Abstract
Dyslexia is a common and complex disorder with evidence for a genetic component. Multiple loci (i.e., quantitative-trait loci [QTLs]) are likely to be involved, but the number is unknown. Diagnosis is complicated by the lack of a standard protocol, and many diagnostic measures have been proposed as understanding of the component processes has evolved. One or more genes may, in turn, influence these measures. To date, little work has been done to evaluate the mode of inheritance of individual component—as opposed to composite—phenotypes, beyond family or twin correlation studies that initially demonstrate evidence for a genetic basis of such components. Here we use two approaches to segregation analysis in 102 nuclear families to estimate genetic models for component phenotypes associated with dyslexia: digit span and a nonword-repetition task. Both measures are related to phonological skills, one of the key component processes in dyslexia. We use oligogenic-trait segregation analysis to estimate the number of QTLs contributing to each phenotype, and we use complex segregation analysis to identify the most parsimonious inheritance models. We provide evidence in support of both a major-gene mode of inheritance for the nonword-repetition task, with ∼2.4 contributing QTLs, and for a genetic basis of digit span, with ∼1.9 contributing QTLs. Results obtained by reciprocal adjustment of measures suggest that genes contributing to digit span may contribute to the nonword-repetition score but that there are additional QTLs involved in nonword repetition. Our study adds to existing studies of the genetic basis of composite phenotypes related to dyslexia, by providing evidence for major-gene modes of inheritance of these single-measure component phenotypes.
Introduction
Dyslexia, or specific reading disability, is a common and complex disorder. As one of several learning disabilities it is characterized by difficulties in single-word decoding, resulting in failure to acquire reading proficiency. Although initial manifestation and diagnosis usually occurs during childhood, with 5%–10% of school-age children affected (Shaywitz et al. 1990), dyslexia may have long-term educational, economic, and social repercussions. Diagnosis is complicated by the educational experience and the lack of a standard protocol regarding which measures should be used for diagnosis (Berninger 1994). Many measures have been proposed, as understanding of the component processes has evolved. Two of the component processes increasingly recognized as being important to making the diagnosis include (1) orthographic coding—that is, ability to code written words into short-term memory and to represent them in long-term memory, and (2) phonological coding—that is, the ability to code spoken words into short-term memory, manipulate component sounds, and reproduce words, without the aid of meaning cues (Berninger et al., in press). Finally, dyslexia is believed to be a language-based disorder (Vellutino 1979; Decker and DeFries 1981; Liberman and Karlin 1984; Pennington et al. 1990), rather than a visual or perceptual disorder; however, this language-based disorder may also involve deficits in physiological mechanisms of the visual system (e.g., see Eden et al. 1996).
A genetic basis of dyslexia has long been suspected. Although the earliest evidence was observational (Fisher 1905; Hinshelwood 1907; Stephenson 1907), evidence for a genetic basis has since been obtained by more-systematic twin and family studies. Increased concordance rates in MZ versus DZ twin pairs, for a diagnosis of dyslexia (Stevenson et al. 1987); stronger correlations for MZ than for DZ pairs, for continuous-trait measures (Olson et al. 1994; Bishop et al. 1996); and more regression to the mean for DZ than for MZ pairs, for continuous-trait measures (DeFries et al. 1987), all provide evidence for a genetic basis of both a dichotomous diagnosis and associated continuous traits. Family studies provide further support for this conclusion: the risk of reading impairment in first-degree relatives of probands exceeds that in the general population (Hallgren 1950; Pennington et al. 1991); phenotypes of probands are predictive of those of siblings (Decker and DeFries 1981); the risk and severity of the disorder increase with the number of affected parents (Wolff and Melngailis 1994); and correlations among first-degree relatives are significantly greater than 0, for some continuous-component measures (Raskind et al., in press). Finally, results of two complex segregation analyses (CSAs) provide evidence not only for positive correlations among relatives but also for a pattern of inheritance consistent with a Mendelian mode of inheritance, for both a dichotomous definition of dyslexia (Pennington et al. 1991) and a phenotype based on factor scores from multiple continuous measures (Gilger et al. 1994). A third, older segregation analysis (Lewitter et al. 1980) gave equivocal results, but it was based on a small sample size and predated development of CSA methods; thus these results may have suffered from lack of power and from inability to accommodate a more complex model.
There is also consensus, from a number of observations, that there is familial and etiological heterogeneity in dyslexia. First, heterogeneity is typical of complex traits: among the complex traits for which genes have been identified, there are, as yet, no exceptions to this finding. It is worth noting that accompanying this heterogeneity are sometimes subtle phenotypic differences associated with the different genetic forms of the diseases—such as differences in age at onset or presence/absence of other disease phenotypes, as is observed in Alzheimer disease and breast cancer (Hall et al. 1990; Schellenberg et al. 1994a; Schellenberg et al. 1994b; Wooster et al. 1994; Levy-Lahad et al. 1998). Second, for dyslexia there have now been a number of reports of positive evidence for linkage to different genomic regions (Smith et al. 1983; Cardon et al. 1994; Grigorenko et al. 1997; Fagerheim et al. 1999; Fisher et al. 1999; Gayan et al. 1999; Petryshen et al. 1999). Several of these studies have provided moderately strong evidence for linkage to the same genomic region on chromosome 6p, with similar phenotypic measures providing, in more than one study, evidence for linkage; however, the significance levels are sometimes sensitive to the method of analysis used (Grigorenko et al. 1997, 2000) and remain low for a complex trait (Lander and Kruglyak 1995), and there are also some reports of failure to confirm results (Bisgaard et al. 1987; Field and Kaplan 1998; Petryshen et al. 2000), as well as an earlier report (Cardon et al. 1995) of one correction that weakened the significance level. In addition, highly correlated phenotypes do not necessarily provide similar support for linkage to the same region (Grigorenko et al. 2000). Third, even in regions for which more than one group has reported evidence for linkage, in some cases the evidence for linkage is found only in a specific subset of phenotypically defined families. Finally, the diagnosis of dyslexia and the interpretation of results among studies is complicated by use of variable measures and procedures both among different research groups and among the same and different research groups over time. These differences in how the phenotype has been measured may account for some of the observed heterogeneity in results reported by mapping studies. The resultant diagnostic heterogeneity is likely to be one of the most serious obstacles in mapping—and eventually cloning—relevant genes contributing to dyslexia.
One approach to reduction of this heterogeneity for subsequent mapping studies is to perform a detailed evaluation of the genetic basis of phenotypic components of dyslexia. This can lead both to increased understanding of the heterogeneity and to the possibility of identification, on the basis of phenotypic criteria, of pedigree subgroups that are genetically more homogeneous. This strategy of careful phenotypic analysis may lead to improved prognoses for linkage detection, as well as to more-accurate fine-scale mapping of genes contributing to this complex trait (Wijsman and Amos 1997). The phenotypes associated with dyslexia are multivariate—several correlated language measures are predictive of affected status and/or are used for diagnosis (Berninger et al., in press). However, there are technical difficulties in the use of multivariate distributions in the full range of analyses that are involved in genetic studies. Consequently, one approach that may be an efficient way of eventually identifying the genes contributing to complex traits is to focus on individual univariate components of the phenotype and to adjust for correlated measures as covariate effects (Goddard et al. 1995; Wijsman and Amos 1997; Hokanson et al. 1999). For example, a focus on single-component phenotypes permits use of CSA (Morton and MacLean 1974) for construction of genetic models for the phenotypes. With a genetic model, it is then possible to use model-based gene-mapping methods, which not only are more powerful than model-free methods when the trait model is reasonably well estimated (Vieland et al. 1992; Greenberg et al. 1996; Wijsman and Amos 1997) but also provide a framework in which to estimate and refine gene location once evidence for linkage is found. This general approach to covariate adjustment followed by genetic modeling and linkage analysis was recently shown to be one of the most successful approaches in the analysis of simulated complex traits (Wijsman and Amos 1997) and recently has proved to be helpful in real studies of component phenotypes related to some other complex continuous-trait phenotypes in which strong evidence for linkage was eventually found (Hokanson et al. 1999; Knoblauch et al. 2000). Therefore, it seems likely that it could also prove to be a useful approach for the study of the genetic basis of dyslexia.
