Abstract
We have developed a strategy for the isolation of terminal deletion breakpoints from any chromosome that has been healed by de novo addition of a telomere repeat array. Breakpoints at 7q32 and 22q13.3 have been isolated and characterized in two patients (patients FB336R and AJ). Both truncated chromosomes have been healed by the addition of a novel telomere, with such an addition possibly mediated by the enzyme telomerase. The breakpoint at 7q32 in patient FB336R shows a structure similar to that of breakpoints on other chromosomes that have been healed in this way. However, the breakpoint at 22q13.3 in patient AJ has 10 nucleotides of unknown origin inserted between the sequence unique to chromosome 22q and the start of the telomere repeat array. This unusual structure is suggestive of a multistep healing event resulting in de novo telomere addition at this breakpoint, and possible mechanisms are discussed.
Introduction
Human telomeres, which are essential for chromosome stability, function as a protective “cap” at the chromosome terminus, preventing fusion and degradation and helping cells to distinguish intact chromosome ends from damaged DNA (Muller 1938; McClintock 1941). Telomeres consist of tandem-repeat arrays of TTAGGG (Moyzis et al. 1988), although variant repeats—such as TTGGGG, TGAGGG, and TCAGGG—are common and tend to be clustered at the proximal ends of telomeres (Allshire et al. 1989; Baird et al. 1995, 2000; Coleman et al. 1999). Telomere repeat arrays are extended and maintained in the germline by the enzyme telomerase, which synthesizes the G-rich strand of the array in a 5′→3′ direction. Telomerase uses an integral telomere-complementary RNA template to generate repeats (Greider and Blackburn 1987, 1989; Morin 1989; Shippen-Lentz and Blackburn 1990). Telomerase can add repeats, de novo, to nontelomeric DNA during developmentally programmed chromosome healing in ciliates (Zakian et al. 1990; Yu and Blackburn 1991). This process does not take place in mammalian cells; however, telomerase is thought to heal and stabilize broken chromosomes sporadically in the human germline. This potentially gives rise to chromosomes carrying terminal deletions and a new telomere, but it allows the truncated chromosome to replicate and segregate normally. It is not known how frequently breakage and healing events resulting in terminal deletions occur, but such events are thought to contribute to a number of different disease syndromes, including some cases of idiopathic mental retardation (Flint et al. 1995). Small or cryptic deletions of some chromosome ends may not give a distinct clinical phenotype and may often be an unidentified cause of human genetic disease. De novo telomere addition has been identified in vivo by characterization of six terminal deletions at the tip of chromosome 16p (resulting in α-thalassemia) (Flint et al. 1994) and one 130-kb terminal microdeletion (causing mild mental retardation and speech difficulties) at 22q13.3 (Wong et al. 1997). In all these cases, the truncated chromosomes have been healed by the direct addition of a telomere onto nontelomeric sequence. Subterminal sequences were not found in the vicinity of these breakpoints, and only TTAGGG repeats were detected at the proximal end of the array, suggesting that the telomere addition was mediated by telomerase. In five of the six breakpoints at chromosome 16p, the 3–4 nucleotides at the telomere junction were complementary to and in phase with the RNA template of human telomerase. By use of in vitro assay, human telomerase was shown to recognize the sequence at the breakpoint of one of these deletions and to add TTAGGG repeats, generating the same junction sequence (Morin 1991). This suggests that 3′ termini with only minimal complementarity to the RNA template of telomerase are sufficient for recognition and repeat addition (Morin 1991).
Alternative mechanisms for the healing of broken chromosomes in the germline also exist. Characterization, at chromosome 16p, of a subtelomeric rearrangement giving rise to α-thalassemia suggested that the broken chromosome was stabilized by capture of an existing telomere via illegitimate recombination between closely related Alu elements (Flint et al. 1996). In this case, subterminal sequences were transferred to the breakpoint with the telomere.
Cloning and sequencing of human telomeres have enabled the characterization of adjacent subterminal sequences (Cross et al. 1989; Brown et al. 1990; de Lange et al. 1990; Cheng et al. 1991; Royle et al. 1992; Flint et al. 1997a, 1997b). These regions are usually composed of several members of low-copy repetitive sequence families that are often shared by a number of chromosome ends. The chromosome distribution and copy number of these repeats can be polymorphic between unrelated individuals. A simple telomere-anchored PCR method has been used to isolate telomere-junction fragments containing a short stretch of nontelomeric subterminal DNA adjacent to repeat sequences from the proximal end of a telomere (Royle et al. 1992). Understanding the processes that lead to terminal deletions is limited by the low number of terminal deletion breakpoints isolated and their narrow distribution within the genome, and so we aimed to adapt the telomere-anchored PCR method for the isolation of breakpoints healed by the de novo addition of a telomere, irrespective of their chromosomal location. The combination of size fractionation, telomere-anchored PCR, hybridization selection, and directional cloning has resulted in the isolation of two terminal deletion breakpoints in patients FB336R (Gurrieri et al. 1993) and AJ (Nesslinger et al. 1994). Patient FB336R has a cytogenetically visible deletion of 7q32-qter and mild holoprosencephaly, including mental retardation and mild facial dysmorphia. Patient AJ has a visible maternal deletion of 22q13.3 and showed general developmental delay and mild facial dysmorphia (Nesslinger et al. 1994). Both breakpoints have been stabilized by the addition of a novel telomere to nonterminal sequence. The presence of an additional 10 nucleotides at the breakpoint in patient AJ suggests that stabilization of the chromosome break with a new telomere was a multistep process.
Material and Methods
Genomic DNAs
The lymphoblastoid cell line of patient AJ and genomic DNAs of patient AJ's parents (individuals KJ and FJ) were a gift from Heather McDermid (University of Alberta, Edmonton, Canada). The fibroblast cell line (GM07412) of patient FB336R was obtained from M. Muenke (Department of Pediatrics, The Children's Hospital of Philadelphia, University of Pennsylvania School of Medicine, Philadelphia). Tissue-culture manipulation and DNA extraction were performed using standard techniques. All studies conducted had received prior approval from the Leicestershire Health Authority’s Committee on the Ethics of Clinical Research Investigation.
