Abstract
The t(11;22)(q23;q11) translocation is the only non-Robertsonian rearrangement for which there are a large number of unrelated families, apparently with the same breakpoints. These families most often have been ascertained through an abnormal child with the karyotype 47,XX or XY, +der(22) t(11;22)(q23;q11). To explain the high incidence of 3:1 segregants, rarely seen in offspring of carriers of other reciprocal translocations, a number of theoretical models have been suggested. We have used both electron microscope analysis of the synaptonemal complex (SC) and dual-color FISH to investigate the meiotic chromosome behavior in a male carrier of the translocation who has the karyotype 46,XY, t(11;22)(q23;q11). Chromosome synapsis, first-meiotic chiasma configuration, and segregation behavior of this translocation have been analyzed directly. Examination of SCs by electron microscopy showed pachytene-cross formation in 49/50 nuclei. Approximately 50% (26/50) revealed a classical fully synapsed quadrivalent. A proportion of these (10/26), however, showed some central asymmetry, suggesting heterologous synapsis. The remaining cells appeared to have incomplete synapsis. FISH analysis showed only quadrivalents in all 100 metaphase I nuclei. The chiasma frequency was increased within the interstitial segments, in comparison with the same region in normal bivalents. All types of segregation category were found in metaphase II nuclei. There was no indication of preferential 3:1 anaphase I segregation. We conclude that the +der(22) constitution in offspring of carriers of t(11;22)(q23;q11) is not likely to be due to meiotic 3:1 segregation being especially common. Rather, the +der(22) constitution is more likely to be the result of postzygotic selection against other unbalanced karyotypes.
Introduction
Most constitutional reciprocal translocations in humans are unique with respect both to chromosomes and to the breakpoints involved. The only outstanding exception concerns the translocation t(11;22)(q23; q11); >100 unrelated families that have this translocation have been identified (Fraccaro et al. 1980; Iselius et al. 1983). Ascertainment has usually been through children with the karyotype 47,XX or XY, +der(22) t(11;22)(q23;q11), interpreted to result from parental first-meiotic 3:1 segregation.
It has become generally accepted that carriers of this balanced translocation may show preferential 3:1 first-meiotic segregation, and a number of hypotheses have been put forward to explain why this should be the case. Zackai and Emanuel (1980) suggested that the asymmetry of the pachytene-cross configuration would result in discordant orientation of the quadrivalent at first metaphase, predisposing it to 3:1 anaphase segregation. Koduru and Chaganti (1989) proposed that there would be a failure of chiasma formation within the short 22q segment, producing a der(22) univalent segregating randomly at first-meiotic anaphase. To our knowledge, no direct meiotic analysis has been performed on any carrier of t(11;22) to confirm or refute these hypotheses.
Here, we report the results of electron microscopic analysis of synaptonemal complex (SC) formation and the use of dual-color FISH to investigate the meiotic chromosome behavior in a male carrier of a translocation who has the karyotype 46,XY, t(11;22)(q23;q11). Contrary to the accepted expectations, there was no preferential 3:1 first-meiotic segregation. Neither was there any evidence of failure of chiasma formation and univalence of the der(22). In fact, there is no substantial difference in meiotic segregation patterns in this carrier of t(11;22), in comparison with other carriers of translocations, in whom meiotic segregation analysis has been made possible by use of FISH on first-metaphase (MI) and second-metaphase (MII) spermatocytes (Goldman and Hultén 1992, 1993a, 1993b; Armstrong et al. 1995; Armstrong and Hultén 1998).
We conclude that the common occurrence of children with the extra der(22) in families with t(11;22) is likely to be due to postzygotic selection against other unbalanced-translocation derivatives, rather than to a preferential first-meiotic 3:1 segregation.