Phonological skills appear to play a major role in the development of reading skills and thus, in the study of the genetic basis of dyslexia, are candidates for component phenotypes. Several studies have reported that most individuals with dyslexia exhibit impairments in phonological skills, including phonological coding and phonological awareness (Pennington et al. 1987; Olson et al. 1989; Field and Kaplan 1998; Berninger et al., in press). Previous twin and family studies also provide strong evidence that phonological awareness (Olson et al. 1994) and phonological coding (Olson et al. 1991, 1994; Raskind et al., in press) are correlated among family members of dyslexic probands, as would be predicted if there is a genetic basis to these phenotypes. In addition, recent studies have shown that phonological short-term memory, as assessed by verbal repetition of nonwords (Wagner and Torgesen 1999), and verbal short-term memory, as assessed by digit span, both show strong correlations with each other (correlation coefficient [ρ] .45) and with other measures used to diagnose dyslexia (ρ=.27–.47, depending on the measure); these correlations are found both in probands and in family members of dyslexic probands (Berninger et al., in press; Raskind et al., in press). Additional interest in phonological short-term memory as a candidate for a genetically influenced trait also comes from three observations: (1) measures of phonological short-term memory and verbal short-term memory show evidence for a role in specific language impairment (Gathercole and Baddeley 1990) as well as in reading disabilities (Kamhi and Catts 1986); (2) there is evidence for a genetic basis of performance on phonological short-term–memory tasks, from twin studies in families ascertained through a child with a language impairment (Bishop et al. 1996); and (3) there is a recent report of successful gene localization for an autosomal dominant form of language impairment in which a phonological short-term–memory measure discriminates between normal and affected individuals (Fisher et al. 1998). Although specific language impairment and dyslexia are different disorders, the observation that phonological short-term memory may have a genetically influenced contribution for both disorders suggests that, even if the underlying mechanisms prove to be different, the phenotype may be of interest for an understanding of several language-based disorders.
These observations suggest that a deeper evaluation of the genetic basis of phonological short-term memory may be useful in the study of dyslexia. Therefore, here we present the results of segregation analyses for (1) an operational measure of phonological short-term memory defined by a nonword-repetition task and (2) an operational measure of verbal short-term memory—that is, digit span—that, elsewhere, we have shown to be correlated with, although not identical to, this nonword-repetition task (Raskind et al., in press). Our working hypothesis in considering these two phenotypes together is that both measures are related to phonological skills—digit span through ability to recall a series of highly familiar names for numbers, thus representing a measure of verbal short-term memory, and the nonword-repetition task through ability to recall a novel made-up word, thus combining short-term memory with ability to precisely represent, in phonological short-term memory, all the phonemes in such a nonword. Evidence for shared genetic effects could suggest common pathways to phonological short-term memory, whereas unique genetic effects may reflect differences in stimulus properties of these two measures. Elsewhere, we have shown that both measures demonstrate an aggregation pattern in families that is consistent with an inherited basis (Raskind et al., in press), but our previous study did not further examine possible modes of inheritance. Our goals in the present study were, first, to estimate the number of genes that play a role in each phenotype; second, to determine whether a major-gene model is compatible with inheritance of each phenotype in pedigrees ascertained through a dyslexic proband and to obtain estimates for the parameters of the most parsimonious genetic model(s); and, third, to obtain insights into the relationship between the effects that genetic factors have on each of these two correlated phenotypes. We provide evidence in support of a major-gene mode of inheritance for scores on the nonword-repetition task, with a small number of contributing loci. There is also evidence for a genetic basis of digit span, with somewhat fewer contributing loci than for the nonword-repetition task. The results also suggest that genes contributing to digit span may contribute also to the nonword-repetition task but that, beyond those that influence digit span, there are additional genetic factors involved in nonword repetition.
Subjects and Methods
Subjects
A sample of 102 nuclear families consisting of a total of 409 individuals were used in the analyses. Because we have elsewhere given extensive descriptions of the sample and its ascertainment (Berninger et al., in press; Raskind et al., in press), only a brief description will be supplied here. Potential probands who struggled in learning to read were identified through referral by parents, school psychologists, or special educators, without regard to family size or family history of difficulty in learning to read. Subsequent eligibility for proband status required meeting the researcher-defined criteria for learning disabilities, as based on test scores on a battery of measures administered by the research team. Proband exclusion criteria consisted of developmental history or diagnosis of mental retardation, developmental delay, primary language disorder, neurological disorder such as traumatic brain injury or seizure disorder, or psychiatric disorder including attention-deficit–hyperactivity disorder (ADHD) without a reading problem. Subjects qualified as probands if their reading level was below the age-specific population mean, their prorated verbal IQ was ⩾90, and there was ⩾1 SD discrepancy between verbal IQ and outcome on at least 1 of 10 reading or writing measures, with a score, on the measure, that was below the population mean (Berninger et al., in press; Raskind et al., in press).
The average proband met this inclusion criterion on 7.5 measures, and 25 of the 102 probands met inclusion criteria on all 10 measures. All of the measures used are conventional measures used in schools and research: the Woodcock Reading Mastery Test–Revised, for word identification or word attack (Woodcock 1987); the prepublication version of the real and pseudoword subtests of the Test of Word Reading Efficiency (Torgesen et al. 1999), for real-word or pseudoword reading efficiency; Gray Oral Reading Test–Third Edition, for rate or accuracy; the Wechsler Individual Achievement Test (Wechsler 1992); the Wide Range Achievement Test–Third Edition (Wilkinson 1993); The Woodcock-Johnson Psychoeducational Battery–Revised (Woodcock and Johnson 1990), for writing-fluency subtest; and an alphabet task (Berninger and Rutberg 1992). Most of the probands were severely impaired on most of the measures given, with an average discrepancy of 1.47–2.14 SD below the verbal IQ on the different diagnostic measures. Use of IQ-discrepancy measures to diagnose dyslexia has been shown to result in higher estimates of heritabilities than does use of IQ-nondiscrepancy measures (Olson et al. 1999) and, therefore, is likely to result in a sample that, for genetic studies, is more useful than alternative diagnostic schemes. All first-degree relatives of all probands were subsequently invited to participate in the study if they were age >6.5 years. All participating family members were given the same test battery as was the proband, with the exception that family members age ⩾17 years were administered an adult version rather than a juvenile version of the verbal IQ test (see below). The ethnic background of the 102 probands was 89 white, 5 Native American, 3 Hispanic, 2 Asian, 2 African American, and 1 East Indian. The study was approved by the University of Washington institutional review board, and informed consent was obtained from participants.
Measures
The present study used a subset of the 24 psychometric measures collected on the sample (Berninger et al., in press; Raskind et al., in press). Two of the continuous measures that previously had been shown to correlate significantly among relatives were used in the segregation analysis described here: the nonword-memory task of the prepublication version of the Comprehensive Test of Phonological Awareness (Wagner and Torgesen 1999), hereafter referred to as “nonword memory,” and the digit span subtest of the Wechsler Adult Intelligence Scale–Revised (Wechsler 1981) and the Wechsler Intelligence Scale for children–Third Edition (Wechsler 1992), hereafter referred to as “digit span.” These two measures also had been shown to be strongly positively correlated both within the probands and within their first-degree relatives (Raskind et al., in press). Nonword memory was scored on a unit-normal scale; digit span was scored on a scale with mean 10 and SD 3. In addition to these measures, the remainder of the verbal scale of the Wechsler Intelligence Scale was administered to each subject (with the exception of the Arithmetic subtest), with the WISC-III (Wechsler 1992) used for children age <17 years and the WAIS-R (Wechsler 1992) used for individuals age ⩾17 years. Verbal IQ was scored with a mean of 100 and SD 15. Note that digit span contributes to verbal IQ in adults but not to that in children.
Statistical Analyses
The primary goal of the analyses was to determine whether there is evidence for the existence of a major-gene pattern of inheritance for each of the two continuous measures—nonword memory and digit span. Segregation analysis was used to evaluate evidence for such a possible genetic basis, with or without the influence of additional genes. In all cases, the continuous-trait level obtained from an individual item from the test battery was used as the phenotype, rather than as a dichotomous affected/unaffected diagnosis. This should increase power to detect major-gene effects, compared with use of the qualitative diagnosis, because use of a continuous trait substantially increases the available information with which to estimate genetic-model parameters and to discriminate among models (Goddard et al. 1995; Wijsman and Amos 1997).
Two types of segregation analysis were carried out. The first was based on an oligogenic segregation-analysis approach that is implemented via a Bayesian Monte Carlo Markov-chain (MCMC) method, as described elsewhere (Heath 1997; Daw et al. 1999) and implemented in the Loki computer program. The second approach was a CSA (Morton and MacLean 1974), as parameterized by the class D model of the logistic-regression approach (Bonney 1986) and implemented in the REGC program in the S.A.G.E. (1997) package. The two approaches differ in how multiple trait loci are considered. CSA consolidates the effects of loci beyond the single, modeled, Mendelian locus, into the polygenic component of the mixed model. Parameters of the model are estimated by maximum likelihood, and the parameter space in any one analysis run is fixed. Different models are fitted to the data, and, ideally, a single, most parsimonious model can be obtained by choosing among models, with likelihood-ratio tests being used to compare nested models. It is also possible to correct for ascertainment, when ascertainment is through a single proband within each family without regard to family history. In contrast, the MCMC approach is an oligogenic approach, which models several loci, simultaneously, as Mendelian loci, but which does not include a polygenic component. The number of parameters describing the mode of inheritance, including the number of loci in the model, is allowed to vary, but estimation involves a stochastic element associated with use of Monte Carlo methods. No single model is identified to the exclusion of others, but a series of models with associated posterior probabilities can be obtained. Currently, it is not possible to apply ascertainment corrections within the MCMC analyses. The MCMC analyses were used primarily to estimate the number of loci contributing to each trait, whereas the CSAs were used to identify the most parsimonious model(s) and to estimate parameters for such models.
