Abstract
Down syndrome is a complex genetic and metabolic disorder attributed to the presence of three copies of chromosome 21. The extra chromosome derives from the mother in 93% of cases and is due to abnormal chromosome segregation during meiosis (nondisjunction). Except for advanced age at conception, maternal risk factors for meiotic nondisjunction are not well established. A recent preliminary study suggested that abnormal folate metabolism and the 677C→T polymorphism in the methylenetetrahydrofolate reductase (MTHFR) gene may be maternal risk factors for Down syndrome. The present study was undertaken with a larger sample size to determine whether the MTHFR 677C→T polymorphism was associated with increased risk of having a child with Down syndrome. Methionine synthase reductase (MTRR) is another enzyme essential for normal folate metabolism. A common polymorphism in this gene was recently associated with increased risk of neural tube defects and might also contribute to increased risk for Down syndrome. The frequencies of the MTHFR 677C→T and MTRR 66A→G mutations were evaluated in DNA samples from 157 mothers of children with Down syndrome and 144 control mothers. Odds ratios were calculated for each genotype separately and for potential gene-gene interactions. The results are consistent with the preliminary observation that the MTHFR 677C→T polymorphism is more prevalent among mothers of children with Down syndrome than among control mothers, with an odds ratio of 1.91 (95% confidence interval [CI] 1.19–3.05). In addition, the homozygous MTRR 66A→G polymorphism was independently associated with a 2.57-fold increase in estimated risk (95% CI 1.33–4.99). The combined presence of both polymorphisms was associated with a greater risk of Down syndrome than was the presence of either alone, with an odds ratio of 4.08 (95% CI 1.94–8.56). The two polymorphisms appear to act without a multiplicative interaction.
Introduction
Trisomy 21, or Down syndrome, is the most common genetic cause of human mental retardation, with an incidence of ∼1/600–1,000 live births (Smith and Berg 1976). In addition, trisomy 21 is a major cause of premature pregnancy failure. It is estimated that 1/150 conceptions have trisomy 21 and that 80% of these are lost during early pregnancy (Boue et al. 1975; Hassold and Jacobs 1984; Freeman et al. 1991). The nondisjunction event that results in two copies of chromosome 21 takes place in anaphase of meiosis I, during oocyte maturation before ovulation, and/or in anaphase of meiosis II, around the time of fertilization in the adult female (Lemaire-Adkins et al. 1997; Picton et al. 1998; Hunt and Lemaire-Adkins 2000). Despite the prevalence and health consequences of Down syndrome, the biochemical and molecular basis for meiotic nondisjunction is not understood.
Folic acid is essential for the de novo synthesis of nucleotide precursors for normal DNA synthesis and is also essential for normal cellular methylation reactions. Chronic folate/methyl deficiency in vivo and in vitro has been associated with abnormal DNA methylation (Balaghi and Wagner 1993; Pogribny et al. 1995; Fowler et al. 1998; Jacob et al. 1998), DNA strand breaks (Blount et al. 1997; Pogribny et al. 1997; Duthie 1999), altered chromosome recombination (Knuutila et al. 1978; MacGregor et al. 1997), and aberrant chromosome segregation (Libbus et al. 1990; Leyton et al. 1995; Chen et al. 1998b; Titenko-Holland et al. 1998; Xu et al. 1999) On the basis of this evidence, James et al. (1999) suggested the possibility that gene-nutrient interactions associated with abnormal folate metabolism and DNA hypomethylation might increase the risk of chromosome nondisjunction. In this preliminary study, mothers of children with Down syndrome were found to have mildly elevated plasma homocysteine levels and a 2.6-fold increased frequency of the 677C→T polymorphism in the methylenetetrahydrofolate reductase (MTHFR) gene, compared with control mothers (95% CI 1.2–5.8). The data from this study further suggested that polymorphisms in other genes in the folate pathway may increase the risk of having a child with Down syndrome (James et al. 1999; Rosenblatt 1999).