DNA Amplification and PCR Primers
PCR was performed in the buffer described elsewhere (Jeffreys et al. 1991), in the presence of 1 μM each primer and 0.1 U Taq polymerase (Advanced Biotechnologies) per μl, by use of a PTC200 DNA Engine (MJ Research) or a Cetus 9600 thermal cycler (PE Biosystems). Sequences for SauL-A, TelC have been reported elsewhere (Royle et al. 1992). Primer sequences for D22S274, D22S282, and D22S1169 are available from the Genome Database. The following primers and annealing temperatures were used: Tel1, 5′-TTAGGGTTAGGGTTAGGG-3′ (60°C); Tel2, 5′-CTAACCCTAACCCTAACC-3′ (60°C); TelG, 5′-CCCTCACCCTCACCCTCACCCTC-3 (60°C); TelGcomp, 5′-AGGGTGAGGGTGAGGGTG-3′ (60°C); TelJ, 5′-ACCCCAACCCCAACCCCAACCCC-3′ (60°C); TelJcomp, 5′-GGGTTGGGGTTGGGGTTG-3′ (60°C); TelK, 5′-CCCTGACCCTGACCCTGACCCTG-3′ (60°C); TelKcomp, 5′-AGGGTCAGGGTCAGGGTC-3′ (60°C); 6a8fa, 5′-GATCACTCATTGGCTCTCTCC-3′ (65°C); 6a8fb, 5′-TACCTGACATTATAGCTTCC-3′ (65°C); AJ111b1, 5′-GCACTTTAAATGGGCGACAGAGG-3′ (60°C); AJ111c, 5′-GGAATGAGTTAATACCAGAGTG-3′ (65°C); AJ111d, 5′-CTTCATGGATGTGGACAACTAG-3′ (65°C); AJ111e, 5′-CTGCCCTGGAGCACCCTGCAT-3′ (65°C); AJ111f, 5′-CACAGTGAGACCCTGTTTC-3′ (62°C); and AJ111g, 5′-CCCTAACCCTACATTCCTAAG-3′ (56°C).
Southern Blot Analysis and Preparation of Probes
Preparation of Southern blots and hybridization were performed, as described elsewhere (Wong et al. 1990), in either nonfat dried-milk solution or phosphate-SDS-EDTA. Hybridization probes were generated by means of agarose-gel purification of PCR products, and 5–10 ng of probe DNA were labeled with α[32P]-dCTP, by use of the random-primed-labeling reaction (Feinberg and Vogelstein 1983, 1984).
Telomere Repeat Probes
Double-stranded telomere repeat probe (TTAGGG)n was generated in a PCR reaction with complementary primer pair Tel1 and Tel2 (Royle et al. 1992), without target DNA. The PCR conditions were 25 cycles at 96°C for 1 min, 60°C for 1 min, and 70°C for 5 min. Product sizes ranged from 20 bp to ∼12 kb. Fragments between 300 bp and 10 kb in size were purified by electroelution from an agarose gel. Double-stranded telomere variant-repeat (TVR) probes TelG (TGAGGG), TelJ (TTGGGG), and TelK (TCAGGG) were generated in a similar manner with complementary primer pairs TelG/TelGcomp, TelJ/TelJcomp, and TelK/TelKcomp.
Construction and Screening of Telomere-Anchored PCR Libraries
Telomere-anchored PCR libraries from individual patients were constructed essentially as described by Royle et al. (1992). In brief, MboI-digested genomic DNA fragments >5 kb were ligated to Sau3AI linker amplimers and were amplified in a telomere-anchored PCR reaction with linker amplimer SauL-A and telomere primer TelC, which anneals to the G-rich strand of the telomere repeat array. Digested PCR products were cloned into the EcoRI and KpnI sites of the pBluescriptII SK+ vector, picked into 96-well plates, and replicated onto nylon filters. Filters were screened by colony hybridization (Buluwela et al. 1989) with 32P-labeled (TTAGGG)n probe, to identify clones positive for telomere repeat arrays, and with probes that identify known subterminal-repeat sequence families. Clones in the telomere-anchored PCR library containing variant-repeat sequences were identified, in patient AJ, by hybridization to (TCAGGG)n, (TGAGGG)n, and (TTGGGG)n probes.
Hybridization Selection to Construct Telomere-Anchored PCR Libraries
Genomic DNA from each patient was prepared for telomere-anchored PCR as described in the Construction and Screening of Telomere-Anchored PCR Libraries section. Approximately 50 ng prepared DNA were PCR amplified, for 10 cycles, with primers SauL-A and TelC and were recovered using a QIAquick PCR purification spin column (Qiagen). Nylon filters (3 mm × 3 mm) bearing either mixed synthetic variant-repeat sequences (TCAGGG), (TGAGGG), and (TTGGGG), or synthetic (TTAGGG) telomere repeat sequences only, were generated for filter hybridization as described elsewhere (Armour et al. 1994). Anchored-PCR products were alkali denatured, were added to phosphate-SDS-EDTA hybridization buffer containing the variant-repeat filter, and were incubated overnight at 65°C to remove telomere repeat arrays containing variant-repeat types. The variant filter was removed, and the buffer containing unhybridized DNA was heat denatured and was incubated overnight with the TTAGGG filter at 65°C, to select for TTAGGG repeat arrays. After hybridization, the TTAGGG filter was thoroughly washed with 0.5 × SSC/0.1% SDS, and the bound PCR products were recovered from the filter in 10 mM KOH/0.01% SDS and then were neutralized in 0.5 M Tris-HCl (pH 7.5)/0.01% SDS at room temperature. DNA was purified using QIAamp spin columns (Qiagen). This fraction, which was enriched for amplicons containing arrays of (TTAGGG)n repeats only, was reamplified with linker primer SauL-A and telomere primer TelC in a telomere-anchored PCR reaction and was cloned.
Sequencing Telomere-Junction Clones
DNA sequencing from single-stranded plasmid templates was performed manually by use of the dideoxy-chain-termination method (Sanger et al. 1977) with a protocol devised by Stratagene (Sequenase protocol). Sequencing of PCR-amplified double-stranded DNA was done using a model 377 DNA sequencing system (PE Biosystems) and the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Biosystems).
Computer-Aided Sequence Analysis
Computer-aided sequence analysis was performed using Wisconsin Package, version 10, sequence analysis software (Genetics Computer Group). Searches of the EMBL Nucleotide Sequence Database and GenBank were conducted using the FASTA and BLAST programs. Analysis of automated sequencing data was done using Factura, release 1.2.0 (PE Biosystems), and Autoassembler, release 1.4.0 (PE Biosystems), on Apple Macintosh computers.
Microsatellite Marker Typing
Primers were labeled by phosphorylation of the 5′ termini with γ[32P]-ATP by T4 polynucleotide kinase (Sambrook et al. 1989). PCR reactions were performed in 10 × buffer (Advanced Biotechnologies) with 200 μM each dNTP, 1–1.5 mM MgCl2, and 0.2 U Taq polymerase. The following cycling conditions were used: 26 cycles at 96°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by 72°C for 5 min. Products were resolved by electrophoresis on a 6% denaturing polyacrylamide gel and were detected by autoradiography.
FISH
The cosmid clones from the breakpoint region in patient FB336R were labeled with biotin-16-dUTP in a random-primed-labeling reaction and were recovered by ethanol precipitation. After hybridization to metaphase spreads of normal human lymphocytes from whole-blood cultures, the bound probe was detected using fluorescein isothiocyanate (FITC)–avidin DN, and further amplification of the signal was achieved with biotinylated anti–avidin D and FITC-avidin DN (Vector Laboratories) (Royle et al. 1994). Verification that the cosmid probes hybridized to chromosome 7 was achieved using the primed in situ labeling (PRINS) method (Koch et al. 1989) with oligonucleotides specific to chromosome 7 α-satellite.