MCMC segregation analyses
A complete description of the Bayesian MCMC method of analysis used here is beyond the scope of the current paper. Descriptions of the underlying method, with some guidelines for use, can be found elsewhere (Heath 1997; Daw et al. 1999, 2000). However, a brief description of the MCMC approach is as follows. We assume a model that relates the phenotype vector, y, to an additive function of covariate and genotype effects: y=μ+Xβ+Σki=0Qiαi+e. In this model, μ is an overall population “baseline” phenotype level, X is a matrix containing the values for all covariates in all individuals, β is the vector of covariate effects, Qi is a matrix indicating the genotypes in all individuals for quantitative-trait locus (QTL) i, αi is the vector of effects for QTL i, e is a normally distributed residual effect, and k is the number of QTLs currently considered in the model. With the exception of the elements of y and X, which are observed, all of these parameters, including the value of k in a particular iteration of the Monte Carlo sampler, are estimated via the MCMC process, as detailed elsewhere (Heath 1997). In brief, this MCMC process uses importance sampling (Hammersley and Handscomb 1964) in which values for model parameters are selected at random and then either accepted or not, according to a Metropolis-Hastings acceptance ratio (Metropolis et al. 1953; Hastings 1970). After many iterations, the sampled model configurations provide an estimate of the posterior probability distribution over the space of possible parameter configurations, including the number of QTLs in the model. In the analyses presented here, this process was run for 100,000 iterations, with every iteration used to estimate the posterior distributions.
In the absence of linked markers, it is not generally useful to try to obtain, from the MCMC segregation analyses, parameter values for specific QTLs: the birth-death process by which the sampler changes the number of parameters of the model space means that individual QTLs among iterations of the MCMC process cannot be individually related to each other. However, it is possible to obtain composite estimates of certain parameters of interest, including the number of QTLs, total genetic-variance contribution, and covariate effects.
The MCMC analyses require specification of prior distributions for unknown model parameters. The number of QTLs in the model was assumed to be drawn from either a uniform distribution on 0–16 or a Poisson distribution with mean 2. The results were unaffected by this choice of prior distribution, so only the results from the Poisson distribution will be shown. The variance contribution for each QTL was assumed to be distributed as N(0,τβVe), where Ve is the residual variance, and the value for τβ was chosen by successively doubling and halving an initial value based on the total variance, until estimates for the overall mean in the model stabilized. The frequencies for the two alleles for each QTL were drawn from a uniform distribution on 0–1.
CSAs
The basic model for a CSA can be written as y=μ+g+a+e, where y is the phenotype, μ is an overall mean, g is the major-gene effect (the modeled Mendelian locus), a is the effect of other familial components (other genes and shared environment), and e is a random environmental effect. Up to four classes of models were considered in each segregation analysis: the environmental model assumed that there were no familial correlations and, therefore, that y=μ+e; the Mendelian models assumed that the familial correlation could be explained by a single, Mendelian locus, so that y=μ+g+e; the polygenic models assumed that there were familial correlations but that these could be modeled by a polygenic component, so that y=μ+a+e; and the mixed models assumed that all components were needed to explain the observed pattern of phenotypic data on the pedigrees, so that y=μ+g+a+e.
All genetic models fitted to the data included one Mendelian submodel and/or one polygenic submodel. The mixed model included one submodel from both the Mendelian and polygenic classes of submodels, whereas the Mendelian and polygenic models included only one submodel. Within each class of models that included a Mendelian component, three submodels were considered (table 1). Each assumed a diallelic Mendelian locus with alleles A and B and with allele frequencies pA and pB=1-pA. The genotype effects for the three genotypes AA, AB, and BB are gAA, gAB, and gBB. For the dominant/recessive submodel, MD, gAB=gAA. (The dominant and recessive models are symmetric and equivalent for a continuous trait, so that, henceforth, we will refer only to the dominant model.) For the additive submodel, MA, gAB=(gAA+gBB)/2. For the general submodel, MG, all three mean genotype effects (gAA, gAB, and gBB) were estimated: no constraints were placed on the position of the heterozygote relative to the homozygotes. By reparameterizing the model, the difference between homozygous means can be described as δ, δ=(gAA-gBB), and the dominance, d, can be described as d=(gAB-gBB)/δ. The dominant and additive models therefore require estimation of three parameters—pA, gAA, and gBB—whereas the general model requires, in addition, the estimation of a fourth parameter, gAB. Up to five submodels (table 2) were considered within each class of models that included a polygenic component. A sixth model, the environmental model, in which no family correlations were estimated, also was considered. For this model, only the residual variance was estimated. The polygenic models included up to three types of familial correlations: sibling correlations, ρSS; parent-offspring correlations, ρPO; and spouse correlations, ρFM. In addition, for some models, the possibility of different mother-offspring correlations, ρMO, and different father-offspring correlations, ρPO, was considered.
Table 1.
Mendelian Submodels
|
Parametera |
||||
| Model | pA | gAA | gAB | gBB |
| MD | + | + | (gAA) | + |
| MA | + | + | (gAA+gBB/2) | + |
| MG | + | + | + | + |
A plus sign (+) denotes that the parameter is estimated in model; parentheses indicate that the value is constrained by other parameters.
Table 2.
Polygenic Submodels[Note]
|
Parameter |
|||||
| Model Symbol | ρFM | ρPO | ρMO | ρFO | ρSS |
| P1 | (0) | (ρSS) | − | − | + |
| P2 | (0) | + | − | − | + |
| P3 | + | + | − | − | + |
| P4 | (0) | − | + | + | + |
| P5 | + | − | + | + | + |
Note.— Data are as defined in the footnote to table 1.
For most analyses with a Mendelian submodel, the transmission probabilities were fixed at Mendelian proportions. In such analyses, the probabilities of transmitting an A allele from genotypes AA, AB, or BB were assumed to be the Mendelian probabilities of τAA=1, τAB= 1/2, or τBB=0, respectively. For model validation, when the most parsimonious model included a Mendelian component, τAB was no longer fixed at 1/2 but was estimated along with the parameters of the most parsimonious model.
CSA models were fitted both with and without a correction for ascertainment of the proband. Because the proband was ascertained through a complex phenotype rather than through the value on a single phenotypic measure, an optimally efficient ascertainment correction based on a threshold value of a single test value for the proband was not possible. Because of both the complex phenotype used for proband ascertainment and the limitations in the analysis package for the choice of ascertainment correction, the most appropriate ascertainment correction that was available as part of the analysis package was that which was based on the trait value of the proband.
Likelihood-ratio tests were used to compare and choose among nested models. For two models, 1 and 2, with likelihoods L1 and L2, respectively, where model 1 is nested within model 2, −2 ln (L1/L2) is asymptotically distributed as a χ2 distribution with f degrees of freedom, where f is the difference, in number of estimated parameters, between the two models. The environmental model plus all single submodels in tables 1 and 2 were fitted, plus all models consisting of one submodel from table 1 and one submodel from table 2. To thoroughly explore the likelihood surface, multiple starting configurations spanning the full range of legal parameter values were always tried. Only when each local maximum was identified more than once was it assumed that the likelihood surface had been adequately explored. Where no clear discrimination between models could be made, the Akaike information criterion (AIC), AIC = −2ln(likelihood) + 2(parameters fit) (Akaike 1974), was used to compare nonnested models. A smaller AIC indicates better fit.
Estimates of heritability
For the major-gene components in models fitted to the data, heritability was computed as the ratio of the additive genetic variance, σ2a, to total variance, σ2t, or h2=σ2a/σ2t. For the CSAs, since there was only one major gene in the model, this required computation of only σ2a from the major-gene–model parameters. For the MCMC analyses, heritability was averaged over all iterations, with the additive genetic variance, σ2a(k), in an iteration with k loci, computed as  , where σ2ai is the contribution to the additive genetic variance of QTL i.
, where σ2ai is the contribution to the additive genetic variance of QTL i.
Covariate adjustments
A number of covariates were included in all analyses. We have noted elsewhere the effects of age, sex, and verbal IQ on many of the studied measures (Raskind et al., in press). Therefore, in all analyses, these three variables were included as covariates. Digit span also was included as a covariate in some analyses of nonword memory. The rationale for this was that (1) the two variables were correlated in both probands and family members (Raskind et al., in press), and (2) preliminary aggregation analyses suggested that there were shared genetic components to these two traits, by showing that correlations among family members were reduced when each variable was used as a covariate in the aggregation analysis of the other variate (results not shown).