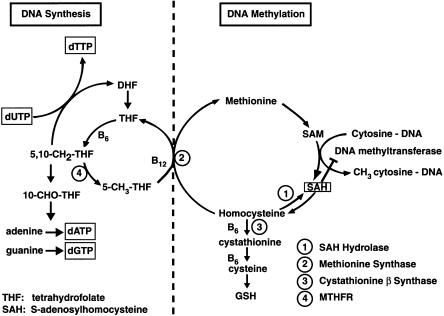
MTHFR catalyzes the synthesis of 5-methyltetrahydrofolate, the methyl donor for the B12-dependent remethylation of homocysteine to methionine via the methionine synthase reaction (fig. 1). Methionine is the precursor for the synthesis of S-adenosylmethionine (SAM), the major cellular methyl donor for DNA, RNA, protein, and phospholipid methylation. The reduction in enzyme activity associated with the 677C→T MTHFR polymorphism raises the dietary requirement for folic acid to maintain normal remethylation of homocysteine to methionine (Bailey and Gregory 1999). Consequently, low folate status in individuals with the MTHFR polymorphism results in an increase in homocysteine and a decrease in methionine levels. Chronic elevation in intracellular homocysteine can lead to a decrease in the ratio of SAM to S-adenosylhomocysteine (SAH) that is associated with inhibition of the DNA methyltransferase and DNA hypomethylation (Balaghi and Wagner 1993; De Cabo et al. 1995; Melnyk et al. 2000) Methionine synthase reductase (MTRR) is a related flavoprotein that maintains the methionine synthase enzyme in an active state for the remethylation of homocysteine to methionine. The cloning of the cDNA for MTRR led to the identification of a polymorphism, 66A→G, that was recently associated with increased risk for spina bifida (LeClerc et al. 1998; Wilson et al. 1999). Because of the importance of the methionine synthase reaction in maintaining normal folate metabolism and DNA methylation, we hypothesized that this polymorphism could be a second maternal genetic risk factor for Down syndrome.
Figure 1.
Overview of the interactive and interdependent reactions involved in cellular one-carbon metabolism, with emphasis on the two major functions of these pathways in DNA metabolism: normal DNA synthesis and normal DNA methylation. These two major functions intersect at the folate/B12–dependent methionine synthase reaction, which generates metabolically active tetrahydrofolate for DNA nucleotide precursor synthesis, and, at the same time, regenerates methionine from homocysteine. Both DNA synthesis and DNA methylation are negatively affected by inadequate folate or B12 intake and/or by mutations in these pathways. Note that an elevation in homocysteine induces the reversal of the SAH hydrolase reaction and causes an elevation in SAH, a potent product inhibitor of the DNA methyltransferase reaction (De Cabo et al. 1995).
The association between folate deficiency and DNA hypomethylation suggests that genetic and/or nutritional deficiencies that negatively affect folate metabolism may be mechanistically related to increased risk of nondisjunction and Down syndrome. In the present study, we have extended the initial work of James et al. with an analysis of the association between the MTHFR 677C→T polymorphism and the maternal risk of Down syndrome in a larger study population, and we have evaluated, for the first time, the association between the MTRR 66A→G polymorphism and Down syndrome risk.
Subjects and Methods
Study Population
Blood samples from women who had a pregnancy affected by karyotypically confirmed full trisomy 21 (case mothers) were obtained from three study sites: metropolitan Atlanta, California, and the National Center for Toxicological Research (NCTR) in Jefferson, Arkansas. For the purposes of our analyses, case and control mothers were limited to whites, to reduce the ethnic variation between groups. Of 157 case mothers, 77 were derived from a population-based case-control study of trisomy 21 in the five-county area of metropolitan Atlanta. Mothers of live-born infants with trisomy 21 were ascertained with the assistance of the Metropolitan Atlanta Congenital Defects Program of the Centers for Disease Control from 1989 to 1998; details regarding the methods and sources have been described elsewhere (Edmonds and Oakley 1981). An additional 23 case mothers were also ascertained from the California Birth Defects Monitoring System, which is an active birth defects–surveillance program covering counties that include more than half of the births in California. Case mothers were identified at genetic laboratories that record all abnormal karyotypes of live births in the monitored counties (Torfs and Christianson 1999). Recruitment of mothers participating in the NCTR study was done through Down syndrome support-group newsletters and the Internet; thus, this population (57/157 case mothers) was geographically diverse, representing 16 states and Canada (James et al. 1999). Each case mother was asked to recruit a control mother who was approximately the same age, who resided in the same geographic area (with a similar socioeconomic status), and who had experienced no miscarriages or abnormal pregnancies (46/140 control mothers). In addition, 94 control mothers were recruited through the Pediatric Test Centre at Montreal Children’s Hospital, from a previous case-control study (Christensen et al. 1999; Wilson et al. 1999). Informed consent was obtained from all participants.
Specimen Collection and Genotype Analyses
Genomic DNA was isolated from lymphocytes in whole blood, by use of standard chloroform/phenol extraction, and was stored at −20°C until the genotype analysis was performed. DNA samples obtained from case and control mothers were analyzed at NCTR for the MTHFR genotype and at the Montreal Children’s Hospital Research Institute for the MTRR genotype. For both polymorphisms, genotyping was done by PCR followed by appropriate restriction enzyme digestion and has been described in detail elsewhere (James et al. 1999; Wilson et al. 1999). Laboratory personnel who performed the genotyping were unaware of whether the samples came from case or control mothers.