Subjects, Material, and Methods
Subjects
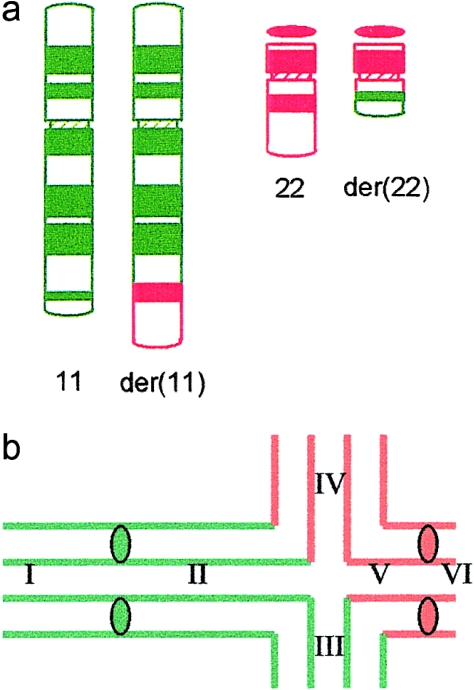
The subject (J.B.) was ascertained through his dysmorphic, handicapped brother with the unbalanced karyotype 47,XY, +der(22) t(11;22)(q23;q11) and was found to be a carrier with the karyotype 46,XY, t(11;22)(q23; q11) (fig. 1a). J.B. volunteered to provide testicular material for research purposes. The procedures were in agreement with the Ethics Committee of Birmingham Heartlands Hospital. Testicular material, of size ∼1 mm3, was obtained by open incision under local anesthesia when the patient was 29 years old. The tissue was collected in isotonic solution (1% sodium citrate) and was split into two samples, one for electron microscopic (EM) analysis and one for light microscopic (LM) work. The former sample was sent to Edinburgh and was used the following day.
Figure 1.
a, Ideogram showing the balanced translocation in J.B. b, Expected pairing configuration at pachytene. The chromosome segments have been designated I–VI, and they are defined as follows: I, 11p-11cen; II, 11 cen-11q23 (11q interstitial segment); III, 11q23-11qter (11q translocated segment); IV, 22qter-22q11 (22q translocated segment); V, 22q11-22cen (22q interstitial segment); and VI, 22cen-22pter.
EM
Spreads for EM analysis were prepared according to the method of Speed and Chandley (1990) and were used immediately. SCs were stained with silver nitrate (Howell and Black 1980) and were examined on G200HS copper grids (Gilder), in a Phillips CM10 electron microscope, at 100 KV.
LM
Meiotic spreads for LM analysis were prepared according to the method of Hultén et al. (1992) and were stored in air at −70°C for up to 18 mo before examination. Prior to hybridization, slides were stained with Leishman's stain by a standard technique. They were screened, and both MI and MII cells were photographed. The position of the metaphases was recorded with an England Finder Grid (Graticules). Slides were subsequently destained in 3:1 methanol:glacial acetic acid prior to FISH. Photographs were produced for each metaphase, as an aid to classification and analysis, after FISH. After FISH, a Nikon Labophot Fluorescence microscope was used with a UV2A filter set for location of DAPI-stained metaphases and with a triple-band bypass filter (DAPI, Texas Red, FITC), to visualize the chromosome paints. Photographs were taken using Fujichrome 400 ASA film.
FISH Analysis
Dual-color FISH was carried out with commercially produced chromosome paints (Cambio). The combinations used were FITC directly labeled chromosome 11 paint (green) and biotinylated chromosome 22 paint detected by the fluorochrome Texas Red. Hybridization, posthybridization, and signal detection were according to the manufacturer's instructions. Slides were counterstained with DAPI (10 μg/ml) and briefly were dehydrated through an ethanol series and were mounted in Vectorshield (Vectastain).
Results
SC Formation
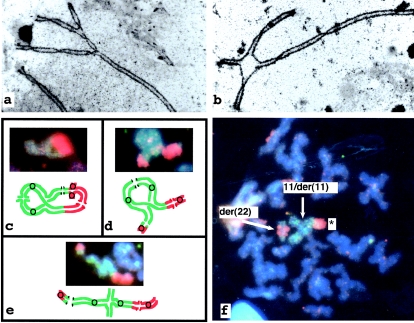
SC spreads of 50 nuclei were examined. Almost all (49/50) nuclei contained a quadrivalent. Approximately 50% (26/50) had a classical pachytene cross (fig. 1b). Within this group, (16/26) were fully synapsed. Some (10/26), however, showed central asymmetry, suggesting heterologous synapsis (fig. 2a). Incomplete synapsis of the quadrivalent was seen in 42% (21/50) of cells with an asynaptic region around the breakpoints (fig. 2b). In a few cells (4% [2/50]), an asynaptic arm was in contact with the XY bivalent. One nucleus (1/50) appeared to contain 23 SCs, without any indication of quadrivalent formation.
Figure 2.
Electron micrograph of a surface-spread and silver-stained pachytene nuclei. a, Quadrivalent fully synapsed but with some asymmetry in the region of the breakpoints. b, Incomplete synapsis, with an asynaptic region around the breakpoints. c–e, FISH and diagram representing quadrivalents of metaphase I nuclei. Chromosome 11 hybridized with directly labeled (with FITC) chromosome 11 paint, and chromosome 22 hybridized with biotinylated paint detected with Texas Red. Panels c–e show quadrivalents. c, Complete ring with five chiasmata. d, “Branched” ring with four chiasmata. e, Chain with three chiasmata. f, FISH of a metaphase II cell (probes and detection as for c–e): alternate/adjacent I segregant, containing asymmetric 11/der(11) (denoted by a star) and der(22).