Results
Number of Contributing Genes
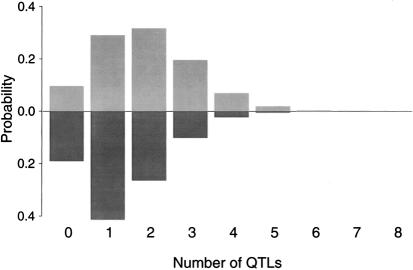
There is evidence that least one gene contributes to phenotypic variance, for both nonword memory and digit span. Figures 1 and 2 give estimates obtained from the MCMC segregation analyses of the posterior probability that different numbers of QTLs contribute to each of the phenotypes analyzed. The number of QTLs affecting nonword memory is estimated as a mean of 2.36, with a posterior probability of .99 that at least one gene contributes to nonword memory, adjusted for age, sex, and verbal IQ (fig. 1). The posterior probability of at least two genes contributing to the nonword-memory phenotype is .77. The number of QTLs estimated to affect digit span is estimated as a mean of 1.93, with a posterior probability of .90 that at least one gene contributes to digit span, adjusted for age, sex, and verbal IQ (fig. 2). The posterior probability of at least two genes contributing to digit span is .61.
Figure 1.
Estimated number of underlying QTLs contributing to nonword memory, adjusted for age, sex, and verbal IQ, (gray-shaded bars, above the baseline) or for age, sex, verbal IQ, and digit span (black bars, below the baseline).
Figure 2.
Estimated number of underlying QTLs contributing to digit span, adjusted for age, sex, and verbal IQ (gray-shaded bars, above the baseline) and for age, sex, verbal IQ, and nonword memory (black bars, below the baseline).
Estimates for the number of genes contributing to nonword memory and to digit span decrease when each phenotype is used as a covariate in the analysis of the other phenotype. When nonword memory is adjusted for digit span, the number of genes estimated to affect the nonword phenotype drops to a mean of 1.93, and the posterior probabilities of at least one and at least two genes falls to .93 and .62, respectively (fig. 1). When digit span is adjusted for the effects of nonword memory, the estimate of the number of contributing QTLs for digit span drops to 1.37, and the estimated posterior probabilities of at least one and at least two contributing QTLs fall to .81 and .40, respectively (fig. 2).
Mixed-Model Segregation Analyses
Nonword memory
Segregation analysis of nonword memory, adjusted for the basic covariates only, suggested that the most parsimonious model is a general Mendelian model with an intermediate heterozygous phenotype with dominance of ∼.8 but with no residual polygenic background. The parameter values and likelihoods for a subset of the models considered in CSA of nonword memory are given in table 3.
Table 3.
Model Parameter Estimates Obtained from CSA of Nonword Memory, Adjusted For Age, Sex, and Verbal IQ
|
Parametera |
|||||||||||
| Model | Ve | pA | τAB | δ | d | ρFM | ρPO | ρMO | ρFO | ρSS | −2lnL |
| Environmental | .65 | … | … | … | … | … | … | … | … | … | 741.30 |
| MG | .42 | .84 | (.5) | 2.45 | .8 | … | … | … | … | … | 710.84 |
| MD | .49 | .84 | (.5) | 2.17 | (1) | … | … | … | … | … | 718.15 |
| MA | .49 | .92 | (.5) | 1.70 | (.5) | … | … | … | … | … | 726.11 |
| P1 | .64 | … | … | … | … | (0) | (ρSS) | … | … | .13 | 730.26 |
| P5 | .64 | … | … | … | … | −.14 | … | .07 | .11 | .18 | 727.61 |
| MG+P1 | .48 | .86 | (.5) | 2.5 | .89 | (0) | (ρSS) | … | … | .11 | 708.59 |
| MG+P5 | .48 | .86 | (.5) | 2.54 | .9 | −.27 | … | .01 | .17 | .13 | 703.14 |
| MG+τ | .42 | .85 | .38 | 2.42 | .79 | … | … | … | … | … | 709.20 |
Parentheses indicate that the value is constrained by other parameters.
Initial comparisons among models suggested that a transmissible component is necessary to explain the inheritance pattern of nonword memory, adjusted for the basic covariates. Comparison between the environmental model and the simplest polygenic model (P1) gave strong evidence that the polygenic model provided a better fit to the data than did the environmental model (χ2=11.04, 1 df, P<.001). A comparison of the environmental model and the general Mendelian model, MG, also provided strong evidence that the Mendelian model provided the better fit (χ2=30.46, 4 df, P<.001), as did both more-restricted Mendelian models compared with the environmental model (χ2=23.00, 3 df, for the Mendelian dominant model, MD, vs. the environmental model, P<.001; χ2=15.19, 3 df, for the Mendelian additive model, MA, vs. the environmental model, P<.005).
A Mendelian component appeared to be important. A mixed model containing both a polygenic component and a major-gene component (MG+P1) fitted the data significantly better than did the submodel model containing only P1 (χ2=21.67, 4 df, P<.001). Comparison among Mendelian models suggested that the general major-gene model, MG, was superior to both the additive major-gene model, MA, (χ2=15.27, 1 df, P<.001), and to the dominant major-gene model, MD, (χ2=7.31, 1 df, P<.01). Within the class of models containing only a polygenic component, the most parsimonious model, P1, had equal sibling and parent-offspring correlations. This model was not significantly worse than the next most complicated polygenic model, P2 (χ2=1.34, 1 df), which allowed the sibling correlation to differ from parent-offspring correlation.
A mixed model was not significantly better than a major-gene model without a polygenic component. When MG was present, the polygenic component, P1, of the mixed model was unnecessary, compared with a model that contained only MG (χ2=2.25, 1 df). Mixed models with more structure also did not significantly improve the fit: the mixed model with separate sibling and parent correlations was not significantly better than the simplest mixed model with shared correlations (MG+P2 vs. MG+P1; χ2=0.06, 1 df); a model with a nonzero spouse correlation was not significantly better than one without a spouse correlation (MG+P3 vs. MG+P2; χ2=1.19, 1 df); and a model with different father-offspring and mother-offspring correlations was not better than one with a shared parent-offspring correlation (MG+P5 vs. MG+P3; χ2=0.12, 1 df). Finally, there was no evidence for non-Mendelian transmission: estimation of the heterozygous transmission rate, τAB, rather than restriction to .5, did not significantly improve the fit of the major-gene model, MG (χ2=1.64, 1 df).
Nonword memory adjusted for digit span
Segregation analysis of nonword memory, adjusted for digit span in addition to the basic covariates, suggested that, for this phenotype, the best model among those considered is a dominant Mendelian model with no residual polygenic background. However, it is also possible that a mixed model with separate mother-offspring and father-offspring correlations should be considered also. The parameter values and likelihoods for a subset of the models considered in CSA are given in table 4.
Table 4.
Model Parameter Estimates Obtained from CSA of Nonword Memory, Adjusted for Age, Sex, Verbal IQ, and Digit Span
|
Parametera |
|||||||||||
| Model | Ve | pA | τAB | δ | d | ρFM | ρPO | ρMO | ρFO | ρSS | −2lnL |
| Environmental | .57 | … | … | … | … | … | … | … | … | … | 691.62 |
| MG | .41 | .87 | (.5) | 2.57 | .84 | … | … | … | … | … | 661.97 |
| MD | .44 | .87 | (.5) | 2.46 | (1) | … | … | … | … | … | 664.47 |
| MA | .45 | .96 | (.5) | 2.11 | (.5) | … | … | … | … | … | 679.02 |
| P1 | .56 | … | … | … | … | (0) | (ρSS) | … | … | .09 | 686.84 |
| P2 | .56 | … | … | … | … | (0) | .07 | … | … | .11 | 686.53 |
| P3 | .56 | … | … | … | … | −.12 | .07 | … | … | .11 | 685.56 |
| P4 | .56 | … | … | … | … | (0) | … | .01 | .14 | .11 | 684.73 |
| P5 | .56 | … | … | … | … | −.12 | … | .0 | .14 | .11 | 683.81 |
| MD+P1 | .44 | .88 | (.5) | 2.48 | (1) | (0) | (ρSS) | … | … | .07 | 661.87 |
| MD+P2 | .44 | .88 | (.5) | 2.49 | (1) | (0) | .05 | … | … | .09 | 661.59 |
| MD+P3 | .44 | .87 | (.5) | 2.48 | (1) | −.18 | .05 | … | … | .10 | 659.30 |
| MD+P4 | .43 | .87 | (.5) | 2.46 | (1) | (0) | … | −.08 | .17 | .09 | 655.01 |
| MD+P5 | .44 | .87 | (.5) | 2.45 | (1) | −.17 | … | −.10 | .18 | .09 | 652.99 |
| MD+τ | .44 | .88 | .41 | 2.44 | (1) | … | … | … | … | … | 664.01 |
| MD+P4+τ | .43 | .87 | .41 | 2.43 | (1) | (0) | … | −.08 | .18 | .09 | 654.48 |
Parentheses indicate that the value is constrained by other parameters.