Statistical Analyses
Allele frequencies were calculated for each genotype, and the differences in allele frequencies between case and control mothers were determined using a Pearson χ2 test. Expected genotype frequencies were calculated from the allele frequencies under the assumption of Hardy-Weinberg equilibrium. Odds ratios for both the heterozygous and homozygous mutant genotypes, as compared with the wild types, were calculated as a measure of the association between the MTHFR and MTRR genotype and a Down syndrome–affected pregnancy. These odds ratios were used as an estimate of relative risk. The interaction between the two genotypes was evaluated by calculation of the odds ratio for the exposure genotype of either MTHFR or MTRR, while the other was controlled for, followed by calculation of the odds ratio for the presence of both genotypes compared with the absence of both. For analysis of gene-gene interactions, the MTRR homozygous AA genotype and the heterozygous AG genotypes were combined because neither genotype was associated with increased risk of neural tube defects (Wilson et al. 1999) or of Down syndrome. For MTHFR, the heterozygous CT and homozygous TT genotypes were combined for the gene-gene interaction analysis, because both genotypes reduce MTHFR enzyme activity and because both have been associated with an increased risk of neural tube defects (Ou et al. 1996) and Down syndrome (James et al. 1999). Throughout, a two-tailed P value of .05 was interpreted as indicating a statistically significant difference. All statistical analyses were done with SAS software, version 8 (SAS Institute).
Results
Allele Frequencies
The distribution of the MTHFR genotypes in the control population was found to be in Hardy-Weinberg equilibrium. Table 1 indicates the MTHFR and MTRR allele frequencies for women with pregnancies affected by Down syndrome and for control women, using the combined data from the three sources. The frequencies of the MTHFR 677T allele and the MTRR 66G allele were both significantly higher among case mothers than among control mothers. The MTHFR mutant allele frequency was .41 (128/314 alleles) among the case mothers and .31 (87/280 alleles) among the control mothers (P<.01). The mean distribution of the T allele frequency in the combined control group was .31, which is in the range of that recently reported in a meta-analysis of several studies of North American whites (Botto and Yang 2000). For the MTRR gene, the mutant allele frequency was .60 (174/290 alleles) among case mothers, compared with .47 (132/278 alleles) among the control mothers (P<.003). These data confirm that, in the current study population, the mutant allele frequencies for both MTHFR and MTRR genes are significantly higher among case mothers than among control mothers.
Table 1.
Allele Frequencies of MTHFR 677C→T and MTRR 66A→G in Women with Down Syndrome–Affected Pregnancies (Case Mothers) and Control Mothers
|
Frequency in Mothers |
|||||||
| Case |
Control |
||||||
| Genotype | Allele | Alleles | % | Alleles | % | χ2 | P |
| MTHFR | C | 186 | .59 | 193 | .69 | ||
| T | 128 | .41 | 87 | .31 | 6.02 | .01 | |
| MTRR | A | 116 | .40 | 146 | .53 | ||
| G | 174 | .60 | 132 | .47 | 8.95 | .003 | |
Maternal MTHFR Genotype and Risk of Down Syndrome
The composite data in table 2 show that the frequencies of the CC, CT, and TT genotypes among all control mothers were 48% (67/140), 42% (59/140), and 10% (14/140), respectively. The corresponding frequencies among all case mothers were 32% (51/157), 54% (84/157), and 14% (22/157). The presence of the 677C→T substitution in one or both alleles was associated with a 1.91-fold increase in the risk of having a child with Down syndrome (95% CI 1.19–3.05). The odds ratio for the heterozygous C/T genotype was 1.87 (95% CI 1.14–3.06), whereas for the homozygous TT genotype, the odds ratio was 2.06 (95% CI 0.96–4.43). Analysis of the combined samples from the Atlanta and California populations, without the previously reported NCTR data, yielded an odds ratio of 1.63 (95% CI 0.91–2.90) for the CT and TT mutant genotypes.
Table 2.
Association between Maternal MTHFR Genotype and Down Syndrome–Affected Pregnancies (Case Mothers) and Control Mothers
| Genotype | No. (%)of CaseMothers(n = 157) | No. (%)of ControlMothers(n = 140) | OddsRatio | 95% CI | P |
| CC | 51 (32) | 67 (48) | 1.0 | ||
| CT | 84 (54) | 59 (42) | 1.87 | 1.14–3.06 | .02 |
| TT | 22 (14) | 14 (10) | 2.06 | .96–4.43 | .09 |
| CT or TT | 106 (68) | 73 (52) | 1.91 | 1.19–3.05 | .01 |
Maternal MTRR Genotype and Risk of Down Syndrome
In table 3, the distribution of MTRR genotypes in the control population was 28% (39/139), for the homozygous wild type, 49% (68/139) for the heterozygous genotype, and 23% (32/139) for the homozygous mutant genotype. The corresponding frequencies in the case mothers were 18% (26/145) homozygous wild type, 44% (64/145) heterozygous, and 38% (55/145) homozygous mutant. Stratified by genotype, the data show that homozygosity for the 66A→G mutation was associated with a 2.57-fold increased risk of having a Down syndrome–affected pregnancy (95% confidence interval [CI] 1.33–4.99) compared with homozygous normal subjects. The heterozygous mutation, however, was not associated with a significant increase in risk (odds ratio 1.41; 95% CI 0.77–2.56).