Results
Isolation of Candidate Terminal Deletion Breakpoints in Patients FB336R and AJ
The sequence at individual terminal deletion breakpoints is unknown, but breakpoints that have been healed by the addition of a novel telomere are likely to share three features: (1) they will lack subterminal sequences adjacent to the new telomere, (2) the length of the new telomere will be similar to that of the other telomeres, and (3) the new telomere will not contain variant telomere repeat sequences that are commonly found in normal telomeres (Allshire et al. 1989; Baird et al. 1995; Coleman et al. 1999; Baird et al. 2000; H. Varley and N. J. Royle, unpublished observations). The telomere-anchored PCR strategy has been adapted to facilitate the isolation of terminal deletion breakpoints from any chromosome. Initially, enrichment for all terminal restriction fragments in a patient is achieved by means of size selection of MboI-digested genomic DNA fragments >5 kb. This step enriches (50–100-fold [N. J. Royle, unpublished observation]) for tandemly repeated DNA sequences that are devoid of MboI sites. The majority of internal telomere-like repeat arrays are <1 kb (Flint et al. 1997a) and therefore are excluded during the size-fractionation step. Telomere-anchored PCR amplifies all telomeres from the size-fractionated DNA but excludes other tandem repeats. This enrichment of telomere-junction fragments is facilitated further in a directional cloning step using restriction sites within the PCR primers. Subsequent screening of the ordered array library allows for normal chromosome ends to be identified by the presence of known subterminal sequences adjacent to the telomere repeat array or by the presence of variant repeats (such as TCAGGG, TGAGGG, and TTGGGG) within the arrays. The breakpoint in patient FB336R was isolated from a library of telomere-junction clones, by use of this approach. The library contained 376 clones, 128 (34%) of which contained telomere repeat arrays; 39 of these 128 clones were excluded by screening with subterminal repeat probes. The remaining clones were sequenced; clones FB336R-6a8f and FB336R-5a3h were selected for further investigation because they had the expected structure of a terminal deletion breakpoint healed by novel telomere addition. These clones had identical structure that differed by one base pair, and the consensus sequence, FB336R-6a8f (EMBL Nucleotide Sequence Database accession number AJ277168), was used for further study (see the Characterization of the Breakpoint in Patient FB336R section, below).
There was considerable variation in the proportion of TTAGGG-positive clones in the ordered array libraries from other patients. This necessitated the inclusion of hybridization selection steps (fig. 1) based on a method developed to isolate human STR loci (Armour et al. 1994). After telomere-anchored PCR was performed, telomere-junction fragments containing variant telomere repeats were removed from the pool of amplified products by hybridization to a filter containing TGAGGG, TCAGGG, or TTGGGG repeats. This step partially removes “normal” telomeres from the amplified products. The remaining amplicons containing TTAGGG repeats were then selected by hybridization to a filter of TTAGGG repeats only. This step enriches for telomere-junction fragments, including breakpoint fragments. Rescue of the amplicons bound to the TTAGGG filter was achieved by denaturation, PCR amplification, and cloning into a plasmid. This strategy was used to generate an ordered array library from patient AJ. The library contained a total of 752 clones, 383 (51%) of which were positive for telomere repeats. Screening with subterminal repeat probes and variant-repeat probes TGAGGG, TCAGGG, and TTGGGG excluded 238 of these telomere-positive clones. Of the remaining clones, 51 were sequenced. Clone AJ-111B was selected for further investigation, because it comprised 170 bp of nontelomere DNA extending from an MboI restriction site adjacent to an array of 26 TTAGGG repeats (EMBL Nucleotide Sequence Database accession number AJ277167) resulting in an insert size of 326 bp. Two additional independently isolated clones were sequenced; they contained identical flanking DNA but only 24 TTAGGG repeats. Screening the library with the flanking sequence identified nine additional clones containing this sequence, with insert sizes ranging from 230–600 bp. The basis of this size variation is likely to be the result of differences in the telomere repeat-array length, although this was not investigated further.
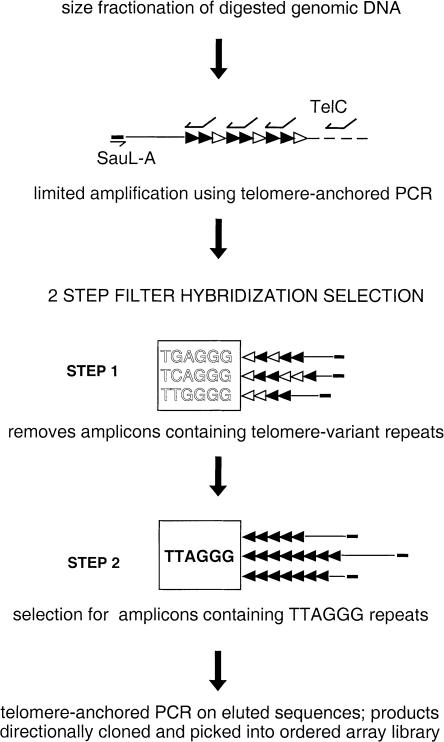
Figure 1.
Diagram representing hybridization selection enrichment for amplicons containing telomere-junction fragments with homogenous arrays of TTAGGG repeats only. Size fractionation for MboI-digested genomic DNA >5 kb gives a 50–100-fold enrichment for long MboI fragments, including telomeres. After amplification of these products by telomere-anchored PCR with linker primer SauL-A and telomere primer TelC, a two-step hybridization selection is done. In step 1, telomere repeat arrays containing variant-repeat types TGAGGG, TCAGGG, and TTGGGG are partially removed by hybridization to the variant-repeat filter. In step 2, the TTAGGG filter is used to select amplicons containing (TTAGGG)n repeats, and the selected amplicons are reamplified by telomere-anchored PCR, directionally cloned, and picked into an ordered array library. The combined effect of each step is to enrich for homogenous arrays of TTAGGG repeats. Blackened arrowheads denote TTAGGG repeats; unblackened arrowheads, variant repeats TGAGGG, TCAGGG, and TTGGGG.