Initial comparisons suggested that a transmissible component was necessary, when digit span was included as a covariate in the segregation analysis of nonword memory. Comparison between the environmental model and P1 gave strong evidence that the polygenic model provided a better fit to the data than did the environmental model (χ2=4.78, 1 df, P<.05). A comparison of the environmental model and MG also provided strong evidence that the Mendelian model provided the better fit (χ2=29.65, 4 df, P<.001), as did both more-restricted Mendelian models compared with the environmental model (χ2=27.15, 3 df, for MD vs. environmental model, P<.001; χ2=12.6, 3 df, for MA vs. environmental model, P<.01).
As for the previous analysis of nonword memory, the Mendelian component appeared to be important when digit span was included as a covariate. A mixed model containing both a polygenic component and a major-gene component (i.e., MD+P1) fitted the data significantly better than did the submodel model containing only P1 (χ2=24.97, 3 df, P<.001). Comparison among Mendelian models suggested that MG was superior to MA (χ2=17.05, 1 df, P<.001) but not to MD(χ2=2.5, 1 df). Within the class of models containing only a polygenic component, the most parsimonious model, P1, was also the simplest. This model was not significantly worse than any of the other polygenic models (i.e., P2–P5) considered.
A mixed model with a simple residual correlation structure was not significantly better than a major-gene model without the polygenic component, although, with a more complex correlation structure, there was some evidence for a better fit. When the major-gene component of the model MD was present, the polygenic component of the mixed model was unnecessary (MD+P1 vs. MD, χ2=2.6, 1 df; MD+P3 vs. MD, χ2=5.17, 3 df). However, when a more complicated mixed model with separate mother-offspring and father-offspring residual correlations (i.e., MD+P4) was compared with the dominant Mendelian model, there was evidence for shared, familial effects (χ2=9.46, 3 df, P<.025), although the evidence for a nonzero spouse correlation was insignificant (MD+P4 vs. MD + P5, χ2=2.02, 1 df). Finally, within the dominant Mendelian and mixed Mendelian models (i.e., MD, and MD+P4, respectively) there was no evidence for non-Mendelian transmission: estimation of the heterozygous transmission rate, τAB, did not significantly improve the model over restriction of τAB to .5, (MD+P4 vs. MD+P4+τ, χ2=0.53, 1 df; MD vs. MD+τ, χ2=0.46, 1 df).
Digit span
Segregation analysis of digit span adjusted for age, verbal IQ, and sex suggested that the most parsimonious model among those considered is a Mendelian dominant model, with no residual polygenic background. The parameter values and likelihoods for a subset of the models considered in CSA are given in table 5.
Table 5.
Model Parameter Estimates Obtained from CSA of Digit Span Adjusted for Age, Sex, and Verbal IQ
|
Parametera |
|||||||||||
| Model | Ve | pA | τAB | δ | d | ρFM | ρPO | ρMO | ρFO | ρSS | −2lnL |
| Environmental | 5.33 | … | … | … | … | … | … | … | … | … | 1,376.00 |
| MG | 3.29 | .21 | (.5) | 1.21 | 2.68 | … | … | … | … | … | 1,361.73 |
| MD | 3.51 | .22 | (.5) | 2.95 | (1) | … | … | … | … | … | 1,362.71 |
| MA | 3.80 | .23 | (.5) | 4.55 | (.5) | … | … | … | … | … | 1,365.40 |
| P1 | 5.33 | … | … | … | … | (0) | (ρSS) | … | … | .13 | 1,367.69 |
| P2 | 5.34 | … | … | … | … | (0) | .12 | … | … | .20 | 1,366.95 |
| MD+P1 | 3.51 | .22 | (.5) | 2.95 | (1) | (0) | (ρSS) | … | … | 0 | 1,362.71 |
| MD+τ | 3.30 | .25 | .36 | 3.04 | (1) | … | … | … | … | … | 1,361.96 |
Parentheses indicate that the value is constrained by other parameters.
Initial comparisons suggested that a transmissible component was necessary to explain the inheritance pattern of digit span. Comparison between the environmental model and P1 gave strong evidence that the polygenic model provided a better fit to the data than did the environmental model (χ2=8.31, 1 df, P<.005). A comparison of the environmental model and MG also provided strong evidence that the Mendelian model provided the better fit (χ2=14.27, 4 df, P<.01), as did MD compared with the environmental model (χ2=13.29, 3 df, P<.005). However, MA did not provide a significantly better fit than did the environmental model (χ2=0.98, 3 df).
Comparison among Mendelian, mixed, and polygenic models suggested that MD is the most parsimonious model. MG is only marginally superior to MA (χ2=3.67, 1 df, P<.1), but is not significantly better than MD (χ2=0.98, 1 df). In addition, MG was an overdominant model, suggestive of problems with the model. Among the Mendelian models, therefore, a Mendelian dominant model is most reasonable. Within the class of models containing only a polygenic component, P1 was the most parsimonious model: P1 was not significantly worse than either P2 (χ2=0.74, 1 df) or than P3 (χ2=1.17, 2 df). When MD and P1 were combined in a mixed model, the parameters converged to the boundary of the parameter space at MD, so that it was not possible to formally compare this mixed model with P1. When a mixed model consisting of MD and P2 was fitted, this slightly more complex mixed model was not significantly better than either MD (χ2=1.82, 2 df) or P2 (χ2=6.06, 3 df). Finally, there was no evidence for non-Mendelian transmission: estimating the heterozygous transmission rate, τAB, rather than restricting it to .5, did not significantly improve the fit of MD (χ2=0.75, 1 df). Thus it was not possible, with likelihood-ratio tests, to exclude either a Mendelian dominant or a polygenic model as being the model of inheritance. However, when the AIC is used, the Mendelian dominant model provides better fit (AIC = 1,370.71, for MD, vs. AIC = 1,371.69, for model P1).
Digit span adjusted for nonword memory
Segregation analysis of digit span adjusted for nonword memory in addition to the basic covariates did not provide evidence for an inherited basis of the adjusted phenotype. The parameter values and likelihoods for a subset of the models considered in the CSA are given in table 6. Comparison between the environmental model and P1 gave little evidence that the polygenic model provided a better fit to the data than did the environmental model (χ2=1.85, 1 df). A comparison of the environmental model and MG also provided little evidence that the Mendelian model provided the better fit (χ2=7.07, 4 df), nor did either MD compared with the environmental model (χ2=5.98, 3 df) or MA compared with the environmental model (χ2=5.01, 3 df). Comparison of more-complex mixed models with either the simpler Mendelian or polygenic models or with the simplest environmental model also failed to provide significant evidence that a model that included transmitted components fitted the data better than did an environmental model (results not shown).
Table 6.
Model Parameter Estimates Obtained for a Subset of Models from CSA of Digit Span for Age, Sex, Verbal IQ, and Nonword Memory
|
Parametera |
|||||||||||
| Model | Ve | pA | τAB | δ | d | ρFM | ρPO | ρMO | ρFO | ρSS | −2lnL |
| Environmental | 4.82 | … | … | … | … | … | … | … | … | … | 1,345.43 |
| MG | 3.2 | .82 | (.5) | .52 | 6.75 | … | … | … | … | … | 1,338.36 |
| MD | 3.49 | .5 | (.5) | 2.83 | (1) | … | … | … | … | … | 1,339.45 |
| MA | 3.93 | .11 | (.5) | 4.86 | (.5) | … | … | … | … | … | 1,340.42 |
| P1 | 4.83 | … | … | … | … | (0) | (ρSS) | … | … | .06 | 1,343.58 |
| P2 | 4.83 | … | … | … | … | (0) | .07 | … | … | .05 | 1,343.55 |
| MD+P1 | 3.49 | .5 | (.5) | 2.83 | (1) | (0) | (ρSS) | … | … | 0 | 1,339.45 |
Parentheses indicate that the value is constrained by other parameters.
Heritability and Covariate Effects
The combined fraction of the total phenotypic variance explained by the modeled Mendelian loci is 21%–35% of the total phenotypic variance (table 7), depending on the phenotype, model, and ascertainment criterion used for estimation. In general, the estimates obtained with the MCMC method were similar to those obtained with CSA, although some differences are also apparent. Estimates of heritability from the two methods of analysis tended to be more similar for the general major-gene model than for the dominant major-gene model; for example, for nonword memory adjusted for digit span, the dominant-locus heritability from CSA was only .21, whereas that for the general major-gene–model can be computed as .28, which is closer to the .30 estimated by the MCMC methods. Adjustments for the phenotypic measures resulted in an increase in the estimated heritabilities from the MCMC analyses, an effect not observed in CSA.
Table 7.