Table 3.
Association between Maternal MTRR Genotype and Down Syndrome–Affected Pregnancies (Case Mothers) and Control Mothers
| Genotype | No. (%)of CaseMothers(n = 145) | No. (%)of ControlMothers(n = 139) | OddsRatio | 95% CI | P |
| AA | 26 (18) | 39 (28) | 1.0 | ||
| AG | 64 (44) | 68 (49) | 1.41 | .77–2.56 | .33 |
| GG | 55 (38) | 32 (23) | 2.57 | 1.33–4.99 | .01 |
| AG or GG | 119 (82) | 100 (72) | 1.78 | 1.02–3.13 | .06 |
Interaction between MTHFR Genotype and MTRR Genotype
To evaluate potential gene-gene interactions, both MTHFR and MTRR polymorphisms were evaluated in a two-by-four table, as suggested by Botto and Mastroiacovo (1998). Using this approach, it was possible to evaluate the risk associated with each genotype independently and, also, to assess the combined risk when both polymorphisms are present. As is shown in table 4, case mothers were more likely to be heterozygous or homozygous for the MTHFR mutant genotype and not have the MTRR mutant GG genotype (63/145) than were control mothers (odds ratio 2.37; 95% CI 1.31–4.31). Case mothers who were homozygous for the MTRR mutant polymorphism but were negative for the MTHFR mutant allele (19/145) were at a 2.44-fold increased risk (CI 1.07–5.54). The presence of both MTHFR mutant alleles and the MTRR homozygous mutant allele (36/145) was associated with a 4.08-fold increased risk of having a child with Down syndrome (95% CI 1.94–8.56). Although the presence of both polymorphisms consistently conferred a greater risk of Down syndrome than did the presence of either alone, the polymorphisms appear to act without a multiplicative gene-gene interaction.
Table 4.
Interaction between MTHFR and MTRR Genotype in Women with Down Syndrome–Affected Pregnancies (Case Mothers) and Control Mothers
| MTHFR | MTRR | No. (%)of Cases (n=145) | No. (%)of Controls(n=135) | Odds Ratio | 95% CI | P |
| CC | AA or AG | 27 (19) | 52 (39) | 1.0 | ||
| TT or CT | AA or AG | 63 (43) | 51 (38) | 2.37 | 1.31–4.31 | .01 |
| CC | GG | 19 (13) | 15 (11) | 2.44 | 1.07–5.54 | .05 |
| TT or CT | GG | 36 (25) | 17 (12) | 4.08 | 1.94–8.56 | .001 |
Discussion
Maternal age is the only well-established risk factor for Down syndrome, and the associated risk increases exponentially at age >30 years (Hassold and Jacobs 1984). It has been estimated that 15%–20% of all human conceptions are chromosomally abnormal because of errors in meiotic division; however, the majority of these errors are embryonic lethal and result in fetal loss (Hunt 1998). The high frequency of maternal nondisjunction is now thought to be due to the absence of a meiotic checkpoint in the oocyte, and this provides a plausible biological explanation for the predominance of maternal nondisjunction (Lemaire-Adkins et al. 1997). In the human female, primordial oocytes enter meiosis I during fetal development, undergo DNA replication and homologous recombination, and then remain arrested in prophase I (diplotene stage) for several decades until initiation of oocyte maturation and ovulation in the adult female (Hunt and Lemaire-Adkins 2000)
Recent preliminary studies have implicated the MTHFR 677C→T polymorphism (James et al. 1999) and excessive smoking (Yang et al. 1999) as maternal risk factors for Down syndrome. Interestingly, low folate status has been associated with each of these potential risk factors (Piyathilake et al. 1994; Lewis et al. 1998; James et al. 1999). A recent clinical study has more directly implicated folate deficiency as a risk factor for human aneuploidy. Lymphocytes from women consuming a controlled folate-deficient diet were found to have a significantly increased frequency of kinetochore-positive micronuclei, which are surrogate markers for abnormal chromosome segregation (Titenko-Holland et al. 1998). Folate supplementation after the folate-depletion phase in this metabolic study was associated with a significant decrease in these centromeric fragments. Taken together, these studies support the possibility that multifactorial gene-environment interactions that compromise maternal folate status may promote meiotic nondisjunction and the risk of a Down syndrome conception.
