 2000).
Novel OTOF Mutation Affecting the Long Isoforms in an Indian Family Affected by DFNB9
The nonsense mutation detected in the Lebanese consanguineous families (Yasunaga et al. 1999) is predicted to truncate the 500 C-terminal amino acids common to the short and long otoferlin putative isoforms. This does not permit a conclusion about the implication of one or the other types of otoferlin isoforms in the hearing loss.
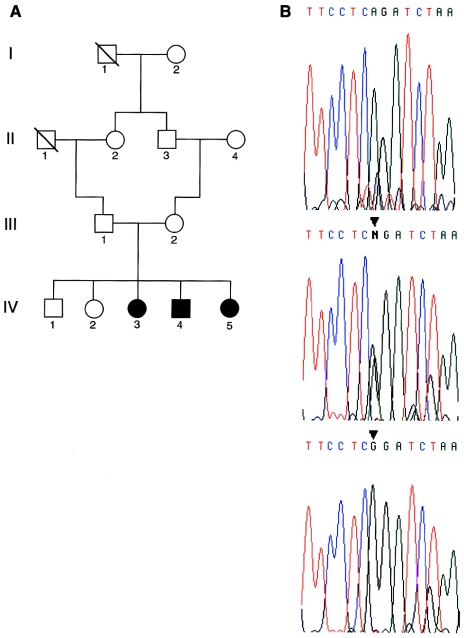
We analyzed a consanguineous family originating from India (family J2) in which three siblings suffered from severe-to-profound hearing loss (fig. 6A). Segregation analysis with polymorphic markers of the DFNB9 chromosomal region (see Subjects and Methods) allowed us to conclude that an OTOF mutation was likely to underlie deafness in this family (data not shown). The 48 OTOF coding exons were sequenced in the three affected individuals (IV-3, IV-4, IV-5), their unaffected sister (IV-2), and their parents (III-1, III-2). A homozygous A→G substitution at the intron 8/exon 9 junction (IVS8-2A→G) was detected in the three affected siblings. The same substitution was found at the heterozygous state in the parents and the unaffected siblings (fig. 6B). This mutation affects the invariant A of the AG acceptor splice site and was not detected in 109 DNAs (218 alleles) collected from random individuals in southwestern India. This establishes that IVS8-2A→G is the causative mutation for deafness in family J2. The mutation is expected to lead to aberrant splicing, such as the skipping of the 55-bp-long exon 9, creating a premature stop codon in exon 10. This result demonstrates that the expression of the long otoferlin isoforms is required for the auditory function in humans. We have previously shown that, in the inner ear, Otof is only expressed by the sensory hair cells and, in adults, predominantly by the inner hair cells. The next step will be to clarify the role of each of the predicted otoferlin isoforms—and, in particular, the long ones—with regards to the functioning of the hair cells.
Figure 6.
A, Pedigree of the nuclear family J2. B, Sequence analysis of the OTOF intron 8–exon 9 junction showing the A→G substitution (ISV8-2A→G) at the invariant AG dinucleotide of the acceptor splice-site (arrowhead). Top, middle, and bottom electrophoregrams correspond to a control individual, heterozygous carrier (III-1), and affected individual (IV-3), respectively.