Characterization of the Breakpoint in Patient FB336R
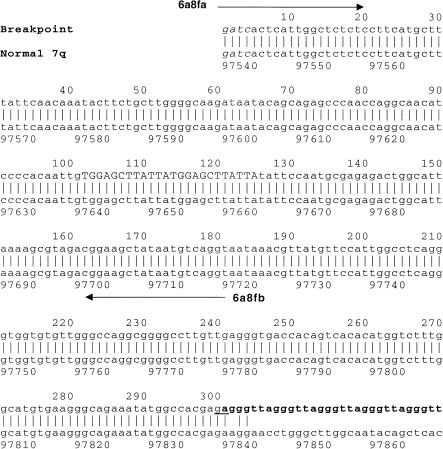
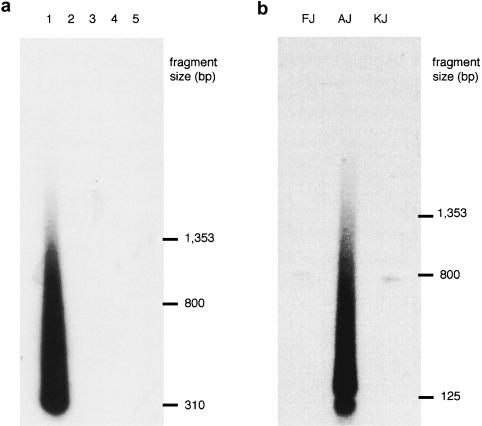
Clone FB336R-6a8f (fig. 2) contained 304 bp of DNA extending from an MboI site, followed by six TTAGGG repeats. The telomere-adjacent DNA did not show homology either to previously identified subterminal sequences or to other genomic sequences available in the EMBL Nucleotide Sequence Database and GenBank at the time of isolation. Primers 6a8fa and 6a8fb (fig. 2) were designed to amplify a 182-bp product from the telomere-adjacent region. PCR amplification of this sequence on the National Institute of General Medical Sciences human/rodent somatic-cell hybrid mapping panel (Drwinga et al. 1993) indicated that this region was unique to chromosome 7. DNA from patient FB336R's family members was not available for testing. To determine whether this sequence was found adjacent to an array of telomere repeats in other individuals, PCR amplification with primer 6a8fa and telomere primer TelC was performed on DNA from 79 unrelated individuals, mainly comprising parents from the CEPH panel. Products were detected by Southern hybridization with the use of probe 6a8f, generated by PCR with primers 6a8fa and 6a8fb. A smear of hybridizing products characteristic of amplification from a telomere was detected only in patient FB336R (fig. 3a)—that is, in only 1/158 chromosomes tested—confirming that this is a novel location for a telomere. TVR mapping by PCR (Baird et al. 1995) can be used to map the interspersion patterns of TTAGGG repeats with variant-repeat types TGAGGG and TCAGGG at the proximal end of a telomere. TVR-PCR mapping at the breakpoint telomere in patient FB336R indicated that this telomere contains only TTAGGG repeat types for ⩾50 repeat units into the array. The absence of variant repeats was further verified by the appearance of a homogenous smear of TTAGGG hybridizing sequences up to 1.5 kb into the telomere (fig. 3a).
Figure 2.
Alignment of the breakpoint sequence FB336R-6a8f (EMBL Nucleotide Sequence Database accession number AJ277168) with sequence from normal chromosome 7 (EMBL Nucleotide Sequence Database accession number AC018643). The MboI site at the start of clone 6a8f is indicated by italic type. A 12-bp direct repeat at base positions 102–125 is indicated by uppercase letters. The telomere repeat array at the breakpoint starts at position 301 and is indicated by boldface type. The two possible nucleotides to which the telomere was added are underlined. Black arrows denote primers 6a8fa and 6a8fb.
Figure 3.
PCR amplification of the novel telomeres in patients FB336R and AJ. a, PCR amplification was performed, with primers 6a8fa and TelC, on genomic DNA from the following individuals: patient FB336R (1) and CEPH parental DNAs 02-01 (2), 02-02 (3), 12-01 (4), and 12-02 (5). The products were detected by means of Southern hybridization with the 6a8f probe. A smear of hybridizing products characteristic of amplification from a telomere repeat array was detected only in patient FB336R. b, PCR amplification was performed, with primers AJ111b1 and TelC, on genomic DNA from patient AJ and parents KJ and FJ, and products were detected by Southern hybridization with the AJ111B probe. A smear of hybridizing products was detected in patient AJ only. The faint, discrete products of ∼800 bp that were detected in both parents are due to amplification of related Alu elements by primer AJ111b1.
FISH Mapping
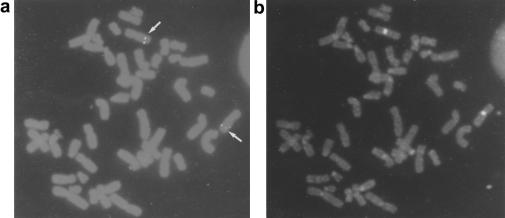
Eight cosmid clones containing the chromosome 7–unique breakpoint region (detected by probe 6a8f) seen in patient FB336R were isolated from human genomic DNA cosmid library HL1095m (Clontech). Figure 4 shows FISH of one cosmid, cDIZ, to normal human chromosomes. Both copies of chromosome 7 show a discrete hybridization signal at an interstitial site at 7q32.
Figure 4.
FISH of the cosmid cD1Z to normal human chromosomes. a, The cD1Z cosmid hybridizes to a unique interstitial site at 7q32 (white arrows). b, Verification that the hybridizing chromosome was chromosome 7 was obtained by use of the PRINS technique with primers specific for chromosome 7 α-satellite sequences.
Location, on the Chromosome 7 Physical Map, of the Breakpoint in Patient FB366R
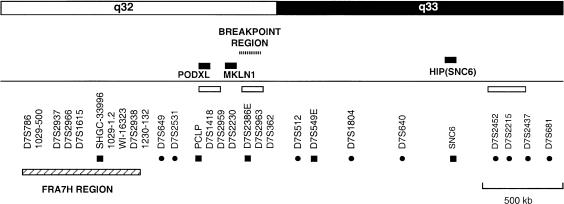
The breakpoint region was mapped to 7q32-33 on the chromosome 7 physical map, by hybridization to a YAC clone contig of this region (fig. 5). The common fragile site FRA7H lies within this region and shows unusual chromatin organization, suggesting undercondensation of this region (Mishmar et al. 1998). These sequence characteristics may extend through the region around the fragile site and predispose toward chromosome breakage. For example, FRA11B—a rare inherited fragile site at 11q23 (Voullaire et al. 1987)—has been linked with ∼10% of cases of Jacobsen syndrome (Michaelis et al. 1998; Tunnacliffe et al. 1999), which is normally associated with loss of 11q23 to 11qter, severe mental retardation, and dysmorphic features (Jacobsen 1973; Schinzel et al. 1977). However, the breakpoint in patient FB336R maps ⩾500 kb distal to FRA7H, and it seems unlikely that this chromosome break is directly linked to the fragile site. The breakpoint in patient FB336R was localized between genetic markers D7S2531 and D7S512 and was closest to DNA markers D7S2386E and D7S2963 (fig. 5).
Figure 5.
Location, on the chromosome 7q physical map, of the breakpoint region in patient FB336R. The breakpoint region in patient FB336R, detected by probe 6a8f, is indicated by a dotted bar. The breakpoint lies ⩾500 kb distal to fragile-site region FRA7H, which is indicated by a hatched bar. Unblackened bars denote groups of markers for which the local order has not been determined; blackened circles, genetic markers; blackened squares or bars, ESTs or genes. Other markers shown are STSs or unique DNA probes.