FISH
Hybridization using dual-color fluorescence allowed unequivocal identification of all the chromosome segments involved in the reciprocal translocation. Chromosome 11 material was labeled green, with FITC, and chromosome 22 was labeled with Texas Red.
MI
A total of 106 MI cells were initially recorded from Leishman's stained slides, 100 of which were subsequently found, by FISH, to show adequate signals. Only quadrivalents involving chromosome 11 and chromosome 22 and the translocation chromosomes t(11;22) were seen (table 1). There was no evidence of any other arrangement (e.g., univalent + trivalent or bivalent + bivalent). Figure 1b illustrates the expected pairing configuration at pachytene and defines the pairing segments involved.
Table 1.
Distribution of Different Configurations for the Translocation Chromosomes at MI
| Configuration | Segments Containing Chiasmata | % of Quadrivalents |
| Ring | I–V | 62 |
| “Branched” ring | II–V | 2 |
| I–IV | 33 | |
| Chains | I–III | 3 |
For definition of chromosome segments I–V, see fig. 1b.
Nearly two-thirds of quadrivalents (64% [64/100]) formed rings with five chiasmata, as illustrated in figures 1b and 2c. Chiasmata then occurred in regions 11p (I), the 11q interstitial segment (II), the 11q23-qter segment (III), the 22q-qter segment (IV), and the 22q interstitial segment (V). Two further rings (2% [2/100]) were similar but lacked a chiasma in 11p (i.e., no chiasma in segment I of fig. 1b). None of the quadrivalents had a chiasma in 22p (region VI).
One-third of quadrivalents (33% [33/100]) formed “branched” rings with four chiasmata (fig. 2d), in regions I–IV and lack a chiasma in the interstitial segment of chromosome 22 segment V. Only a small proportion of quadrivalents (3% [3/100]) were in the form of chains with chiasma formation in regions 11p (I), 11q (III), and 22q (IV) (fig. 2e).
The chiasma frequency distribution for the translocation chromosomes, compared with data for men with a normal 46,XY karyotype that have been published elsewhere (Hultén 1974), is given in table 2. There is an increased chiasma frequency in the interstitial segments II and V in this translocation.
Table 2.
Chiasma Frequencies within the Translocated Chromosomes and Comparison with Control Bivalents[Note]
|
TranslocatedChromosomes |
Control Bivalents |
||||||
| Region | Segment | Mean | Range | No. ofCells | Mean | Range | No. ofCells |
| 11p | I | .98 | 0–1 | 100 | 1.0 | 1 | 24 |
| 11cen-11q23 | II | .98 | 0–1 | 100 | .21 | 0–1 | 24 |
| 11q23-11qter | III | 1.0 | 1 | 100 | 1.0 | 1 | 24 |
| 22q11-22qter | IV | 1.0 | 1 | 100 | 1.0 | 1 | 41 |
| 22cen-22q11 | V | .64 | 0–1 | 100 | .22 | 0–1 | 41 |
Source.— Hultén (1974).
MII
A total of 100 Leishman's stained MII cells were identified by LM and were photographed. After FISH, 87 of these cells were found to be suitable for the detailed analysis. The distribution of MII cells found within each segregation category is summarized in table 3.
Table 3.
Distribution of MII Cells Found within Each Category
|
Proportion (%) of |
|||
| Segregation Category | No. of Cells | Total | Segregation |
| Alternate/Adjacent I: | |||
| 11+22/der(22) | 2 | 2.30 | 3.17 |
| der(11)+22/der(22) | 1 | 1.15 | 1.59 |
| 11/der(11)+22 | 8 | 9.20 | 12.70 |
| 11/der(11)+der(22) | 10 | 11.49 | 15.87 |
| 11/der(11)+22/der(22) | 42 | 48.28 | 66.67 |
| Subtotal | 63 | 72.42 | 100.00 |
| Alternate: | |||
| 11+22 | 1 | 1.15 | 50.00 |
| der(11)+der(22) | 1 | 1.15 | 50.00 |
| Subtotal | 2 | 2.30 | 100.00 |
| Adjacent I: | |||
| 11+der(22) | 2 | 2.30 | 50.00 |
| der(11)+22 | 2 | 2.30 | 50.00 |
| Subtotal | 4 | 4.60 | 100.00 |
| Adjacent II: | |||
| 11+der(11) | 1 | 1.15 | 12.50 |
| 22+der(22) | 1 | 1.15 | 12.50 |
| 11/der(11)+11/der(11) | 3 | 3.45 | 37.50 |
| 22/der(22)+22/der(22) | 3 | 3.45 | 37.50 |
| Subtotal | 8 | 9.20 | 100.00 |
| 3:1 | 10 | 11.49 | 100.00 |
Chiasmata in interstitial segments produces asymmetric dyads, in which one sister chromatid is normal and one is translocated. In this situation, it is not possible to tell whether the underlying MI segregation is alternate or adjacent I because they are morphologically identical (see Armstrong and Hultén 1998). Altogether, 72% (63/87) of MII cells belonged to this category. Asymmetric dyads for both chromosomes [11/der(11) and 22/der(22)], found in 48% (42/87) of MII cells, are interpreted to be products of MI rings containing chiasmata in both interstitial segments. An additional 24% (21/87) of MII cells, containing one asymmetric and one symmetric dyad (fig. 2f), are likely products of the “branched” MI rings,” in which chiasma formation is lacking in one of the interstitial segments.