Acknowledgments
We are grateful to Dominique Weil, for her kind and helpful advice throughout this study, and to Jean-Pierre Hardelin, for critical reading of the manuscript. This work was supported by grants from Association Française contre les Myopathies (AFM), Association Entendre (France), Université Saint Joseph (Lebanon), EEC (QLG2-CT-1999-00988), and the National Institute on Deafness and Communicative Disorders (intramural research projects Z01 DC 00035 [T.B.F.] and Z01 DC 00038 [E.R.W.]). M'hamed Grati was supported by Direction Générale de la Recherche Scientifique de Tunisie (Tunisia) and AFM.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (sequence comparison–analysis tool)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for poly(A)+ cDNA sequences in cochlea and brain [AF183183, AF183184], 3′-UTR sequences of the murine and human otoferlin genes, [AF183184, AF183185], newly discovered cDNA [AF183186], and predicted C2 domains [AF107403])
- FASTA, http://www.ebi.ac.uk/searches/fasta.html (sequence comparison–analysis tool)
- Nonsyndromic Hearing Impairment Autosomal Recessive Loci, http://hgins.uia.ac.be/dnalab/hhh/recessive.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim/ (for DFNB9 [MIM 601071])
- Pfam, http://www.sanger.ac.uk/Software/Pfam/, (protein-motif search tool)
- Pôle Bio-informatique Lyonnais server, http://pbil.ibcp.fr/NPSA/npsa_server.html (prediction tool for secondary structure of protein)
- Prosearch 2.0, http://bioweb.pasteur.fr/seqanal/interfaces/prosearch-simple.html (prediction tool for putative enzyme posttranslational modification sites)
- PSORT II, http://psort.nibb.ac.jp:8800/ (prediction tool for subcellular localization of proteins)
- SMART, http://smart.embl-heidelberg.de/ (search tool for protein motifs)
References
- Achanzar WE, Ward S (1997) A nematode gene required for sperm vesicle fusion. J Cell Sci 110:1073–1081 [DOI] [PubMed] [Google Scholar]
- Bashir R, Britton S, Strachan T, Keers S, Vafiadaki E, Lako M, Richard I, Marchand S, Bourg N, Argov Z, Sadeh M, Mahjneh I, Marconi G, Passos-Bueno MR, de S Moreira E, Zatz M, Beckmann J, Bushby K (1998) A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat Genet 20:37–42 [DOI] [PubMed] [Google Scholar]
- Cohen-Salmon M, El-Amraoui A, Leibovici M, Petit C (1997) Otogelin: a glycoprotein specific to the acellular membranes of the inner ear. Proc Natl Acad Sci USA 94:14450–14455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DB, Delmonte AJ, Ly CT, McNally EM (2000) Myoferlin, a candidate gene and potential modifier of muscular dystrophy. Hum Mol Genet 9:217–226 [DOI] [PubMed] [Google Scholar]
- Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, Mueller RF, Leigh IM (1997) Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 387:80–83 [DOI] [PubMed] [Google Scholar]
- Kozak M (1996) Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome 7:563–574 [DOI] [PubMed] [Google Scholar]
- Li XC, Everett LA, Lalwani AK, Desmukh D, Friedman TB, Green ED, Wilcox ER (1998) A mutation in PDS causes non-syndromic recessive deafness. Nat Genet 18:215–217 [DOI] [PubMed] [Google Scholar]
- Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, Serrano C, Urtizberea JA, Hentati F, Ben Hamida M, Bohlega S, Culper EJ, Amato AA, Bossie K, Oeltjen J, Bejaoui K, McKenna-Yasek D, Hosler BA, Schurr E, Arahata K, de Jong PJ, Brown RH Jr (1998) Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb-girdle muscular dystrophy. Nat Genet 20:31–36 [DOI] [PubMed] [Google Scholar]
- Liu X-Z, Walsh J, Mburu P, Kendrick-Jones J, Cope MJTV, Steel KP, Brown SDM (1997) Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet 16:188–190 [DOI] [PubMed] [Google Scholar]
- Mustapha M, Weil D, Chardenoux S, Elias S, El-Zir E, Beckmann JS, Loiselet J, Petit C (1999) An α-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet 8:409–412 [DOI] [PubMed] [Google Scholar]
- Petit C (1996) Genes responsible for human hereditary deafness: symphony of a thousand. Nat Genet 14:385–391 [DOI] [PubMed] [Google Scholar]
- Rizo J, Südhof TC (1998) C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem 273:15879–15882 [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Davletov BA, Berghuis AM, Südhof TC, Sprang SR (1995) Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell 80:929–938 [DOI] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Petit C (1999) Characterization of otoconin-95, the major protein of murine otoconia, provides new insights into the formation of these inner ear biominerals. Proc Natl Acad Sci USA 96:529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, Morton CC, Morell RJ, Noben-Trauth K, Camper SA, Friedman TB (1998) Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science 280:1447–1451 [DOI] [PubMed] [Google Scholar]
- Weil D, Küssel P, Blanchard S, Lévy G, Levi-Acobas F, Drira M, Ayadi H, Petit C (1997) The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet 16:191–193 [DOI] [PubMed] [Google Scholar]
- Yasunaga S, Grati M, Cohen-Salmon M, El-Amraoui A, Mustapha M, Salem N, El-Zir E, Loiselet J, Petit C (1999) A mutation in OTOF, encoding otoferlin, a FER-1 like protein, causes DFNB9, a nonsyndromic form of deafness. Nat Genet 21:363–369 [DOI] [PubMed] [Google Scholar]