The results of the present study indicate that the simultaneous presence of the MTHFR 677T polymorphism and the homozygous MTRR 66G polymorphism conferred a four-fold increase in the maternal risk of having a child with Down syndrome (95% CI 1.94–8.56). In the combined populations, the MTHFR 677T substitution in one or both alleles was associated with a 1.9-fold increase in risk (95% CI 1.19–3.05), and the presence of the homozygous 66G allele in MTRR was associated with a 2.57-fold increase in risk (95% CI 1.33–4.99). The odds ratio for the mutant 677T allele in the Atlanta and California cohorts did not quite reach statistical significance when analyzed independently from that in the NCTR cohort; however, the point estimate remained >1, indicating increased risk (odds ratio 1.63; 95% CI 0.91–2.90).
A frequent challenge in conducting studies of gene-gene interactions is balancing the need for relatively large study populations against the need to minimize bias that may occur because of population admixture. To acquire sufficient statistical power, case mothers in the present study were drawn from two birth defect registries and from a volunteer sample that included case mothers from 16 states and Canada. Control mothers were limited to women who were documented to have no children with birth defects and were pooled from two previous studies. To minimize ethnic bias, all participants were restricted to North American whites. Because genotypes from appropriate control mothers were not available from the Atlanta and California registries, control samples from the NCTR study and from a previous study of NTDs in Montreal were combined. Thus, an inherent limitation of the current study is the potential bias of population admixture introduced by pooling control mothers from geographical areas different from those that provided the case mothers. To determine whether our pooled control population was a representative North American sample, we compared our control MTHFR 677T allele frequencies with those of a recently published meta-analysis of North American whites (Botto and Yang 2000). The MTHFR 677T allele frequency in our control group was .31, which was within the range of frequencies reported in the meta-analysis. We had more difficulty evaluating potential bias in the association between MTRR and Down syndrome, since there are no published data on allele frequencies or genotype distribution outside of our study sites. Thus, we compared the genotype distribution among our control mothers with that among fathers from the Atlanta and California samples, who may be more ethnically similar to the case mothers than were a convenience sample of control mothers. The frequency of MTRR 66A→G homozygosity was 24.2% among the fathers and 23.0% among the control mothers. Taken together, these observations suggest that our combined control samples were representative of the MTHFR allele frequencies and MTRR homozygosity within North American whites. Nonetheless, the findings of the current study need to be confirmed in a large prospectively designed population-based case-control study.
Because MTHFR and MTRR require folate and B12, respectively, to support the methionine synthase reaction, the metabolic impact of both polymorphisms is magnified by low levels of folate or B12 (Bailey and Gregory 1999; Wilson et al. 1999). Accordingly, risk estimates that stratify mutant genotypes by nutritional status result in more sensitive risk estimates than do those based on genotype alone. For example, stratification of the MTHFR 677C→T genotype by folate status or of the MTRR 66A→G genotype by B12 status has resulted in more sensitive risk estimates for neural tube defects (Chen et al. 1998a; Christensen et al. 1999; Wilson et al. 1999). The ability to analyze the MTHFR and MTRR genotypes in terms of specific metabolic biomarkers—such as plasma homocysteine, folate, and/or B12 levels—would increase the power to detect a significant impact on Down syndrome risk. A limitation of the present study is that we did not have access to plasma samples for evaluation of folate, B12, or homocysteine levels in case and control mothers.
A compromise in the methionine synthase reaction caused by genetic and/or dietary factors could promote abnormal chromosome segregation by an indirect effect on oocyte DNA methylation patterns and higher-order chromatin structure. The secondary structure of pericentromeric heterochromatin, at repetitive satellite sequences, is involved in protein-DNA binding and in cohesion between sister chromatids (Renauld and Gasser 1997; Clarke 1998; Cobb et al. 1999). The abundant methyl-binding protein, MeCP2, preferentially binds to pericentromeric DNA in a complex with histone deacetylase, resulting in local histone deacetylation and chromatin condensation (Nan et al. 1996, 1998; Jones et al. 1998). Recent evidence linking DNA methylation to histone deacetylation and chromatin condensation supports the possibility that DNA methylation patterns may be linked mechanistically to the epigenetic alterations in chromatin structure required for normal chromosome segregation (Bestor and Tycko 1996; Grewal et al. 1998; Vig 1998). In nonreplicating cells, such as oocytes, the loss of methyl groups at methylated CpG sites in DNA can occur during excision repair (Brooks et al. 1996), by spontaneous deamination of 5-methyl cytosine to thymine (Gonzalgo and Jones 1997), or during DNA strand exchange associated with recombination (Bestor and Tycko 1996). The failure of the DNA methyltransferase to remethylate the newly synthesized strands would result in permanent loss of DNA methylation patterns under conditions of localized folate/methyl deficiency.