Sequencing the Breakpoint Region from a Normal Copy of Chromosome 7
The breakpoint was localized to a 5.7-kb EcoRI fragment of cosmid cDIZ. This fragment was subcloned into the EcoRI site of the pBluescriptII SK+ vector (Stratagene) and was sequenced by primer “walking” along the subcloned fragment (EMBL Nucleotide Sequence Database accession number AJ277294). A working-draft sequence of this region of chromosome 7 is now available (EMBL Nucleotide Sequence Database accession number AC018643) and shows 99.5% sequence identity to the sequence obtained from cosmid cDIZ. When the sequence of the clone containing the breakpoint was aligned with the normal sequence of this region, the breakpoint was localized to two possible nucleotides to which the novel telomere may have been added (fig. 2). No TTAGGG repeats were found in the 400 bp around the breakpoint in patient FB336R. A pentanucleotide (G)5 was identified within 150 bp distal to the breakpoint. The NIX (Nucleotide Identify X) search tool from the United Kingdom Human Genome Mapping Project Resource Centre (Hinxton, Cambridge) was used to screen for putative coding sequences in the 5.7-kb sequence around the breakpoint. No significant sequence identity to nucleotide or protein/peptide sequences present in the EMBL Nucleotide Sequence Database and GenBank was found, and no expressed-sequence tags (ESTs), sequence-tagged sites (STSs), or CpG islands were identified. This region contained a number of repeats, including short interspersed nuclear elements (Alu's and mammalian-wide interspersed repeats [MIRs]), a long interspersed nuclear element (L1), and a medium reiterated repeat (MER4), which were detected by the RepeatMasker program (available from the University of Washington's Repeat Masker Web server). In total, interspersed repeats comprised 35% (2,058 bp) of the 5.7-kb sequence.
Characterization of the Breakpoint in Patient AJ
The sequence adjacent to 26 TTAGGG repeats in clone AJ-111B showed 96.2% sequence identity to a region of bacterial-artificial-chromosome (BAC) clone bk268H5 (EMBL Nucleotide Sequence Database/GenBank accession no. AL008718), as shown in figure 6. The sequence identity terminates 10 bp proximal to the beginning of the telomere repeat array. The first 62 bases of AJ-111B show very high sequence identity (61/62 bp) to the Alu-Sx subfamily repeat consensus sequence (Jurka and Milosavljevic 1991). The differences between AJ-111B and bk268H5 in the poly(A) tail of this Alu (bases 47–62) may be due to simple polymorphism. However, sequence from the normal copies of chromosome 22 in the family trio including patient AJ and parents KJ and FJ shows 100% identity to the bk268H5 sequence (data not shown).
Figure 6.
Alignment of clone AJ-111B with a region of BAC clone bk268H5. Sequence from clone AJ-111B shows 96.2% identity, over the proximal 160 bp, to a region of BAC clone bk268H5 (EMBL Nucleotide Sequence Database accession number AL008718). Matches between the two sequences are represented by vertical lines. Gaps (dashes [–]) have been inserted in the AJ-111B sequence to maximize the alignment. The telomere repeat array in clone AJ-111B starts at position 167 and is shown in boldface type, and the additional 10 nucleotides that represent the distal limit of homology between clone AJ-111B and bk268H5 are shown in boldface type and are underlined. The region of homology to the Alu-Sx element is underlined. Black arrows denote primers AJ111b1 and AJ111c. The telomere repeat array (TTAGGG)n is shown in boldface type, and the Sau3AI site at the start of the clone is shown in italicized type.
BAC clone bk268H5 maps to chromosome 22q. To investigate whether the sequence at the breakpoint is unique to chromosome 22, PCR amplification was performed on the National Institute of General Medical Sciences human/rodent somatic-cell hybrid mapping panel, with use of primers AJ111b1 and AJ111c (shown in fig. 6). An amplicon of the expected size was detected from chromosome 22, and sequence data obtained from this amplicon confirmed that the telomere-adjacent sequence in clone AJ-111B was derived from genomic DNA. Amplification with primers AJ111c (fig. 6) and AJ111d (located 845 bp proximal to AJ111c) confirmed that the putative breakpoint region is unique to chromosome 22.
To determine whether the putative breakpoint region is found adjacent to a telomere repeat array in other individuals, PCR amplification was done, using primers AJ111b1 and TelC, on genomic DNA from 87 unrelated individuals, a sample mainly comprising parental DNAs from the CEPH panel. PCR products were detected by Southern hybridization using a PCR probe generated from clone AJ-111B with primers AJ-111b1 and AJ111c. As for the breakpoint in patient FB336R, a smear of hybridizing products characteristic of amplification into a telomere repeat array was detected only in patient AJ—that is, in only 1/174 chromosomes investigated. More significant is the finding that neither of the parents (individuals KJ and FJ) had a telomere repeat array at this location (fig. 3b), indicating that this novel telomere repeat array was not inherited from one parent.
To determine whether the novel telomere repeat array in patient AJ contains variant-repeat types, TVR mapping (but with the lower resolution of agarose-gel fractionation) was used to detect TGAGGG, TCAGGG, and TTGGGG variant-repeat types. No variant-repeat types were detected in the proximal 2–3 kb (data not shown); it is therefore likely that the novel telomere repeat array in patient AJ consists entirely of TTAGGG repeats. In addition, to verify that the novel telomere repeat array in patient AJ has a terminal location, sensitivity to Bal31 nuclease was investigated. The novel telomere in patient AJ was sensitive to Bal31 digestion under the conditions used; however, the D16309 locus, which is estimated to be 1.3 Mb from the chromosome 16p telomere (May et al. 1996), was not (data not shown).
Location, on the Chromosome 7q Physical Map, of the Breakpoint in Patient AJ
The 22q13.3 breakpoint in patient AJ was originally determined, by means of RFLP/loss-of-heterozygosity (LOH) studies (Nesslinger et al. 1994), to lie between the D22S95 and D22S94 loci. Furthermore, the results of densitometry studies of loci that were monomorphic in patient AJ indicated that the breakpoint was located in the interval D22S95–D22S64, which is ∼350 kb (fig. 7). BAC clone bk268H5, which shows sequence identity with the AJ-111B breakpoint clone, has been placed in the interval D22S40–D22S94 in band 22q13.3 (Dunham et al. 1999). This localization of the breakpoint in patient AJ is consistent with the larger interval defined by LOH but is >2 Mb distal to the interval defined by densitometry studies. To resolve this discrepancy, we have examined the inheritance of alleles at three highly polymorphic STRs—D22S274, D22S2282, and D22S1169. The family trio including patient AJ and parents KJ and FJ was fully informative at all three loci; patient AJ has maternal and paternal alleles at D22S274 and D22S282, but he has only a paternal allele at the D22S1169 locus. This indicates that the breakpoint is located in the interval D22S274–D22S94, consistent with the location of BAC clone bk268H5 on the genetic-framework, physical, and sequence maps, and results in deletion of the terminal 5.5 Mb at chromosome 22q.
