Abstract
We determined the antibiotic susceptibilities of 1,785 enterococcal bloodstream isolates collected over 25 years. Antibiotic resistance emerged at a greater rate in Enterococcus faecium than in other enterococcal species, and E. faecium isolates became proportionally more common over time. Our findings confirm the pattern of emerging antibiotic resistance among enterococci and highlight the increasing importance of E. faecium as a cause of bloodstream infection.
Large surveillance programs have provided considerable data about the antimicrobial susceptibilities of enterococci over the past decade (4, 5). However, few studies have reviewed enterococcal isolates collected prior to 1990 by using current criteria for species identification and susceptibility testing. The principal objective of this study was to document the sequential emergence of antimicrobial resistance among enterococci isolated over the past 25 years, as important acquired resistance mechanisms have emerged. Therefore, we tested enterococcal bloodstream isolates collected from 1975 to 1999 by using current antimicrobial susceptibility testing methodology and interpretative criteria.
(This work was presented in part at the 39th Annual Meeting of the Infectious Diseases Society of America, San Francisco, Calif., 25 to 28 October 2001.)
Sequential isolates were collected at the University of Colorado Hospital, Denver, Colo., from 1975 to 1988 and at Duke University Medical Center, Durham, N.C., from 1989 to 1999. Only one isolate per patient for each bacteremic episode was included (i.e., there was >7 days between episodes unless different species were isolated), and all had been stored at −70°C. Isolates from before 1993 were tested retrospectively, while most other isolates were tested immediately after initial isolation. An additional 100 enterococcal bloodstream isolates collected at the University of Colorado Hospital during the year 2000 by using the same clinical criteria were also tested to determine the present antimicrobial susceptibilities and species profiles of enterococci at this site.
Enterococci were initially identified by their appearance when stained with Gram stain and by their colonial morphology, plus their ability to grow in 6.5% NaCl and to hydrolyze esculin in the presence of 40% bile salts and/or their ability to hydrolyze pyrrolidonyl-β-naphthylamide. Species-level identification was made with the MicroScan Pos Combo Panel type 10 or 13 (Dade Behring, Inc., West Sacramento, Calif.) for all isolates collected before 1993 and the API 20 Strep identification system (bioMérieux Vitek, Inc., Hazelwood, Mo.) for isolates collected since 1993. These methods were supplemented by conventional test procedures (1).
All susceptibility testing was performed according to the methods and interpretative criteria recommended by the National Committee for Clinical Laboratory Standards (8). For isolates collected before 1993, the broth microdilution method (MicroScan Pos Combo Panel type 10 or 13) was used to determine susceptibilities to ampicillin, vancomycin, gentamicin, and streptomycin. For the remaining isolates, the disk diffusion method was used to determine susceptibilities to ampicillin and vancomycin and an agar dilution method (Synergy Quad; Remel, Lenexa, Kans.) was used to detect high-level gentamicin and streptomycin resistance and vancomycin resistance. All isolates were tested for β-lactamase production by using nitrocefin disks (Cefinase; Becton Dickinson Diagnostic Systems, Sparks, Md.). Ampicillin-resistant isolates were further tested by the broth microdilution method to determine precise MICs. Custom-made frozen reference microdilution panels were used for this purpose (Dade Behring, Inc.).
A total of 1,785 enterococcal bloodstream isolates collected from 1975 to 1999 were tested. There were 1,311 Enterococcus faecalis, 410 E. faecium, 17 E. casseliflavus, 12 E. avium, 10 E. gallinarum, 6 E. durans, and 4 E. raffinosus isolates. The other 15 isolates could not be identified to the species level, could not be found, or could not be recovered for testing. Overall, E. faecium isolates became proportionally more common after 1979. The ratios of E. faecalis to E. faecium isolates were 3:1 from 1975 to 1979, 7:1 from 1980 to 1984, 7:1 from 1985 to 1989, 4:1 from 1990 to 1994, and 3:1 from 1995 to 1999. For 1999 alone, the ratio was 2.2:1.