Estimates of Major-Gene Heritabilities and Covariate Effects of the Most Parsimonious Models
|
Mean (SD) for |
||||||||
| Nonword Memory |
Nonword Memory with Digit Span |
Digit Span |
Digit Span with Nonword Memory |
|||||
| MCMC | CSA-MG | MCMC | CSA-MD | MCMC | CSA-MD | MCMC | CSA-MD | |
| Heritabilitya | .28 | .30 | .30 | .21 | .24 | .33 | .35 | .097 |
| Age | .077 | .012 (.003) | .015 | .01 (.00) | .032 | .013 (.008) | .016 | .004 (.008) |
| Sexb | −.21 | −.20 (.08) | −.16 | −.15 (.08) | −.43 | −.003 (.26) | −.25 | .06 (.26) |
| Verbal IQ | .031 | .027 (.003) | .02 | .02 (.00) | .084 | .087 (.011) | .057 | .06 (.01) |
| Digit span | … | … | .11 | .11 (.02) | … | … | … | … |
| Nonword memory | … | … | … | … | … | … | .84 | .81 (.16) |
For MCMC analyses, the results are the total averaged over all loci in the model; for CSA, the results are the total for the single major-gene component of the model.
Sex effect is that of female relative to male.
Covariate effects estimated with the two segregation-analysis approaches also tend to be similar for most variables (table 7). The one apparent exception is the sex effect estimated for digit span; however, the difference seen in table 7 appears to reflect the difference in the way in which the two analysis approaches can (or cannot) take proband ascertainment into account. In table 7, the CSA results are those obtained with use of an ascertainment correction for the proband, whereas, for the MCMC results, no ascertainment correction was possible. In contrast, when CSA of digit span (without adjustment for nonword memory) was performed without an ascertainment correction, the estimated covariate effects were much closer to those obtained with the MCMC methods, with an estimated age effect of .038 and an estimated sex effect of −.42, for a major-gene dominant model, estimates that were very close to the MCMC estimates of .032 and −.43, respectively, in table 7.
Each of the covariates identified in previous aggregation analyses showed some evidence for significant covariate effects in the current segregation analyses. Even though the measures are corrected for age, there appear to be small, residual age effects. Verbal IQ was positively correlated with all the phenotypic measures, with a stronger effect for digit span than for nonword memory. The strong correlation between nonword memory and digit span is apparent in the estimated covariate effects when each measure is used as a covariate in the analysis of the other measure: for nonword memory, the difference between the likelihoods for the two models that was obtained with or without adjustment for digit span was highly significant (χ2=48.87, 1 df, P<.001, for the general major-gene model with vs. without adjustment for digit span), and a similar effect was obtained for digit span with vs. without adjustment for nonword memory (χ2=23.26, 1 df, P<.001).
Discussion
We have provided evidence for a genetic basis for performance on each of two short-term–memory tasks related to dyslexia: the nonword-memory task of the prepublication version of the Comprehensive Test of Phonological Processing (Wagner and Torgesen 1999), which is regarded as an operational measure of phonological short-term memory, and digit span, which is an operational measure of verbal short-term memory (Bishop et al. 1996). Estimation of the number of loci involved in each of these traits suggests that 1.4–2.4 underlying loci contribute to each of the two phenotypes, with somewhat stronger evidence for multiple genes contributing to nonword memory than to digit span. CSA suggests that a Mendelian model with an intermediate heterozygous phenotype best explains the inheritance pattern of nonword memory and that a major-gene dominant model best explains the inheritance pattern for digit span. The results of CSA also suggest that the evidence for a genetic basis of nonword memory is stronger than the evidence for a genetic basis of digit span. This latter conclusion is underscored by comparison of the difference between Mendelian-locus homozygous mean effects relative to the residual variance: for nonword memory, the difference between the homozygous means is ∼3.7 SDs, whereas that for digit span (unadjusted for nonword memory) it is only ∼1.6 SDs. Finally, the results also suggest that there may be genetic components shared by the two phenotypes, with additional genes contributing to nonword memory that do not influence digit span.
It may appear as if the number of genes estimated to contribute to each of the phenotypes evaluated here is lower than might be expected for a complex trait such as dyslexia. However, there are a number of issues to consider in interpreting the results. First, the phenotypes under investigation are component phenotypes, not dyslexia itself. As for other complex traits, the number of genes involved in any one component may be considerably lower than the total involved in all components related to the final phenotype. Second, CSA involves fitting an inherently single-Mendelian-locus, or major-locus, model with possibly a polygenic component, to absorb the remaining familial effects. Power to detect the polygenic component, especially in nuclear families, is not high (MacLean et al. 1975). No tools exist for easy estimation of power in this context, so it is not possible to state exactly what this would be in the current data set; however, the observation that we could discriminate among several possible genetic models, especially for nonword memory, means that there was considerable information in the data set. Third, the MCMC approach indicates that at least a small number of genes is involved, but, in terms of strength of evidence favoring specific numbers of loci, this approach is still limited by the sample. Again, there is currently no way to evaluate the power of a data set to detect loci with small contributions to the trait. Finally, although different estimates of the number of genes obtain from CSA and the MCMC analyses, it is important to remember that this is the direct consequence of the differences between the underlying models used in the two approaches.
The two segregation-analysis approaches used here are based on different underlying models and estimation procedures, yet they result in very similar overall conclusions. Both methods suggest that there is stronger evidence for a genetic basis of nonword memory than of digit span. This greater evidence for genetic factors contributing to nonword memory compared with digit span is unlikely to be an artifact of the reliability of the two measures: nonword memory is slightly less reliable (.8) (Berninger et al., in press) than digit span (.85) (Wechsler 1981) and, all else being equal, therefore could be expected to show less, rather than more, evidence for a genetic basis than does digit span. For CSA, the evidence for a genetic basis is suggested by the evidence for one class of genetic model identified with likelihood-ratio tests, for nonword memory, compared with the inability to clearly differentiate between the mixed versus Mendelian and polygenic models, for digit span. The MCMC methods' stronger evidence for an inherited basis of nonword memory is obtained from the posterior probability that no genes contribute to each phenotype: this probability is much smaller for nonword memory than for digit span. Where the results from the two methods differ is in the evidence (or lack thereof) for more than one contributing gene. In the CSA, there was no evidence for residual family correlations in addition to major-gene effects, with the possible exception of nonword memory with digit span as a covariate. In contrast, the MCMC analyses gave fairly strong evidence for more than one contributing locus, especially in the case of nonword memory; however, the evidence for a second locus was not overwhelming, leaving open the possible interpretation that this estimate of the posterior probability of two (or more) genes simply reflects the MCMC sampling process, which, by necessity, will spend some time in improbable parts of the sample space. The relatively modest estimates of heritability obtained in this study are consistent with this weak evidence for multiple genes. One other study, of twins, reported higher estimates for heritability (.64–1.00) of a similar nonword-repetition task (Bishop et al. 1996), but it did not adjust for ascertainment and did not use the identical test.
Both nonword memory and digit span have been shown to tap phonological deficits in individuals with reading disabilities (Wagner and Torgesen 1987). In our sample, both of these tasks are correlated with some measures used in the diagnosis of dyslexia, with particularly strong correlations, both within probands and within relatives, for measures of real-word reading and spelling and for pseudoword reading (Raskind et al., in press). Other studies also have reported correlations, in unrelated individuals, between digit span and measures of dyslexia, including spelling (Newman et al. 1993; Shaywitz et al. 1999). Evidence for a genetic basis of phonological deficits in reading disabilities has been reported elsewhere (Olson et al. 1994). Our results provide the first evidence that at least part of the genetic basis of the phonological core deficit in dyslexia may be related to phonological short-term memory rather than to phoneme deletion (also see Raskind et al., in press). The observation that a recent intervention study to improve phonological short-term memory resulted in both improved reading skill and changes in brain lactate activation during phonological processing (Richards et al. 2000) suggests that phonological short-term memory may be fundamental. However, the results in the present study do not imply that other measures are unimportant, nor do they provide estimates of the relative contribution of each of the components to the phonological core deficit. Segregation analyses that include reciprocal adjustments for these additional measures and that begin to address the interrelationships of the latter will be undertaken in future studies. The finding that the evidence for a genetic basis was somewhat stronger for nonword memory than for digit span may reflect the fact that reading requires manipulation of verbal—rather than numeric—information in phonological short-term memory and that ascertainment of the families was through probands with dyslexia, which would tend to select for variation in related language processes in these families.
The measures investigated here relate most strongly to phonological skills. Both tasks are purely oral. Whereas the nonword-memory task requires the ability to perceive, remember, and reproduce successively more-difficult nonsense words, the digit-span tasks require the ability to perceive, remember, and reproduce increasingly longer strings of familiar names of numbers (both forward and backward). Because holding either the novel series of phonemes or the names of the numbers in short-term memory is necessary for successful execution of the tasks, it is not surprising that the two measures are correlated and that both measures therefore show evidence for a genetic basis if at least one measure shows such evidence.