Figure 7.
Location, on the chromosome 22 map, of the breakpoint in patient AJ. Genetic markers are shown in the correct order, but distances are not to scale. The positions of the markers on the sequence map are according to the start position of the clone in which they are found. The breakpoint in patient AJ was originally mapped, by LOH and densitometric analysis, to the interval D22S95–D22S94. “2” denotes two copies of a marker; “1,” one copy of a marker; and “c,” loci that were typed by RFLP/LOH analysis. The breakpoint sequence (AJ-111b) in patient AJ shows strong homology to a region of BAC clone bk268H5 (vertical bar on sequence map) which is within the interval originally defined by LOH but which is >2 Mb distal to the interval defined by densitometry studies. Analysis of three highly polymorphic STR loci showed that patient AJ had two alleles at the D22S282 and D22S274 loci but only one allele at the D22S1169 locus, consistent with the location of the breakpoint in patient AJ that was identified on the sequence map of chromosome 22. Some of the information shown in this figure was obtained from the Sanger Centre Chromosome 22 Database.
Characterization of Sequence Across the Breakpoint Region from a Normal Copy of Chromosome 22
The sequence identity between the breakpoint in patient AJ and BAC clone bk268H5 terminates 10 bp before the beginning of the telomere repeat array. Results of sequencing across the breakpoint region on normal copies of chromosome 22 from patient AJ and parents KJ and FJ showed that the additional 10 nucleotides were absent from these chromosomes. The presence of the additional 10 nucleotides on the truncated chromosome 22 in patient AJ was verified by PCR amplification of genomic DNA from patient AJ, with primer AJ111d, which anneals to a unique sequence 823 bases proximal to the chromosome 22q breakpoint, and primer AJ111g, which spans the breakpoint. Primer AJ111g terminates at the 3′ end, with sequence complementary to the additional 10 nucleotides. A discrete PCR product of the expected size was generated from the genomic DNA of patient AJ but not from that of her parents (data not shown).
Normal sequence (6 kb) of chromosome 22 in the breakpoint region was obtained from BAC clone bk268H5 (bases 4,102–10,102) and screened for interesting or unusual features. No TTAGGG repeats were found in 400 bp around the breakpoint in patient AJ. A pentanucleotide (G)5, like pentanucleotides detected close to breakpoints on chromosomes 16p and 22q and at the breakpoint on chromosome 7q in patient FB336R, was identified 159 bp distal to the breakpoint in patient AJ. No candidate exons or CpG islands were identified, and the 6-kb sequence did not show any significant matches to other sequences present in either the EMBL or GenBank nucleotide, protein, EST, or STS databases. A high number of repeat elements were identified, including four Alu subfamilies, MIRs, and an MLT1D (mammalian long terminal repeat–transposon) repeat. In total, interspersed repeats comprised 56% (3,347 bp) of the sequence. Alu repeats occur frequently in the 6-kb sequence, and, as indicated in figure 6, the 3′ end of an Alu-Sx repeat was identified 100 bp proximal to the breakpoint in patient AJ. In addition, the 3′ end of an Alu-Jb repeat was identified only 53 bp distal to the breakpoint.
Discussion
A Strategy for Isolating Novel Telomeres
Sequence data and telomere maps generated from many different chromosome ends have shown that most telomeres at normal chromosome ends contain many variant and degenerate repeats at the proximal end of the array. A breakpoint healed by de novo addition of a telomere is likely to be free of variant repeats; therefore, we have developed a strategy to isolate chromosome ends containing only TTAGGG repeats. We used the strategy to isolate two terminal deletion breakpoints that have the structure expected of a chromosome healing event involving novel telomere addition. This method could be used to isolate healed terminal deletion breakpoints anywhere in the genome.
Timing of Chromosome Healing Events
It is hypothesized that de novo telomere addition is mediated by the enzyme telomerase. Telomerase is inactive in oocytes and mature spermatozoa but is active during the blastocyst stage of development. It remains active in most somatic tissues at 16–20 wk development; however, by the neonatal stage, it is inactive again (Wright et al. 1996). In the absence of any evidence to suggest otherwise, it is generally assumed that healing of truncated chromosomes takes place in the germline—for example, in patient AJ. However, despite having a large deletion, patient FB336R has a relatively mild phenotype, in comparison with that of other individuals with chromosome 7q–associated holoprosencephaly. The structure and sequence at the breakpoint in patient FB336R show that the chromosome break was stabilized by de novo telomere addition. This healing event may have occurred in the germline of one parent, but, alternatively, it could have occurred early in development, so that patient FB336R is mosaic for the deletion. Mosaicism has not been examined in this study; however, if it is present, it might explain the relatively mild phenotype seen in patient FB336R.
Mechanisms of Chromosome Healing
Two mechanisms for healing of terminal deletions have been proposed. The first mechanism is the de novo addition of a telomere mediated by telomerase, and the second is capture of an existing telomere by a recombination-based mechanism. Examples that suggest that both mechanisms operate in the human germline have been described. De novo telomere addition by telomerase is likely to result in repeat addition to nonterminal sequence lacking subterminal repeats and a telomere containing only TTAGGG repeats. In contrast, telomere capture is likely to involve transfer of subterminal sequence with an existing telomere that will most likely contain variant repeats, and this type of healing event should be not be cloned using the strategy described here. The breakpoints in patients FB336R and AJ have both been healed by novel telomere addition. Binding and elongation of a telomere substrate by telomerase usually involves Watson-Crick base pairing between the substrate and telomerase RNA residues (Gilley and Blackburn 1996), with binding of at least four nucleotides at the 3′ end of the substrate (Collins and Greider 1993). Human telomerase can utilize some oligonucleotides in vitro, with 3′ ends showing as few as two bases complementary to the template, suggesting that 3′ end pairing is not essential for repeat addition (Harrington and Greider 1991; Morin 1991). Furthermore, minimal complementarity (2–4 bases) has been identified in vivo at five chromosome 16p breakpoints and at one chromosome 22q breakpoint in the human genome (Flint et al. 1994; Wong et al. 1997). At the breakpoint in patient FB336R that is described here, there are two possible nucleotides (GA) to which the repeats may have been added. The second nucleotide (A) is in phase with the template and the novel repeat array. However, the novel telomere junction in patient AJ, described here, is more complex, and there is no complementarity to the telomerase RNA template.
The presence of a pentanucleotide (G)5 in the normal sequence, ⩽200 bp distal to the breakpoint, is a common feature of all the previously reported breakpoints (Flint et al. 1994; Wong et al. 1997), and they have also been identified at the breakpoints in patients FB336R and AJ (data not shown). It has been suggested that these G-rich motifs may be involved in the recruitment of telomerase to the broken end. Other motifs that might be involved in recruitment of telomerase have been identified immediately distal to the breakpoints in patients FB336R and AJ, but none are common to all eight breakpoints characterized to date.