In only a minority of 2:2 segregants was it possible to determine whether they were the products of alternate or adjacent I segregation. Thus, only 2/87 (2.3%) of MII cells contained two normal or two derived chromosomes, representing alternate MI segregation of chains without chiasma formation in either interstitial segment. A further 4/87 (4.6%) of MII nuclei had one normal and one derivative chromosome, representing adjacent I segregation and likely to be products of chains without interstitial chiasmata. An additional 8/87 (9.2%) of MII cells were products of adjacent II segregation, represented by either a normal chromosome plus one (der) chromosome or two asymmetric dyads.
Finally, 3:1 segregation was seen in only 10 (11.5%) of 87 cells (table 4). Nine of these contained asymmetric dyads of chromosome 11, likely being MII products of MI rings (or “branched” rings) with an interstitial chiasma in chromosome 11. It is noteworthy that the common unbalanced karyotype 24,XorY, +der(22) could be produced by only 3.4% (3/87) of these MII cells and are likely to be the products of segregation from the “branched” ring or chain. The rare segregation product 24,XorY, t(11;22)(q24;q11) +der(22) can be produced only from a complete ring and was not seen in this sample (see the Discussion section below).
Table 4.
Different 3:1 Segregation Types Found and Proportion of +der(22) Expected
| Chromosomes | No.(% of All MII) | Proportion of All Gametes That Could Lead to +der(22) |
| 11/der(11) | 3 (3.4) | .0 |
| 22 | 1 (1.1) | .0 |
| 11/der(11), 22,der(22) | 3 (3.4) | 3.4 |
| 11/der(11),11/der(11),der(22) | 2 (2.3) | .0 |
| 11/der(11),11/der(11),22/der(22) | 1 (1.1) | .0 |
| 10 (11.5) | 3.4 |
Gametic Output
The proportions of gametes expected to be normal, balanced, and unbalanced within each segregation type have been used to predict the overall gametic output (table 5). Under the assumption that all types of MII cells are equally likely to develop into mature spermatozoa, the predicted gametic output is 19% normal, 19% balanced, and 62% unbalanced. For the unbalanced gametes, we predict that 40.8% will be products of the alternate/adjacent I category, 9.2% will be products of adjacent II segregation, and 11.5% will be products of 3:1 segregation.
Table 5.
Proportion of Normal, Balanced, and Unbalanced Gametes Expected To Be Produced by Each Segregation Category[Note]
|
Gametes Expected from (%) |
|||||||
| Theoretical Segregation Products |
Expected Proportions |
||||||
| Segregation category | Found at MII ( n = 87)(%) | Normal | Balanced | Unbalanced | Normal | Balanced | Unbalanced |
| Alternate/Adjacent I: | |||||||
| 11+22/der(22) | 2.30 | 50 | 0 | 50 | 1.15 | 0 | 1.15 |
| der(11)+22/der(22) | 1.15 | 0 | 50 | 50 | 0 | .58 | .58 |
| 11/der(11)+22 | 9.20 | 50 | 0 | 50 | 4.60 | 0 | 4.60 |
| 11/der(11)+der(22) | 11.49 | 0 | 50 | 50 | 0 | 5.75 | 5.75 |
| 11/der(11)22/der(22) | 48.28 | 25 | 25 | 50 | 12.07 | 12.07 | 24.14 |
| Alternate: | |||||||
| 11+22 | 1.15 | 100 | 0 | 0 | 1.15 | 0 | 0 |
| der(11)+der(22) | 1.15 | 0 | 100 | 0 | 0 | 1.15 | 0 |
| Adjacent I: | |||||||
| 11+der(22) | 2.30 | 0 | 0 | 100 | 0 | 0 | 2.30 |
| der(11)+22 | 2.30 | 0 | 0 | 100 | 0 | 0 | 2.30 |
| Adjacent II: | |||||||
| 11+der(11) | 1.15 | 0 | 0 | 100 | 0 | 0 | 1.15 |
| 22+der(22) | 1.15 | 0 | 0 | 100 | 0 | 0 | 1.15 |
| 11/der(11)+11/der(11) | 3.45 | 0 | 0 | 100 | 0 | 0 | 3.45 |
| 22/der(22)+22/der(22) | 3.45 | 0 | 0 | 100 | 0 | 0 | 3.45 |
| 3:1 | 11.49 | 0 | 0 | 100 | 0 | 0 | 11.49 |
| Total | 100.00 | 18.97 | 19.55 | 61.51 | |||
Note.— Cols. (3–5) show theoretical outcomes of gametes from each segregation category. Cols. (6–8) use information from cols. (2–5) to extrapolate proportion of sperm expected.