The importance of stable pericentromeric DNA methylation for normal chromosome segregation has been underscored by several recent discoveries. For example, lymphocytes from individuals with the rare autosomal disorder ICF (immunodeficiency, centromeric instability, and facial anomalies) exhibit profound chromosomal abnormalities, including selective undermethylation of pericentromeric satellite DNA, chromosome decondensation, and complex multiradiate chromosomes (Miniou et al. 1997). The genetic origin of this disorder was recently discovered to be a mutation in the de novo DNA methyltransferase 3B (DNMT3B) and supports a causal association between DNA hypomethylation, pericentromeric decondensation, and abnormal chromosome segregation (Hansen et al. 1999; Xu et al. 1999). Treatment of cultured cells with 5-azacytidine, a potent demethylating agent, similarly results in pericentromeric decondensation and profound chromatid missegregation in anaphase (Leyton et al. 1995; Vig and Hallet 2000). Chromosomal deletions, translocations, and instability were recently reported in murine embryonic stem cells nullizygous for the major DNA methyltransferase (Dnmt1) gene (Chen et al. 1998b). Aneuploidy and chromosomal instability are present in most human cancers, and chromosomal instability has recently been shown to be directly related to the extent of DNA hypomethylation (Vilain et al. 1999). These data strongly suggest that stable methylation in pericentromeric DNA is an essential prerequisite for normal chromosome organization, stabilization, and segregation.
The association between folate deficiency and DNA hypomethylation lends support to the possibility that the increased frequency of the MTHFR and MTRR polymorphisms observed in the present study may be associated with chromosomal nondisjunction and Down syndrome. Further research is needed to evaluate the paradigm suggested by the present results: an association between maternal folate metabolism and Down syndrome. Additional studies of other candidate genes in the folate pathway, as well as a systematic study of interactions with other micronutrients involved in folate/methyl metabolism in women with Down syndrome–affected pregnancies, may suggest opportunities to improve public health strategies for the primary prevention of Down syndrome.
Acknowledgments
This work was supported, in part, by the following: the Food and Drug Administration Office of Women’s Health; the National Heart, Lung, and Blood Institute (HL 58955-01); the Medical Research Council of Canada Group Grant (GR-13297); National Institutes of Health Grant #P01-HD32111; the Tobacco-Related Disease Research Program of the University of California, Grant #2RT0080; and the Center for Disease Control and Prevention Grant (U50/CCU613236-02). We also thank Qing Wu and Nelly Sabbaghian for technical assistance and Caroline Simmons for help in manuscript preparation.
References
- Bailey LB, Gregory J (1999) Polymorphisms of methylenetetrahydrofolate reductase and other enzymes: metabolic significance, risks, and impact on folate requirement. J Nutr 129:919–922 [DOI] [PubMed] [Google Scholar]
- Balaghi M, Wagner C (1993) DNA methylation in folate deficiency—use of CpG methylase. Biochem Biophys Res Commun 193:1184–1190 [DOI] [PubMed] [Google Scholar]
- Bestor TH, Tycko B (1996) Creation of genomic methylation patterns. Nat Genet 12:363–367 [DOI] [PubMed] [Google Scholar]
- Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wickremasinghe RG, Everson RB, Ames BN (1997) Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA 94:3290–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto LD, Mastroiacovo P (1998) Exploring gene-gene interactions in the etiology of neural tube defects. Clin Genet 53:456–459 [DOI] [PubMed] [Google Scholar]
- Botto LD, Yang Q (2000) 5,10-Methylenetetrahydrofolate reductase gene variants and congenital anomalies: a HuGE review. Am J Epidemiol 151:862–877 [DOI] [PubMed] [Google Scholar]
- Boue J, Boue A, Lazar P (1975) Retrospective and prospective epidemiological studies of 1,506 karotyped spontaneous abortions. Teratology 12:11–26 [DOI] [PubMed] [Google Scholar]
- Brooks PJ, Marietta C, Goldman D (1996) DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci 16:939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Giovannucci E, Hankinson SE, Ma J, Willett WC, Spiegelman D, Kelsey KT, Hunter DJ (1998a) A prospective study of methylenetetrahydrofolate reductase and methionine synthase gene polymorphisms, and risk of colorectal cancer. Carcinogenesis 19:2129–2132 [DOI] [PubMed] [Google Scholar]
- Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R (1998b) DNA hypomethylation leads to elevated mutation rates. Nature 395:89–93 [DOI] [PubMed] [Google Scholar]
- Christensen B, Arbour L, Tran P, Leclerc D, Sabbaghian N, Platt R, Gilfix BM, Rosenblatt DS (1999) Genetic polymorphisms in methylenetetrahydrofolate reductase and methionine synthase, folate levels in red blood cells, and risk of neural tube defects. Am J Med Genet 84:151–157 [DOI] [PubMed] [Google Scholar]
- Clarke L (1998) Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr Opin Genet Dev 8:212–218 [DOI] [PubMed] [Google Scholar]
- Cobb J, Miyaike M, Kikuchi A, Handel MA (1999) Meiotic events at the centromeric heterochromatin: histone H3 phosphorylation, topoisomerase II alpha localization and chromosome condensation. Chromosoma 108:412–425 [DOI] [PubMed] [Google Scholar]
- De Cabo SF, Santos J, Fernández-Piqueras J (1995) Molecular and cytological evidence of S-adenosyl-L-homocysteine as an innocuous undermethylating agent in vivo. Cytogenet Cell Genet 71:187–192 [DOI] [PubMed] [Google Scholar]
- Duthie SJ (1999) Folic acid deficiency and cancer: mechanisms of DNA instability. Br Med Bull 55:578–592 [DOI] [PubMed] [Google Scholar]
- Edmonds LD, Oakley GP (1981) Congenital malformations surveillance: two American systems. Int J Epidemiol 10:247–252 [DOI] [PubMed] [Google Scholar]
- Fowler BM, Giuliano AR, Piyathilake C, Nour M, Hatch K (1998) Hypomethylation in cervical tissue: is there a correlation with folate status? Cancer Epidemiol Biomarkers Prev 7:901–906 [PubMed] [Google Scholar]
- Freeman S, Grantham M, Hassold T, Herbert M, Hersey J, Nuccio J, Pettay D, Takesu N, Phillips C (1991) Cytogenetic and molecular studies of human spontaneous abortions. Am J Hum Genet Suppl A49:916 [Google Scholar]
- Gonzalgo ML, Jones PA (1997) Mutagenic and epigenetic effects of DNA methylation. Mutat Res 386:107–118 [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Bonduce MJ, Klar KJ (1998) Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150:563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM (1999) The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA 96:14412–14417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold TJ, Jacobs PA (1984) Trisomy in man. Ann Rev Genet 18:69–97 [DOI] [PubMed] [Google Scholar]
- Hunt PA (1998) The control of mammalian female meiosis: factors that influence chromosome segregation. J Assist Reprod Genet 15:246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PA, Lemaire-Adkins R (1998) Genetic control of mammalian female meiosis. Curr Top Dev Biol 37:359–381 [PubMed] [Google Scholar]
- Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, Henning SM (1998) Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr 128:1204–1212 [DOI] [PubMed] [Google Scholar]
- James SJ, Pogribna M, Pogribny IP, Melnyk S, Hine RJ, Gibson JB, Yi P, Tafoya DL, Swenson DH, Wilson VL, Gaylor DW (1999) Abnormal folate metabolism and mutation in the methylenetetrahydrofolate reductase (MTHFR) gene may be maternal risk factors for Down syndrome. Am J Clin Nutr 70:495–501 [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19:187–191 [DOI] [PubMed] [Google Scholar]
- Knuutila S, Helminen E, Vuopio P, de la Chapelle A (1978) Increased sister chromatid exchange in megaloblastic anaemia—studies on bone marrow cells and lymphocytes. Hereditas 89:175–181 [DOI] [PubMed] [Google Scholar]
- Leclerc D, Wilson A, Dumas R, Gafuik C, Song D, Watkins D, Heng HH, Rommens JM, Scherer SW, Rosenblatt DS, Gravel RA (1998) Cloning and mapping of a cDNA for methionine synthase reductase, a flavoprotein defective in patients with homocystinuria. Proc Natl Acad Sci USA 95:3059–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Adkins R, Radke K, Hunt PA (1997) Lack of checkpoint control at the metaphase/anaphase transition: a mechanism of meiotic nondisjunction in mammalian females. J Cell Biol 139:1611–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DP, Van Dyke DC, Stumbo PJ, Berg MJ (1998) Drug and environmental factors associated with adverse pregnancy outcomes. Part II: Improvement with folic acid. Ann Pharmacother 32:947–961 [DOI] [PubMed] [Google Scholar]
- Leyton C, Mergudich D, de la Torre D, Sans J (1995) Impaired chromosome segregation in plant anaphase after moderate hypomethylation of DNA. Cell Prolif 28:481–496 [DOI] [PubMed] [Google Scholar]