High frequencies of Alu elements have been noted around breakpoints at chromosome 16p (Flint et al. 1994) and at chromosome 22q in patient AJ, but these breakpoints are found within R-bands, which are GC- and Alu-rich. Therefore, the density of Alu elements may be a coincidence rather than an indication that these elements play a role in telomerase-mediated chromosome healing. Other common long-range sequence characteristics may be associated with preferential recruitment of telomerase to a broken end, but no striking similarities were identified in the 6 kb around the breakpoints in patients FB336R and AJ.
Structure of the Breakpoint in Patient AJ
Although the breakpoint in patient AJ appears to have been healed by the addition of a novel telomere, the telomere junction is complex. Ten nucleotides of unknown origin are present between the end of the normal sequence and the start of the telomere repeat array. The possibility that these 10 nucleotides represent an insertion/deletion polymorphism was excluded by sequence analysis of the breakpoint region on the normal copies of chromosome 22 from patient AJ and from AJ's parents (individuals KJ and FJ). There are a number of possible explanations for the presence of these 10 additional nucleotides. First, they may have been generated by telomerase “stuttering,” or slipping, along the template at the start of repeat addition, given that 9 of these 10 nucleotides show similarity to degenerate repeats (TTAGGAATG). Second, the combination of telomerase activity and the action of other enzymes at the breakpoint may have resulted in the generation of the extra 10 bases. Finally, they might have been acquired through a telomere-capture event. Two additional nucleotides were identified at one of the 16p13.3 breakpoints, and it was suggested that this chromosome was healed by telomere capture with consequent transfer of the two nucleotides and a telomere that contained one variant repeat (TTTAGGG) at the proximal end (Flint et al. 1994). However, the lack of variant repeats in the novel telomere of patient AJ tends to suggest that it was not transferred in a telomere-capture event. Instead, a more-complex, multistep healing event may have been involved, in which a telomere-capture event was initiated, allowing for the addition of the 10 nucleotides; however, the process was interrupted, and a novel telomere was then seeded onto the end of the 10 nucleotides. Currently, it is not possible to determine which of these possible mechanisms was involved in healing the breakpoint in patient AJ.
In conclusion, we have developed a method for isolating terminal deletion breakpoints that are located anywhere in the genome and that have been healed by the addition of a novel telomere. We have used this method to isolate and characterize breakpoints at 7q32 and 22q13.3.
Acknowledgments
We thank Heather McDermid, for the lymphoblastoid cell line of patient AJ and the DNA samples from parents KJ and FJ; M. Muenke, for the fibroblast cell line of patient FB336R; and Ian Dunham, for information on the position of genetic markers on the chromosome 22q sequence map. This work was supported by the University of Leicester and the Medical Research Council of Great Britain.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Chromosome 7 Project, The, http://www.genet.sickkids.on.ca/chromosome7/
- EMBL Nucleotide Sequence Database, The, http://www.ebi.ac.uk/embl/index.html (for the breakpoint telomere junction in patient FB336R [accession number AJ277168], the breakpoint telomere junction in patient AJ [accession number AJ277167], a 5.7-kb region of 7q cosmid cDIZ [accession number AJ277294], the chromosome 7 working-draft sequence in the breakpoint region of 7q32 in patient FB337R [accession number AC018643], and BAC clone bk268H5 [accession number AL008718])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html
- Genome Database, The, http://gdbwww.gdb.org/ (for primer sequences for D22S274, D22S282, and D22S1169)
- Repeat Masker Web Server, The, http://ftp.genome.washington.edu/
- Sanger Centre Chromosome 22 Database, The, http://www.sanger.ac.uk/HGP/Chr22/ (for information shown in )
- United Kingdom Human Genome Mapping Project Resource Centre, http://www.hgmp.mrc.ac.uk (for the NIX search tool) [DOI] [PubMed]
References
- Allshire RC, Dempster M, Hastie ND (1989) Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res 17:4611–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour JA, Neumann R, Gobert S, Jeffreys AJ (1994) Isolation of human simple repeat loci by hybridization selection. Hum Mol Genet 3:599–665 [DOI] [PubMed] [Google Scholar]
- Baird DM, Coleman J, Rosser ZH, Royle NJ (2000) High levels of sequence polymorphism and linkage disequilibrium at the telomere of 12q: implications for telomere biology and human evolution. Am J Hum Genet 66:235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird DM, Jeffreys AJ, Royle NJ (1995) Mechanisms underlying telomere repeat turnover, revealed by hypervariable variant repeat distribution patterns in the human Xp/Yp telomere. EMBO J 14:5433–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WRA, Mackinnon PJ, Villasante A, Spurr N, Buckle VJ, Dobson MJ (1990) Structure and polymorphism of human telomere-associated DNA. Cell 63:119–132 [DOI] [PubMed] [Google Scholar]
- Buluwela L, Forster A, Boehm T, Rabbitts TH (1989) A rapid procedure for colony screening using nylon filters. Nucleic Acids Res 17:452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JF, Smith CL, Cantor CR (1991) Structural and transcriptional analysis of a human subtelomeric repeat. Nucleic Acids Res 19:149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J, Baird DM, Royle NJ (1999) The plasticity of human telomeres demonstrated by a hypervariable telomere repeat array that is located on some copies of 16p and 16q. Hum Mol Genet 8:1637–1646 [DOI] [PubMed] [Google Scholar]
- Collins K, Greider CW (1993) Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev 7:1364–1376 [DOI] [PubMed] [Google Scholar]
- Cross SH, Allshire RC, McKay SJ, McGill NI, Cooke HJ (1989) Cloning of human telomeres by complementation in yeast. Nature 338:771–774 [DOI] [PubMed] [Google Scholar]
- de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE (1990) Structure and variability of human chromosome ends. Mol Cell Biol 10:518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drwinga HL, Toji LH, Kim CH, Greene AE, Mulivor RA (1993) NIGMS human/rodent somatic cell hybrid mapping panels 1 and 2. Genomics 16:311–314 [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132:6–13 [DOI] [PubMed] [Google Scholar]
- ——— (1984) “A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity”: addendum. Anal Biochem 137:266–267 [DOI] [PubMed] [Google Scholar]