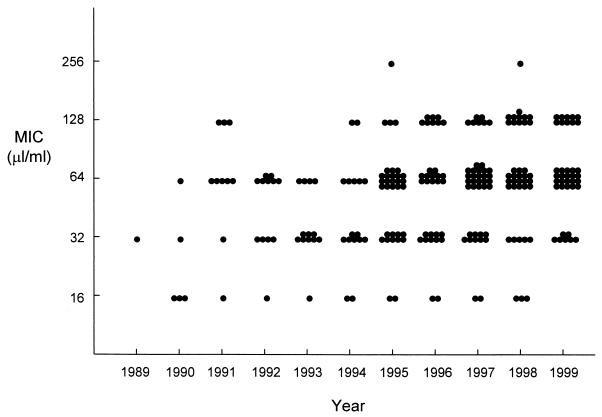
The antimicrobial susceptibilities of the isolates are shown in Table 1. Ampicillin resistance was first detected in E. faecium in 1989, with resistance rates steadily increasing to 80% of E. faecium isolates during 1999. Over this time period there was a gradual temporal rise in MICs for ampicillin-resistant E. faecium isolates, although the median MIC for these isolates remained at 64 μg/ml for most years (Fig. 1). β-Lactamase production was detected in only five E. faecalis isolates, all during an 11-month period (February 1990 to January 1991); at the time, these isolates were found to be part of the large “mid Atlantic” clone (7). The first E. faecalis isolate with ampicillin resistance not due to β-lactamase production was detected in 1997. Vancomycin resistance was first detected in E. faecium in 1992 and in E. faecalis in 1993. High-level gentamicin resistance first appeared in E. faecalis during 1985 and in E. faecium in 1989. Of the 64 enterococcal isolates of species other than E. faecalis and E. faecium, 4 were resistant to ampicillin (3 E. raffinosus isolates and 1 isolate not identified to the species level), 4 had high-level gentamicin resistance (none identified to the species level), and 11 had high-level streptomycin resistance (5 isolates not identified to the species level, 3 E. avium isolates, 2 E. gallinarum isolates, and 1 E. casseliflavus isolate). Reduced susceptibility to vancomycin was detected only in E. casseliflavus and E. gallinarum and was presumed to represent intrinsic vancomycin resistance.
TABLE 1.
Patterns of acquired resistance in enterococcal bloodstream isolates to ampicillin, vancomycin, gentamicin, and streptomycina
| Yr |
E. faecalis isolates
|
E. faecium isolates
|
All isolates
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | No. (%) resistant to:
|
No. | No. (%) resistant to:
|
No. | No. (%) resistant to:
|
||||||||||
| AMP | VAN | GEN | STR | AMP | VAN | GEN | STR | AMP | VAN | GEN | STR | ||||
| 1975-1979 | 57 | 0 | 0 | 0 | 16 (28) | 20 | 0 | 0 | 0 | 9 (45) | 83 | 0 | 0 | 0 | 28 (34) |
| 1980-1984 | 86 | 0 | 0 | 0 | 24 (28) | 12 | 0 | 0 | 0 | 1 (8) | 99 | 0 | 0 | 0 | 25 (25) |
| 1985-1989 | 83 | 0 | 0 | 9 (11) | 28 (34) | 12 | 1 (8) | 0 | 1 (8) | 1 (8) | 98 | 2 (2) | 0 | 9 (9) | 30 (31) |
| 1990-1994 | 448 | 5 (1) | 1 (0.2) | 101 (23) | 135 (30) | 112 | 63 (56) | 18 (16) | 37 (33) | 54 (48) | 594 | 70 (12) | 19 (3) | 141 (24) | 195 (33) |
| 1995-1999 | 637 | 4 (1) | 16 (3) | 161 (25) | 192 (30) | 254 | 189 (74) | 67 (26) | 72 (28) | 120 (47) | 911 | 194 (21) | 83 (9) | 234 (26) | 312 (34) |
| 2000b | 75 | 0 | 1 (1) | 13 (17) | 23 (31) | 22 | 14 (64) | 8 (36) | 7 (32) | 11 (50) | 100 | 14 (14) | 9 (9) | 20 (20) | 34 (34) |
AMP, ampicillin; VAN, vancomycin; GEN, high-level gentamicin; STR, high-level streptomycin.
Colorado isolates only.
FIG. 1.
MICs for ampicillin-resistant E. faecium isolates by year of isolation.
The first multidrug-resistant isolate (i.e., with significant resistance to two or more antibiotics) was an E. faecalis strain from 1985 that exhibited high-level resistance to both gentamicin and streptomycin. In 1999, 59 (35%) of 168 isolates were multidrug resistant. Of these, 33 were identified as E. faecium isolates, 30 were resistant to vancomycin, and 8 (all of which were E. faecium isolates) were resistant to all four antibiotics tested.
The 100 Colorado isolates collected in 2000 comprised 75 E. faecalis isolates, 22 E. faecium isolates, and 1 isolate each of E. casseliflavus, E. gallinarum, and E. durans. The antimicrobial susceptibility patterns of these isolates were similar to those of the most recent isolates from North Carolina (Table 1), indicating similar courses of emergence of antimicrobial resistance in the two centers.
Overall, the sequential appearance of antibiotic resistance in enterococci and the resistance rates determined by our study are similar to the findings from other United States studies during the same time period (4, 5,10, 11). In general, antibiotic resistance has emerged at a greater rate in E. faecium than in other enterococcal species. For the E. faecium isolates that were resistant to ampicillin, we documented a gradual increase in MICs over time, although this was not as marked in our study as in those reported by other investigators (3a).