Discussion
The translocation t(11;22)(q23; q11) is unusual among autosomal reciprocal chromosome rearrangements because it is recurrent in the population, with the same cytological breakpoints in carriers in many biologically unrelated families (Fraccaro et al. 1980; Iselius et al. 1983). Furthermore, Shaikh et al. (1999) have demonstrated that, in 23 unrelated carriers of balanced translocations, the breakpoint was localized within a 400-kb interval on chromosome 22, and that, in 13 of these, the breakpoint was localized within a region of <185 kb on chromosome 11. The breakpoint has now been cloned (B. Emanuel, personal communication).
The translocation is also unusual because the majority of index cases have the same chromosome abnormality, an extra chromosome, der(22). The extra chromosome has long been presumed to be the result of a 3:1 parental meiotic segregation. This interpretation has recently been confirmed by the study by Shaikh et al. (1999), who used short-tandem-repeat polymorphism markers on both chromosome 11 and chromosome 22. From a comparison of markers of parents and offspring, they concluded that 3:1 meiosis I malsegregation in the parent carrying a balanced t(11;22) translocation was the mechanism in all 16 families that they investigated.
The outstanding question is whether this is due to preferential segregation at MI. Our meiotic study of a male carrier (J.B.) of the t(11;22) translocation by FISH clearly demonstrates that 3:1 segregation is not preferential. Rather, direct analysis of meiotic divisions revealed that the predominant mode of segregation is 2:2, with only 11.5% of spermatocytes demonstrating 3:1 segregation. We suggest that our results from this particular case may be extrapolated and that regular 2:2 segregation is the rule in carriers of the t(11;22) translocation.
It has been suggested that the 3:1 segregant progeny are the result of low chiasma numbers leading to univalents plus trivalents at MI (Koduru and Chaganti 1989). In contrast, in our meiotic analysis of a male carrier (J.B.) with the translocation t(11;22)(q23; q11), we found that there is regular quadrivalent formation during pachytene, which is retained through MI. This is likely to be a general phenomenon rather than a peculiarity of our case (see further below).
Under the assumption of no selection during spermatogenesis, extrapolation of all of the chromosome combinations observed leads to the expectation that the spermatozoa in J.B. would be 19% normal, 19% balanced, and 62% unbalanced, of which 50% are due to 2:2 segregation comprising 40.8% from the alternate/adjacent I categories and 9.2% from the adjacent II category. These values are not inconsistent with the observation by Martin (1984), which found, in a small study of 13 sperm nuclei, that only 1 was normal, 2 were balanced, and the remainder (77%) were unbalanced. Of these, only two had a 3:1 meiotic segregation. More recently, Van Assche et al. (1999) studied 1,012 sperm of a carrier of t(11;22), by FISH. They report nearly 34.6% 3:1 segregants. The frequency of 34.6% was derived from 314 nuclei being apparently monosomic and only 36 containing an extra chromosome. On the basis of our own experience (Goldman et al. 1993), it may be that the vast majority of the monosomic sperm nuclei resulted from failed hybridizations. The 36 sperm nuclei with an extra chromosome could represent a total of 7.2% of 3:1 segregation products in their carrier of t(11;22). This latter value is entirely consistent with our direct meiotic analysis at 11.5%.