- Libbus BL, Borman LS, Ventrone CH, Branda RF (1990) Nutritional folate deficiency in CHO cells: chromosomal abnormalities associated with perturbations in nucleic acid precursors. Cancer Genet Cytogenet 46:231–242 [DOI] [PubMed] [Google Scholar]
- MacGregor JT, Wehr C, Hiatt RA, Peters B, Tucker JD, Langlois RG, Jacob RA, Jensen RH, Yager JW, Shigenaga MK, Frei B, Eynon BP, Ames BN (1997) Spontaneous genetic damage in man: evaluation of interindividual variability, relationship among markers of damage, and influence of nutritional status. Mutat Res 377:125–135 [DOI] [PubMed] [Google Scholar]
- Melnyk S, Pogribna M, Pogribny IP, Yi P, James SJ (2000) Measurement of plasma and intracellular S-adenosylmethionine and S-adenosylhomocysteine utilizing coulometric electrochemical detection: alterations with plamsa homocysteine and pyridoxal 5′-phosphate concentrations. Clin Chem 46:265–272 [PubMed] [Google Scholar]
- Miniou P, Jeanpierre M, Bourchis D, Barbosa ACC, Blanquet V, Viegas-Pequignot E (1997) α-satellite DNA methylation in normal individuals and in ICF patients: heterogeneous methylation of constitutive heterochromatin in adult and fetal tissues. Hum Genet 99:738–745 [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD (1998) Transcriptional repression by the methyl CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386–389 [DOI] [PubMed] [Google Scholar]
- Nan X, Tate P, Li E, Bird A (1996) DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol 16:414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou CY, Stevenson RE, Brown VK, Schwartz CE, Allen WP, Khoury MJ, Rozen R, Oakley GP, Adams MJ (1996) 5,10 methylenetetrahydrofolate reductase genetic polymorphism as a risk factor for neural tube defects. Am J Med Genet 63:610–614 [DOI] [PubMed] [Google Scholar]
- Picton H, Briggs D, Gosden R (1998) The molecular basis of oocyte growth and development. Mol Cell Endocrinol 145:27–37 [DOI] [PubMed] [Google Scholar]
- Piyathilake CJ, Macaluso M, Hine RJ, Richards EW, Krumdieck CL (1994) Local and systemic effects of cigarette smoking on folate and vitamin B-12. Am J Clin Nutr 60:559–566 [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Basankian AG, Miller BJ, Lopatina NG, Poirier LA, James SJ (1995) DNA strand breaks in genomic DNA and within the p53 gene are associated with hypomethylation in livers of folate/methyl deficient rats. Cancer Res 55:1894–1901 [PubMed] [Google Scholar]
- Pogribny IP, Muskhelishvili L, Miller BJ, James SJ (1997) Presence and consequence of uracil in preneoplastic DNA from folate/methyl deficient rats. Carcinogenesis 18:2071–2076 [DOI] [PubMed] [Google Scholar]
- Renauld H, Gasser SM (1997) Heterochromatin: a meiotic matchmaker? Trends Biol Sci 7:201–205 [DOI] [PubMed] [Google Scholar]
- Rosenblatt DS (1999) Folate and homocysteine metabolism and gene polymorphisms in the etiology of Down syndrome. Am J Clin Nutr 70:429–430 [DOI] [PubMed] [Google Scholar]
- Smith G, Berg J (1976) Down's anomaly. 2d ed. Churchill Livingstone, Edinburgh and New York [Google Scholar]
- Titenko-Holland N, Jacob RA, Shang N, Balaraman A, Smith MT (1998) Micronuclei in lymphocytes and exfoliated buccal cells of postmenopausal women with dietary changes in folate. Mutat Res 417:101–114 [DOI] [PubMed] [Google Scholar]
- Torfs CP, Christenson RE (1999) Maternal risk factors and major associated defects in infants with Down syndrome. Epidemiology 10:264–270 [PubMed] [Google Scholar]
- Vig BK (1998) Centromere: a candidate for face-lift. Environ Mol Mutagen 32:197–199 [PubMed] [Google Scholar]
- Vig BK, Hallet WH (2000) 5-Azacytidine-induced and Hoechst-induced aneuploidy in Indian muntjac. Mutat Res 466:79–86 [DOI] [PubMed] [Google Scholar]
- Vilain A, Vogt N, Dutrillaux B, Malfoy B (1999) DNA methylation and chromosome instability in breast cancer cell lines. FEBS Lett 460:231–234 [DOI] [PubMed] [Google Scholar]
- Wilson A, Platt R, Wu RK, Leclerc D, Christensen B, Yang HT, Rozen R (1999) A common variant in methionine synthase reductase combined with low cobalamin (vitamin B12) increases risk for spina bifida. Mol Genet Metab 67:317–323 [DOI] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourchis D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu XY, Russo TT, Veigas-Pequignot E (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402:187–191 [DOI] [PubMed] [Google Scholar]
- Yang Q, Sherman SL, Hassold TJ, Allran K, Taft LF, Pettay D, Khoury MJ, Erickson JD, Freeman SB (1999) Risk factors for trisomy 21: maternal cigarette smoking and oral contraceptive use in a population-based case control study. Genet Med 1:80–88 [DOI] [PubMed] [Google Scholar]