As well as becoming more resistant, E. faecium isolates became proportionally more common during the study period. Data from other studies support this trend. In United States studies before 1990, the ratio of E. faecalis to E. faecium was 10:1 to 13:1 for both bloodstream isolates and isolates from other sites (3, 6, 11). Thereafter, the ratio fell to 5:1 in 1992 (4) and to 3:1 from 1997 to 1999 (5). The increasing predominance of E. faecium as a cause of bloodstream and other infections may partly reflect the high rate of antimicrobial resistance in this species and the fact that antibiotic pressure in a hospital environment allows for its selection. The emergence of E. faecium as a cause of bacteremia is of particular concern given that this species is associated with a higher mortality than E. faecalis is (9), independent of the presence or absence of vancomycin resistance (2). Moreover, the large numbers of strains that are resistant to both ampicillin and vancomycin leave few therapeutic options and none with documented synergy (10).
A major limitation of the present study is the fact that the isolates were collected sequentially, rather than concurrently, from the two different medical centers. However, the distribution of species and antimicrobial susceptibility patterns of the enterococcal bloodstream isolates from the University of Colorado Hospital during 2000 were similar to those of recent isolates from Duke University Medical Center, suggesting that resistance has emerged in a similar fashion in both centers.
The results of this study confirm the pattern of emerging antibiotic resistance among enterococci over recent years and highlight the increasing importance of E. faecium as a cause of bloodstream infection.
Acknowledgments
We thank Rachel M. Addison, Julie L. Aldroubi, Natalie M. Coombs, Jacqueline J. Thorpe, Laura K. Smith, and Tamara L. Underwood for technical assistance and Dade Behring for supplying the microtiter panels.
REFERENCES
- 1.Facklam, R. R., and M. D. Collins. 1989. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 27:731-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garbutt, J. M., M. Ventrapragada, B. Littenberg, and L. M. Mundy. 2000. Association between resistance to vancomycin and death in cases of Enterococcus faecium bacteremia. Clin. Infect. Dis. 30:466-472. [DOI] [PubMed] [Google Scholar]
- 3.Gordon, S., J. M. Swenson, B. C. Hill, N. E. Pigott, R. R. Facklam, R. C. Cooksey, C. Thornsberry, Enterococcal Study Group, W. R. Jarvis, and F. C. Tenover. 1992. Antimicrobial susceptibility patterns of common and unusual species of enterococci causing infections in the United States. J. Clin. Microbiol. 30:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Grayson, M. L., G. M. Eliopoulos, C. B. Wennersten, K. L. Ruoff, P. C. De Girolami, M.-J. Ferraro, and R. C. Moellering. 1991. Increasing resistance to β-lactam antibiotics among clinical isolates of Enterococcus faecium: a 22-year review at one institution. Antimicrob. Agents Chemother. 35:2180-2184. [DOI] [PMC free article] [PubMed]
- 4.Jones, R. N., H. S. Sader, M. E. Erwin, S. C. Anderson, and the Enterococcus Study Group. 1995. Emerging multiply resistant enterococci among clinical isolates. I. Prevalence data from 97 medical center surveillance study in the United States. Diagn. Microbiol. Infect. Dis. 21:85-93. [DOI] [PubMed] [Google Scholar]
- 5.Low, D. E., N. Keller, A. Barth, and R. N. Jones. 2001. Clinical prevalence, antimicrobial susceptibility, and geographic resistance patterns of enterococci: results from the SENTRY antimicrobial surveillance program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S133-S145. [DOI] [PubMed] [Google Scholar]
- 6.Maki, D. G., and W. A. Agger. 1988. Enterococcal bacteremia: clinical features, the risk of endocarditis, and management. Medicine 67:248-269. [PubMed] [Google Scholar]
- 7.Murray, B. E., K. V. Singh, S. M. Markowitz, H. A. Lopardo, J. E. Patterson, M. J. Zervos, E. Rubeglio, G. M. Eliopoulos, L. B. Rice, F. W. Goldstein, S. G. Jenkins, G. M. Caputo, R. Nasnas, L. S. Moore, E. S. Wong, and G. Weinstock. 1991. Evidence for clonal spread of a single strain of β-lactamase-producing Enterococcus (Streptococcus) faecalis to six hospitals in five states. J. Infect. Dis. 163:780-785. [DOI] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 9.Noskin, G. A., L. R. Peterson, and J. R. Warren. 1995. Enterococcus faecium and Enterococcus faecalis bacteremia: acquisition and outcome. Clin. Infect. Dis. 20:296-301. [DOI] [PubMed] [Google Scholar]
- 10.Sahm, D. F., M. K. Marsilio, and G. Piazza. 1999. Antimicrobial resistance in key bloodstream bacterial isolates: electronic surveillance with the Surveillance Network Database—USA. Clin. Infect. Dis. 29:259-263. [DOI] [PubMed] [Google Scholar]
- 11.Watanakunakorn, C. 1990. Enterococci from blood cultures during 1980-1989: susceptibility to ampicillin, penicillin and vancomycin. J. Antimicrob. Chemother. 26:602-604. [DOI] [PubMed] [Google Scholar]