The finding that translocation carrier J.B. regularly forms quadrivalents during meiosis is consistent with the behavior normally seen for reciprocal translocations in human males. It has been known for several decades that quadrivalents are regularly formed and that univalents plus trivalents or bivalents plus bivalents are hardly ever seen. Thus, even small translocated chromosome segments are able to sustain adequate homologous synapsis and chiasma formation (Hultén and Lindsten 1970, 1973; Chandley et al. 1976; Laurie et al. 1984; Goldman and Hultén 1993a, 1993b; Armstrong et al. 1995). Furthermore, it has been clearly demonstrated that there is a general tendency for an increase in chiasma formation within the interstitial segment of reciprocal translocations. This phenomenon is also obvious in J.B., the carrier of the t(11;22) translocation. The interstitial segment of chromosome 22 is small in this translocation, representing <1% of the autosomal genome, yet there is still an increase of crossovers in the region. In our case, only 32% of SCs, by EM analysis, revealed complete synapsis around the center of the pachytene cross. It is possible that crossovers were suppressed in the quadrivalents within heterologously synapsed or asynaptic regions.
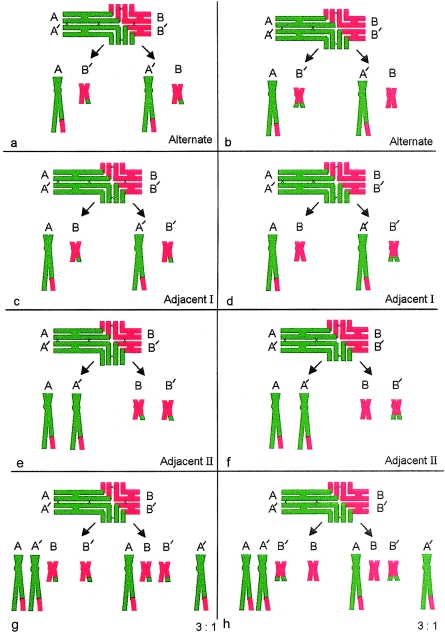
The 47,XX or XY,+der(22) karyotype is by far the most common abnormal outcome found in liveborn offspring of any carriers of t(11;22). The few exceptions to this general rule have attracted special attention—Lockwood et al. (1989), Abeliovitch and Carmi (1990), Lurie and Podleschuk (1992), Simi et al. (1992), and Petovic et al. (1996) have each reported a 47,t(11;22)(q23;q11), +der(22) karyotype in abnormal offspring. The first four groups postulated that this could only be the result of a second meiosis or postzygotic nondisjunction following a 2:2 alternate segregation. Lindenbaum (1990) pointed out that a crossover within the chromosome 22 interstitial segment, followed by 3:1 segregation, would have the same outcome. Petkovic et al. (1996), using evidence of polymorphism for inheritance of the paternal chromosome 22 in their case, supported Lindenbaum's suggestion. Our study clearly supports this view. The rare outcome 47,t(11;22)(q23;q11), +der(22) can be produced only by a 3:1 segregation from a complete ring, a configuration that has interstitial chiasmata in both chromosome 11 and chromosome 22. Our data show why this is a rare outcome. Although complete rings make up 64% of our sample—first, to obtain this karyotype, the anaphase I segregants need to be A, B, and B′ (fig. 3g) and will include both the translocation chromosome t(11;22) and the der(22). Second, there is only a 1/12 likelihood, at anaphase II, for the outcome of 24, t(11;22) +der(22) under the assumption of random cosegregation. In contrast, there is a greater probability for occurrence of the chromosome constitution 24, +der(22) from the “branched” ring (35%; fig. 3h) and chain configurations (3%), neither of which can produce a 24,t(11;22) +der(22) product. It is important to note that neither of these MI quadrivalents can give rise to the translocation +der(22) product.
Figure 3.
a, c, and e, Outcome of first-meiotic segregation from the complete ring configuration with five chiasmata (black crosses), two of which are interstitial in both chromosome 11 (green) and chromosome 22 (red) 2:2 segregation. Note that interstitial chiasmata render alternate (a) and adjacent I (b) identical outcomes. g, 3:1 segregation—only segregants A, B, and B′ have potential to produce either 24,t(11;22) +der(22) or 24,+der(22). b, d, f, and h, Outcome of first-meiotic segregation from the “branched” ring configuration with four chiasmata (no interstital chiasma in chromosome 22). In panel h, only segregants A, B, and B′ have the potential to produce 24,+der(22) but not 24,t(11;22)+der(22) segregants.