- Flint J, Bates GP, Clark K, Dorman A, Willingham D, Roe BA, Micklem G, Higgs DR, Louis EJ (1997a) Sequence comparison of human and yeast telomeres identifies structurally distinct subtelomeric domains. Hum Mol Genet 6:1305–1313 [DOI] [PubMed] [Google Scholar]
- Flint J, Craddock CF, Villegas A, Bentley DP, Williams HJ, Galanello R, Cao A, Wood WG, Ayyub H, Higgs DR (1994) Healing of broken human chromosomes by the addition of telomeric repeats. Am J Hum Genet 55:505–512 [PMC free article] [PubMed] [Google Scholar]
- Flint J, Rochette J, Craddock CF, Dode C, Vignes B, Horsley SW, Kearney L, Buckle VJ, Ayyub H, Higgs DR (1996) Chromosomal stabilization by a subtelomeric rearrangement involving two closely-related Alu elements. Hum Mol Genet 5:1163–1169 [DOI] [PubMed] [Google Scholar]
- Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett NA, King A, Higgs DR (1997b) The relationship between chromosome structure and function at a human telomeric region. Nat Genet 15:252–257 [DOI] [PubMed] [Google Scholar]
- Flint J, Wilkie AOM, Buckle VJ, Winter RM, Holland AJ, McDermid HE (1995) The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat Genet 9:132–140 [DOI] [PubMed] [Google Scholar]
- Gilley D, Blackburn EH (1996) Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol Cell Biol 16:66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1987) The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51:887–898 [DOI] [PubMed] [Google Scholar]
- ——— (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331–337 [DOI] [PubMed] [Google Scholar]
- Gurrieri F, Trask BJ, van den Engh G, Krauss CM, Schinzel A, Pettenati MJ, Schindler D, Dietzband J, Vergnaud G, Scherer SW, Tsui LC, Muenke M (1993) Physical mapping of the holoprosencephaly critical region on chromosome 7q36. Nat Genet 3:247–251 [DOI] [PubMed] [Google Scholar]
- Harrington LA, Greider CW (1991) Telomerase primer specificity and chromosome healing. Nature 353:451–454 [DOI] [PubMed] [Google Scholar]
- Jacobsen P (1973) 18p-syndrome: deletion of the short arm of chromosome 18. Ugeskr Laeger 135:17 [PubMed] [Google Scholar]
- Jeffreys AJ, Macleod A, Tamaki K, Neil DL, Monckton DG (1991) Minisatellite repeat coding as a digital approach to DNA typing. Nature 354:204–209 [DOI] [PubMed] [Google Scholar]
- Jurka J, Milosavljevic A (1991) Reconstruction and analysis of human Alu genes. J Mol Evol 32:105–121 [DOI] [PubMed] [Google Scholar]
- Koch JE, Kolvraa S, Peterson KB, Gregerson N, Bolund L (1989) Oligonucleotide-priming methods for the chromosome-specific labelling of alpha satellite DNA in situ. Chromosoma 98:259–265 [DOI] [PubMed] [Google Scholar]
- May CA, Jeffreys AJ, Armour JA (1996) Mutation rate heterogeneity and the generation of allele diversity at the human minisatellite MS205 (D16S309). Hum Mol Genet 5:1823–1833 [DOI] [PubMed] [Google Scholar]
- McClintock B (1941) The stability of broken ends of chromosomes in Zea Mays. Genetics 26:234–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis RC, Velagaleti GV, Jones C, Pivnick EK, Phelan MC, Boyd E, Tarleton J, Wilroy RS, Tunnacliffe A, Tharapel AT (1998) Most Jacobsen syndrome deletion breakpoints occur distal to FRA11B. Am J Med Genet 76:222–228 [PubMed] [Google Scholar]
- Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B, Kohwi Y, Mandel-Gutfroind Y, Lee JR, Drescher B, Sas DE, Margalit H, Platzer M, Weiss A, Tsui LC, Rosenthal A, Kerem B (1998) Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci USA 95:8141–8146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin GB (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59:521–529 [DOI] [PubMed] [Google Scholar]
- ——— (1991) Recognition of a chromosome truncation site associated with α-thalassemia by human telomerase. Nature 353:454–456 [DOI] [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR (1988). A highly conserved repetitive DNA-sequence, (TTAGGG)n, present at the telomeres of human-chromosomes. Proc Natl Acad Sci USA 85:6622–6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ (1938) The remaking of chromosomes. The Collecting Net 13:181–198 [Google Scholar]
- Nesslinger NJ, Gorski JL, Kurczynski TW, Shapira SK, Siegelbartelt J, Dumanski JP, Cullen RF, French BN, McDermid HE (1994) Clinical, cytogenetic, and molecular characterization of seven patients with deletions of chromosome 22q13.3. Am J Hum Genet 54:464–472 [PMC free article] [PubMed] [Google Scholar]
- Royle NJ, Baird DM, Jeffreys AJ (1994) A subterminal satellite located adjacent to telomeres in chimpanzees is absent from the human genome. Nat Genet 6:52–56 [DOI] [PubMed] [Google Scholar]
- Royle NJ, Hill MC, Jeffreys AJ (1992) Isolation of telomere junction fragments by anchored polymerase chain reaction. Proc R Soc Lond B Biol Sci 247:57–61 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Labeling of synthetic olinucleotides by phosphorylation with bacteriophage T4 polynucleotide kinase. In Sambrook J, Fritsch EF, Maniatis T (eds): Molecular cloning: a laboratory manual. Cold Spring Harbour Laboratory Press, pp 11.31 [Google Scholar]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel A, Auf der Maur P, Moser H (1977) Partial deletion of long arm of chromosome 11[del(11)(q23)]: Jacobsen syndrome. two new cases and review of the clinical findings. J Med Genet 14:438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippen-Lentz D, Blackburn EH (1990) Functional evidence for an RNA template in telomerase. Science 247:546–552 [DOI] [PubMed] [Google Scholar]
- Tunnacliffe A, Jones C, Le Paslier D, Todd R, Cherif D, Birdsall M, Devenish L, Yousry C, Cotter FE, James MR (1999) Localization of Jacobsen syndrome breakpoints on a 40-Mb physical map of distal chromosome 11q. Genome Res 9:44–52 [PMC free article] [PubMed] [Google Scholar]
- Voullaire LE, Webb GC, Leversha MA (1987) Chromosome deletion at 11q23 in an abnormal child from a family with inherited fragility at 11q23. Hum Genet 76:202–204 [DOI] [PubMed] [Google Scholar]
- Wong ACC, Ning Y, Flint J, Clark K, Dumanski JP, Ledbetter DH, McDermid HE (1997) Molecular characterization of a 130-kb terminal microdeletion at 22q in a child with mild mental retardation. Am J Hum Genet 60:113–120 [PMC free article] [PubMed] [Google Scholar]
- Wong Z, Royle NJ, Jeffreys AJ (1990) A novel DNA polymorphism resulting from transfer of DNA from chromosome 6 to chromosome 16. Genomics 7:222–234 [DOI] [PubMed] [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW (1996) Telomerase activity in human germline and embryonic tissues and cells. Dev Genet 18:173–179 [DOI] [PubMed] [Google Scholar]
- Yu GL, Blackburn EH (1991) Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell 67:823–832 [DOI] [PubMed] [Google Scholar]
- Zakian VA, Runge K, Wang SS (1990) How does the end begin? Formation and maintenance of telomeres in ciliates and yeast. Trends Genet 6:12–16 [DOI] [PubMed] [Google Scholar]