The results of our analyses suggest that there are genes contributing jointly to variation both in nonword memory and in digit span and that there are other genes that contribute to variation only in nonword memory. Three observations lead to this conclusion—and to the consequent implications for models of the underlying genetic structure that can be proposed. The first observation is the relative magnitude of covariate effects estimated for digit span versus nonword memory. The estimated covariate effect for nonword memory in the analysis of digit span was considerably larger than the reverse effect. Although the scale of measurement of the two variables also was different, the difference in scale is insufficient to explain the difference of the covariate effects: rescaling the measures to the same measurement scale gives a ratio of ∼2.5:1 for the effect estimated for nonword memory as a covariate in the analysis of digit span, relative to that estimated for digit span in the analysis of nonword memory. The second observation is that the evidence for a genetic basis of digit span declined dramatically when nonword memory was used as a covariate in the analysis. This was apparent in both the reduced estimate of the number of contributing QTLs and in the loss, compared with use of the unadjusted phenotype, of statistically significant evidence for an inherited basis in CSA. The third observation is that, whereas inclusion of digit span as a covariate in the analysis of nonword memory resulted in a reduction of the estimated number of genes involved in nonword memory, it had little effect on the overall evidence for a genetic basis of nonword memory. Together, these observations suggest that there may be some genetic factors that contribute to nonword memory but not to digit span, whereas there may be no factors that contribute substantially to digit span but not to nonword memory. We speculate that the component that appears to be unique to nonword memory represents the ability to recognize and/or represent phonemes, rather than the ability to remember them. Of course, it is not possible to discount entirely the possibility that there still may be genes of smaller effect that contribute to digit span but not to nonword memory but that power to detect their existence in a segregation analysis is insufficient in this data set.
Successful dissection of the genetic components of dyslexia will require careful consideration of evidence for underlying heterogeneity of the phenotype. The approach taken here is a first step in this direction, identifying evidence for a genetic basis of two short-term–memory phenotypes that previously have been implicated in dyslexia. The families used here were selected through a dyslexic proband, so they may be segregating component phenotypes that are specifically related to verbal skills. In this context, nonword memory may provide stronger evidence for a genetic basis than does digit span, because nonword memory employs more verbal-stimulus elements that may be expected to be deficient in the probands. Alternatively, the genetic basis of digit span may be more elusive because it is a complex measure tapping both short-term memory (Digits Forward) and working memory (Digits Backwards), thereby representing a more heterogeneous phenotype whose genetic basis is harder to elucidate than that for nonword memory. The results suggesting that, for nonword memory, there may be an inherited component that is not shared by digit span is particularly tantalizing and suggests that pursuit of genes contributing to dyslexia may be more successful if this latter, unshared component is tackled first. Further research is needed to determine the genetic basis of nonword memory and digit span—and their relationship to dyslexia. However, the results presented here suggest that, for both of these phenotypes, a small number of genes are involved, with consequent implication for mapping and identification of these genes contributing to these component phenotypes.
Acknowledgments
We appreciate the expert help of Department of Educational Psychology graduate students Sylvia Abbott, Allison Brooks, Ana Rueda Brown, Rebecca Brooksher, Julie Busse, Kristina Byrd, Belle Chenault, Gerry Curtin, Julie Gibson, Renee Harman, Linelle Milatchov, Stacy Ogier, Tanya Prather, James Rodriguez, and Jared Taylor, in administering the test battery. We thank Richard Wagner and Joseph Torgesen for allowing us to use prepublication measures of the Comprehensive Test of Phonological Awareness. The database was constructed and maintained by Ted Holzman, and computer support was provided by Minh Cao. We are grateful to the families, for their willingness to devote the time necessary to participate in these studies. Some of the results of this article were obtained by use of the program package S.A.G.E., which is supported by a U.S. Public Health Service Resource Grant 1 P41 RR03655 from the National Center for Research Resources. The research reported here was supported by National Institute of Child Health and Development grant P50 33812.
Electronic-Database Information
The URL for data in this article is as follows:
- Loki (version 2.1), http://www.stat.washington.edu/thompson/Genepi/Loki.shtml (for MCMC analysis of multiple QTLs)
References
- Akaike H (1974) A new look at the statistical model identification. IEEE Trans Automatic Control AC-19:716–723 [Google Scholar]
- Berninger V (1994) Reading and writing acquisition: a developmental neuropsychological approach. WC Brown & Benchmark, Madison, WI [Google Scholar]
- Berninger V, Abbott R, Thomson J, Raskind W. Language phenotype for reading and writing disability: a lifespan approach. Sci Stud Reading (in press) [Google Scholar]
- Berninger V, Rutberg J (1992) Relationship of finger function to beginning writing: application to diagnosis of writing disabilities. Dev Med child Neurol 34:155–172 [DOI] [PubMed] [Google Scholar]
- Bisgaard M, Eiberg H, Moller N, Niebuhr E, Mohr J (1987) Dyslexia and chromosome-15 heteromorphism: negative lod scores in a Danish material. Clin Genet 32:118–119 [DOI] [PubMed] [Google Scholar]
- Bishop D, North T, Donlan C (1996) Nonword repetition as a behavioural marker for inherited language impairment: evidence from a twin study. J Child Psychol Psychiatry 37:391–403 [DOI] [PubMed] [Google Scholar]
- Bonney G (1986) Regressive logistic models for familial disease and other binary traits. Biometrics 42:611–625 [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC (1994) Quantitative trait locus for reading disability on chromosome 6. Science 266:276–279 [DOI] [PubMed] [Google Scholar]
- ——— (1995) Quantitative trait locus for reading disability: correction. Science 268:1553 [DOI] [PubMed] [Google Scholar]
- Daw EW, Heath SC, Wijsman EM (1999) Multipoint oligogenic analysis of age-at-onset data with applications to Alzheimer’s disease pedigrees. Am J Hum Genet 64:839–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, Wijsman EM (2000) The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet 66:196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker S, DeFries J (1981) Cognitive ability profiles in families of reading-disabled children. Dev Med Child Neurol 23:217–227 [DOI] [PubMed] [Google Scholar]
- DeFries J, Fulker D, LaBuda M (1987) Evidence for a genetic aetiology in reading disability of twins. Nature 329:537–539 [DOI] [PubMed] [Google Scholar]
- Eden G, Van Meter J, Rumsey J, Maisog J, Woods R, Zeffiro T (1996) Abnormal processing of visual motion in dyslexia revealed by functional brain imaging. Nature 382:66–69 [DOI] [PubMed] [Google Scholar]
- Fagerheim T, Raeymaekers P, Tønnessen F, Pedersen M, Tranebjærg L, Lubs H (1999) A new gene (DYX3) for dyslexia is located on chromosome 2. J Med Genet 36:664–669 [PMC free article] [PubMed] [Google Scholar]
- Field LL, Kaplan BJ (1998) Absence of linkage of phonological coding dyslexia to chromosome 6p23-p21.3 in a large family data set. Am J Hum Genet 63:1448–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JH (1905) Case of congenital word-blindness (inability to learn to read). Ophthalmol Rev 24:315 [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP (1999) A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet 64:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Vargha-Khadem F, Waitkins KE, Monaco AP, Pembrey ME (1998) Localisation of a gene implicated in a severe speech and language disorder. Nat Genet 18:168–170 [DOI] [PubMed] [Google Scholar]
- Gathercole SE, Baddeley AD (1990) Phonological memory deficits in language disordered children: is there a causal connection? J Memory Lang 29:336–360 [Google Scholar]
- Gayan J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, Olson RK, Pennington BF, DeFries JC (1999) Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet 64:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilger JW, Borecki IB, DeFries JC, Pennington BF (1994) Commingling and segregation analysis of reading performance in families of normal reading probands. Behav Genet 24:345–355 [DOI] [PubMed] [Google Scholar]