Fetal karyotyping in pregnancies of 45 parental carriers of the t(11;22)(q23;q11) revealed 73.3% to be balanced, 20% to be normal, and only 6.6% to be unbalanced with the extra der(22) chromosome (Daniel et al. 1989). For 3:1 segregants, the analysis that we have carried out predicts that 3.4% of fetuses would have the extra der(22) (table 4), which is comparable with the results of the fetal karyotyping study (Daniel et al. 1989). Considering the relatively late time at which amniocentesis is carried out (∼16-wk gestation), we suggest that fetuses with unbalanced 2:2 translocation would have terminated spontaneously. There is a single case of an unbalanced 2:2 segregation product for this translocation, detected after chronic villus sampling. Soler et al. (1993) reported an empty sac with the karyotype 46,XX,−22,+der(22), in a triplet pregnancy; of the two remaining fetuses, one had the karyotype 46,XX and the other had 46,XY,t(11;22). This demonstrates that 3:1 products containing der(22) are not the only gametes to be fertilized and progress to embryonic development and implantation. It seems reasonable to conclude that, during early pregnancy, there is a strong selection against other unbalanced segregants, compared with the +der(22), representing the smallest genetic balance. Finally, it should be mentioned that Fraccaro et al. (1980), Daniel et al. (1989), and Shaikh et al. (1999) showed that more of the carriers with progeny who had unbalanced translocations in their studies were female (76 female carriers, compared with 9 male carriers). It seems likely that this could be due to ascertainment bias, because of reduced fertility in males. However, the proportion of progeny in the Daniel et al. (1989) study who had unbalanced 3:1 translocations was 6.6%, and this is comparable with the proportion of relevant unbalanced gametes (3.4%) in our study. Meiotic behavior in both male and female carriers is likely to share common characteristics.
In conclusion, we believe that a direct analysis of meiosis in a carrier of the balanced translocation t(11;22) has provided detailed information on patterns of chromosome synapsis, chiasma formation, and segregation, which is entirely in line with what we expect from our previous experience with reciprocal carriers of other translocations. In particular, we demonstrate that the +der(22) constitution in offspring is not due to meiotic 3:1 segregation being especially common; rather, the +der(22) constitution is likely to be the result of postzygotic selection against other unbalanced karyotypes.
Acknowledgment
We would like to express our gratitude to the family for their help with this project.
References
- Abeliovich D, Carmi R (1990) The translocation 11q;22q: a novel unbalanced karyotype. Am J Med Genet 37:288 [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Goldman ASH, Hultén MA (1995) Segregation analysis in human reciprocal translocation carriers using FISH. Chromosome Res 3 Suppl 1:109 [Google Scholar]
- Armstrong SJ, Hultén MA (1998) Meiotic segregation analysis by FISH investigations in sperm and spermatocytes of translocation heterozygotes. Eur J Hum Genet 6:430–431 [DOI] [PubMed] [Google Scholar]
- Chandley AC, Seuanez H, Fletcher JM (1976) Meiotic behaviour of five human reciprocal translocations. Cytogenet Cell Genet 17:98–111 [DOI] [PubMed] [Google Scholar]
- Daniel A, Hook EB, Wulf G (1989) Risks of unbalanced progeny at amniocentesis to carriers of chromosome rearrangements: data from United States and Canadian laboratories. Am J Med Genet 31:14–53 [DOI] [PubMed] [Google Scholar]
- Fraccaro M, Lindsten J, Ford CE, Iselius L (1980) The 11q;22q translocation: a European collaborative analysis of 43 cases. Hum Genet 56:21–51 [DOI] [PubMed] [Google Scholar]
- Goldman ASH, Formina Z, Knights PA, Hill CJ, Walker AP, Hultén MA (1993) Analysis of the primary sex ratio, sex chromosome aneuploidy and diploidy in human sperm using dual colour fluorescence in situ hybridisation. Eur J Hum Genet 1:325–334 [DOI] [PubMed] [Google Scholar]
- Goldman ASH, Hultén MA (1992) Chromosome in situ suppression hybridisation in human male meiosis. J Med Genet 29:101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1993a) Analysis of chiasma frequency and first meiotic segregation in a human male reciprocal translocation heterozygote, t(1;11)(p36.3;q13.1), using fluorescence in situ hybridisation. Cytogenet Cell Genet 63:16–23 [DOI] [PubMed] [Google Scholar]
- ——— (1993b) Meiotic analysis by FISH of a human male 46,XY, t(15:20)(q11.2;q11.2) translocation heterozygote: quadrivalent configuration, orientation, and first meiotic segregation. Chromosoma 102:102–111 [DOI] [PubMed] [Google Scholar]
- Howell MW, Black DA (1980) Controlled silver staining of nucleolus organiser regions with a protective colloidal developer: a one step method. Experientia 36:1014–1015 [DOI] [PubMed] [Google Scholar]
- Hultén M (1974) Chiasma distribution at diakinesis in the normal human male. Hereditas 76:55–78 [DOI] [PubMed] [Google Scholar]
- Hultén MA, Goldman ASH, Sadallah N, Wallace BNM, Creasy MR. (1992) Meiotic investigations. In: Rooney DE, Czepulkowski BH (eds) Human cytogenetics: a practical approach. IRL Press, Oxford, Washington DC, pp 193–221 [Google Scholar]
- Hultén M, Lindsten J (1970) The behaviour of structural aberrations at male meiosis: information from man. In: Human population cytogenetics. Pfizer medical monographs 5. Edinburgh University Press, Edinburgh, pp 24–61 [Google Scholar]
- ——— (1973) Cytogenetic aspects of human male meiosis. In: Harris H , Hirschhorn K (eds) Advances in human genetics. Vol l4 . Plenum Press, New York, pp 327–387 [DOI] [PubMed] [Google Scholar]
- Iselius L, Lindsten J, Aurias A, Fraccaro M, Bastard C, Bottelli AM, Bui et al (1983) The 11q;22q translocation: a collaborative study of 20 new cases and analysis of 110 families. Hum Genet 64:343–355 [DOI] [PubMed] [Google Scholar]
- Koduru PRK, Chaganti RSK (1989) Meiotic segregation in human t(11;22)(q23;q11) carriers: a theoretical consideration. Genome 32:24–29 [DOI] [PubMed] [Google Scholar]
- Laurie DA, Palmer RW, Hultén MA (1984) Studies on chiasma frequency and distribution in two fertile men carrying reciprocal translocations: one with a t(9;10) karyotype and one with a t(Y;10) karyotype. Hum Genet 68:235–247 [DOI] [PubMed] [Google Scholar]
- Lindenbaum RH (1990) Unusual segregation of the constitutional 11q;22q translocation may be explained by crossover in interchange segment followed by 3:1 segregation at meiosis I. Hum Genet 85:43 [DOI] [PubMed] [Google Scholar]
- Lockwood DH, Farrier A, Hecht F, Allanson JE (1989) Not all chromosome imbalance resulting from the 11q;22q translocation is due to 3:1 segregation in first meiosis. Hum Genet 83:287–288 [DOI] [PubMed] [Google Scholar]
- Lurie IW, Podleschuk LV (1992) 11q;22q translocation: third case of imbalance not due to 3:1 non-disjunction in first meiosis. Am J Med Genet 42:216 [DOI] [PubMed] [Google Scholar]
- Martin RH (1984) Analysis of human sperm complements from a male heterozygous for a reciprocal translocation t(11;22)(q23;q11). Clin Genet 25:357–361 [DOI] [PubMed] [Google Scholar]
- Petkovic I, de Capoa A, Giancotti P, Barisic I (1996) Unusual segregation of t(11;22) resulting from crossing-over followed by 3:1 disjunction at meiosis I. Clin Genet 50:515–519 [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Budarf ML, Celle L, Zackai EH, Emanuel BS (1999) Clustered 11q23 and 22q11 breakpoints and 3:1 meiotic malsegregation in multiple unrelated t(11;22) families. Am J Hum Genet 65:1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simi P, Ceccarelli M, Barachini A, Floridia G, Zuffardi O (1992) The unbalanced offspring of the male carriers of the 11q;22q translocation: nondisjunction at meiosis II in a balanced spermatocyte. Hum Genet 88:482–483 [DOI] [PubMed] [Google Scholar]
- Soler A, Carrio A, Perez-Vidal MT, Borrell A, Fortuny A (1993) Unusual segregation for 11q:22q parental translocation in a triplet pregnancy: prenatal diagnosis in chorionic villi and amniotic fluid. Prenat Diagn 13:137–141 [DOI] [PubMed] [Google Scholar]
- Speed RM, Chandley AC (1990) Prophase of meiosis in human spermatocytes analysed by EM microspreading in infertile men and their controls and comparisons with human oocytes. Hum Genet 84:547–554 [DOI] [PubMed] [Google Scholar]
- Van Assche E, Staessen C, Vegetti W, Bonduelle M, Vandervorst M, Van Steirteghem A, Liebaers I (1999) Preimplantation genetic diagnosis and sperm analysis by fluorescence in-situ hybridisation for the most common reciprocal translocation t(11;22). Mol Hum Reprod 5:682–690 [DOI] [PubMed] [Google Scholar]
- Zackai EH, Emanuel BS (1980) Site specific reciprocal translocation, t(11;22)(q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am J Med Genet 7:507–521 [DOI] [PubMed] [Google Scholar]