- Goddard KA, Jarvik GP, Graham J, McNeney B, Hsu L, Siegmund K, Grossner S, Olson J, Wijsman EM (1995) Analysis of quantitative risk factors for a common oligogenic disease. Genet Epidemiol 12:759–764 [DOI] [PubMed] [Google Scholar]
- Greenberg DA, Hodge SE, Vieland VJ, Spence MA (1996) Affecteds-only linkage methods are not a panacea. Am J Hum Genet 58:892–895 [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Hart LA, Speed WC, Shuster A, Pauls DL (1997) Susceptibility loci for distinct components of developmental dyslexia on chromosomes 6 and 15. Am J Hum Genet 60:27–29 [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Pauls DL (2000) Chromosome 6p influences on different dyslexia-related cognitive processes: further confirmation. Am J Hum Genet 66:715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC (1990) Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250:1684–1689 [DOI] [PubMed] [Google Scholar]
- Hallgren B (1950) Specific dyslexia (congenital word-blindness): a clinical and genetic study. Acta Psychiatr Neurol Scand Suppl 65:1–287 [PubMed] [Google Scholar]
- Hammersley JM, Handscomb DC (1964) Monte Carlo methods. Methuen & Co, London [Google Scholar]
- Hastings WK (1970) Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57:97–109 [Google Scholar]
- Heath SC (1997) Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet 61:748–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshelwood J (1907) Four cases of congenital word-blindness occurring in the same family. BMJ 2:1229–1232 [Google Scholar]
- Hokanson J, Brunzell J, Jarvik G, Wijsman E, Austin M (1999) Linkage of low-density lipoprotein size to the lipoprotein lipase gene in heterozygous lipoprotein lipase deficiency. Am J Hum Genet 64:608–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhi AG, Catts HW (1986) Toward an understanding of developmental language and reading disorders. J Speech Hear Disord 51:337–347 [DOI] [PubMed] [Google Scholar]
- Knoblauch H, Müller-Myhsok B, Busjahn A, Ben Avi L, Bähring S, Baron H, Heath S, Uhlmann R, Faulhaber H-D, Shpitzen S, Aydin A, Reshef A, Rosenthal M, Eliav O, Mühl A, Lowe A, Schurr D, Harats D, Jeschke E, Friedlander Y, Schuster H, Luft F, Leitersdof E (2000) A cholesterol-lowering gene maps to chromosome 13q. Am J Hum Genet 66:157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E, Tsuang D, Bird TD (1998) Recent advances in the genetics of Alzheimer’s disease. J Geriatr Psychiatry Neurol 11:42–54 [DOI] [PubMed] [Google Scholar]
- Lewitter F, DeFries J, Elston R (1980) Genetic models of reading disability. Behav Genet 10:9–30 [DOI] [PubMed] [Google Scholar]
- Liberman U, Karlin S (1984) Theoretical models of genetic map functions. Theor Popul Biol 25:331–346 [DOI] [PubMed] [Google Scholar]
- MacLean CJ, Morton NE, Lew R (1975) Analysis of family resemblance. IV. Operational characteristics of segregation analysis. Am J Hum Genet 27:365–384 [PMC free article] [PubMed] [Google Scholar]
- Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E (1953) Equations of state calculations by fast computing machines. J Chem Physiol 21:1087–1091 [Google Scholar]
- Morton N, MacLean C (1974) Analysis of family resemblance. III. Complex segregation analysis of quantitative traits. Am J Hum Genet 26:489–503 [PMC free article] [PubMed] [Google Scholar]
- Newman S, Fields H, Wright S (1993) A developmental study of specific spelling disability. Br J Educ Psychol 63:287–296 [DOI] [PubMed] [Google Scholar]
- Olson R, Datta H, Gagan J, DeFries J (1999) A behavioral genetic analysis of reading disabilities and component processes. In: Klein R, McMullen P (eds) Converging methods for understanding reading and dyslexia. MIT Press, Cambridge, MA, pp 133–151 [Google Scholar]
- Olson R, Forsberg H, Wise B (1994) Genes, environment, and the development of orthographic skills, In: Berninger V (ed) The varieties of orthographic knowledge. I. Theoretical and developmental issues. Kluwer Academic, Dordrecht, The Netherlands, pp 27–71 [Google Scholar]
- Olson R, Gillis J, Rack J, DeFries J, Fulker D (1991) Confirmatory factor-analysis of word recognition and process measures in the Colorado reading project. Reading Writing 3:235–248 [Google Scholar]
- Olson R, Wise B, Conners F, Rack J, Fulker D (1989) Specific deficits in component reading and language skills—genetic and environmental influences. J Learning Disabil 22:339–348 [DOI] [PubMed] [Google Scholar]
- Pennington BF, Gilger JW, Pauls D, Smith SA, Smith SD, DeFries JC (1991) Evidence for major gene transmission of developmental dyslexia. JAMA 18:1527–1534 [PubMed] [Google Scholar]
- Pennington BF, Smith SD, Kimberling WJ, Green PA, Haith MM (1987) Left-handedness and immune disorders in familial dyslexics. Arch Neurol 44:634–639 [DOI] [PubMed] [Google Scholar]
- Pennington B, Van Orden G, Smith S, Green P, Haith M (1990) Phonologica1 processing skills and deficits in adult dyslexics. Child Dev 61:1753–1778 [PubMed] [Google Scholar]
- Petryshen TL, Kaplan BJ, Field LL (1999) Evidence for a susceptibility locus for phonological coding dyslexia on chromosome 6q13-q16.2. Am J Hum Genet Suppl 65:A163 [Google Scholar]
- Petryshen TL, Kaplan BJ, Liu MF, Field LL (2000) Absence of significant linkage between phonological coding dyslexia and chromosome 6p23-21.3, as determined by use of quantitative-trait methods: confirmation of qualitative analyses. Am J Hum Genet 66:708–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskind W, Hsu L, Berninger V, Thomson J, Wijsman E. Familial aggregation of dyslexia phenotypes. Behav Genet (in press) [DOI] [PubMed] [Google Scholar]
- Richards T, Corina D, Serafini S, Steury K, Echelard D, Dager S, Marro K, Abbott R, Maravilla K, Berninger V (2000) Effects of a phonologically driven treatment for dyslexia on lactate levels measured by proton MR spectroscopic imaging. Am J Neuroradiol 21:916–922 [PMC free article] [PubMed] [Google Scholar]
- S.A.G.E. (1997) Statistical analysis for genetic epidemiology, ed 3.1. Computer program package available from the Department of Epidemiology and Biostatistics, Rammelkamp Center for Education and Research, MetroHealth Campus, Case Western Reserve University, Cleveland [Google Scholar]
- Schellenberg GD, Bird TD, Wijsman EM (1994a) Genetic heterogeneity and Alzheimer’s disease. Am J Hum Genet Suppl 55:A353 [Google Scholar]
- ——— (1994b) The genetics of Alzheimer’s disease. Neurobiol Aging Suppl 15:S149 [DOI] [PubMed] [Google Scholar]
- Shaywitz S, Fletcher J, Holahan J, Schneider A, Marchione K, Stuebing K, Francis D, Pugh K, Shaywitz B (1999) Persistence of dyslexia: the Connecticut longitudinal study at adolescence. Pediatrics 104:1351–1359 [DOI] [PubMed] [Google Scholar]
- Shaywitz S, Shaywitz B, Fletcher J, Esobar M (1990) Prevalence of reading disability in boys and girls. JAMA 264:998–1002 [PubMed] [Google Scholar]
- Smith S, Kimberling W, Pennington B, Lubs H (1983) Specific reading disability: identification of an inherited form through linkage analysis. Science 219:1345–1347 [DOI] [PubMed] [Google Scholar]
- Stephenson S (1907) Six cases of congenital word-blindness affecting three generations of one family. Ophthalmoscope 5:482–484 [Google Scholar]
- Stevenson J, Graham P, Fredman G, McLoughlin V (1987) A twin study of genetic influences on reading and spelling ability and disability. J Child Psychol Psychiatry 28:229–247 [DOI] [PubMed] [Google Scholar]
- Torgesen JK, Wagner RK, Reshotte CA (1999) Test of word reading efficiency (TOWRE). Pro-Ed, Austin [Google Scholar]
- Vellutino F (1979) Dyslexia, theory, and research. MIT Press, Cambridge, MA [Google Scholar]
- Vieland VJ, Hodge SE, Greenberg DA (1992) Adequacy of single-locus approximations for linkage analyses of oligogenic traits. Genet Epidemiol 9:45–59 [DOI] [PubMed] [Google Scholar]
- Wagner R, Torgesen J (1987) The nature of phonological processing and its causal role in the acquisition of reading skills. Psychol Bull 101:192–212 [Google Scholar]
- ——— (1999) Comprehensive test of phonological processing. Pro-Ed, Austin [Google Scholar]
- Wechsler D (1981) Wechsler adult intelligence scale–revised (WAIS-R). Psychological Corporation, San Antonio [Google Scholar]
- ——— (1992) Wechsler intelligence scale for children–third edition (WISC-III). Psychological Corporation, San Antonio [Google Scholar]
- Wijsman EM, Amos C (1997) Genetic analysis of simulated oligogenic traits in nuclear and extended pedigrees: summary of CAW 10 contributions. Genet Epidemiol 14:719–735 [DOI] [PubMed] [Google Scholar]
- Wilkinson G (1993) Wide range achievement tests–revised (WRAT-R). Wide Range, Wilmington, DE [Google Scholar]
- Wolff PH, Melngailis I (1994) Family patterns of developmental dyslexia: clinical findings. Am J Med Genet 54:122–131 [DOI] [PubMed] [Google Scholar]
- Woodcock R (1987) Woodcock reading master tests–revised (WRMT-R). American Guidance Service, Circle Pines, MN [Google Scholar]
- Woodcock R, Johnson B (1990) Woodcock-Johnson psychoeducational battery-revised tests of achievement. Riverside Publishing, Chicago [Google Scholar]
- Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, Nguyen K, et al (1994) Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science 265:2088–2090 [DOI] [PubMed] [Google Scholar]