Abstract
Although it is clear that errors in genotyping data can lead to severe errors in linkage analysis, there is as yet no consensus strategy for identification of genotyping errors. Strategies include comparison of duplicate samples, independent calling of alleles, and Mendelian-inheritance–error checking. This study aimed to develop a better understanding of error types associated with microsatellite genotyping, as a first step toward development of a rational error-detection strategy. Two microsatellite marker sets (a commercial genomewide set and a custom-designed fine-resolution mapping set) were used to generate 118,420 and 22,500 initial genotypes and 10,088 and 8,328 duplicates, respectively. Mendelian-inheritance errors were identified by PedManager software, and concordance was determined for the duplicate samples. Concordance checking identifies only human errors, whereas Mendelian-inheritance–error checking is capable of detection of additional errors, such as mutations and null alleles. Neither strategy is able to detect all errors. Inheritance checking of the commercial marker data identified that the results contained 0.13% human errors and 0.12% other errors (0.25% total error), whereas concordance checking found 0.16% human errors. Similarly, Mendelian-inheritance–error checking of the custom-set data identified 1.37% errors, compared with 2.38% human errors identified by concordance checking. A greater variety of error types were detected by Mendelian-inheritance–error checking than by duplication of samples or by independent reanalysis of gels. These data suggest that Mendelian-inheritance–error checking is a worthwhile strategy for both types of genotyping data, whereas fine-mapping studies benefit more from concordance checking than do studies using commercial marker data. Maximization of error identification increases the likelihood of linkage when complex diseases are analyzed.
Introduction
Microsatellite-repeat markers are widely used as a powerful tool in genetic mapping (Dixon et al. 1992; Roberts et al. 1999), population genetics (Huges and Queller 1993; Taylor et al. 1994), linkage analysis (Georges et al. 1993), evolutionary studies (Bowcock et al. 1994), and forensic medicine (Herber and Herold 1998; Sacchetti et al. 1999). The accurate measurement of microsatellite fragment sizes is clearly important for linkage studies, and errors must be minimized because incorrect data will reduce the likelihood that linkage can be detected. In addition, as microsatellite measurement finds greater clinical and forensic application, the demand for rigorous estimation of errors will increase.
Although it is clear that errors in genotyping data can lead to severe errors in analysis, there is as yet no consensus as to how genotyping errors should be identified and what appropriate correction steps must be invoked to minimize these errors. Several strategies for identification and removal of incorrect data have been suggested (Ghosh et al. 1997; Pálsson et al. 1999), in order to produce the most error-free data possible for linkage analysis. For the most part, these strategies involve error-rate assumption (Lincoln and Lander 1992; Goldstein et al. 1997). However, a better understanding of what constitutes an error will enable appropriate identification and reduction of errors, resulting in both a more complete data set and an increased likelihood that linkage with a particular phenotype will be identified.
Genotyping errors arising from amplification difficulties have, to some extent, been addressed. For example, there are several commercially available microsatellite-marker linkage sets, comprising di-, tri-, or tetranucleotide repeats, that provide coverage of the entire human genome and that have a resolution range of 5–20 cM (PE Biosystems ABI PRISM Linkage Mapping Sets HD5, MD10, and LD20 and Research Genetics CHLC Human screening set/Weber versions 6–10). These marker sets have helped in the reduction of errors in data because they use primers chosen not only for location but also for fidelity of amplification. Some of the sets also incorporate a consensus sequence (PIG-tailing [Brownstein et al. 1996]), to encourage the addition of an extra A at the end of a PCR product by the terminal transferase activity of Taq polymerase. Such consistency helps make allele identification clearer. In contrast, there are no primer sets available commercially that cover the genome at <5 cM, and therefore fine-mapping studies usually are performed with primers chosen for their location rather than for their reliability or ease of use.
One overriding consideration with regard to microsatellite genotyping is that, even though several programs are available to help streamline the process, it is still a labor-intensive operation requiring manual assessment and correction of genotypes. Although it is predictable that well-defined commercial primer sets will be easier to call than custom-designed fine-mapping sets, they both still require considerable human input. Preferential amplification of competing microsatellite pairs makes multiplexing reactions difficult to balance and, even though it is possible to do so, it is impractical for large-scale high-throughput applications. For this reason, each microsatellite is amplified individually and later is combined into appropriate electrophoretic running panels. Product sizes are predetermined by choice of primers, so that several nonoverlapping products can be run in the same lane. The use of a fluorescent tag on the end of one of the primer pairs for each microsatellite enables three different colors to be used, so that up to 20 PCR products can be combined as one lane, or “panel,” of microsatellite markers (LMSV2; PE Biosystems).
After electrophoresis, data collected from the run are analyzed, first by lane assignment and then by size-standard assignment and size calling. Only then are the data ready to be genotyped. Templates using predefined allele assignments are used for the first round of genotyping, followed by manual checking of each call. Since allele sizes are assigned from a standard curve, the initial results are not in whole base pairs. Each allele is assigned to a “bin,” which is predefined according to the average size of each allele. Once this has been completed, the results are checked for errors. Some microsatellites used are not true di-, tri-, or tetranucleotide repeats but are compound repeats, in which there also may be a single nucleotide polymorphism. These alleles are 1 bp different in size and need extra care when genotyping is performed. Other microsatellites show “null” alleles, in which one of the two alleles fails to amplify (usually because of a mutation in the priming site), and are identifiable on the basis of pedigree information.
There is, therefore, a range of error types that can be introduced during each step associated with the genotyping process—a range that includes human-handling and calling errors, equipment and reagent failures, and errors caused by mutations of the DNA. Strategies have been proposed to decrease or identify these errors. These include the extreme suggestion of genotyping in duplicate and comparing both sets of data as well as having all data viewed separately by two people and then having the allele calling compared. A better understanding of these errors will allow a more rational approach to detection of errors and, therefore, will improve the overall quality of data obtained from microsatellite analysis. In the present study, we assessed both a commercial set and a fine-mapping set of human primers, to determine the frequency and type of errors that arise in each case, using Mendelian-inheritance–error checking for error identification. Duplication methods for error identification also were compared with the Mendelian-inheritance–error approach to error detection. The outcome of this study has enabled us to derive a strategy to reduce overall errors, thereby allowing us to provide the correct balance between quality of data and the cost and time involved in the genotyping process.
Material and Methods
Primer Sets
Two primer sets were used. The first was a commercial set (LMSV2; PE Biosystems) that has a 10-cM density covering all human chromosomes except the Y chromosome. It consists of 400 primer pairs divided into 28 panels, with 10–20 markers per panel. The second primer set (FMS) was custom designed for fine mapping of positive regions identified in a particular project (Levinson et al. 1998). This set covered five chromosomes and comprised 60 markers divided into seven panels with 3–12 markers per panel.
LMSV2 amplifies dinucleotide microsatellite markers, whereas FMS is directed toward di-, tri-, and tetranucleotide microsatellites. For both mapping sets, one primer of each pair was fluorescently labeled with one of three fluorescein-derived fluorophores, which were designated “6-FAM,” “HEX,” and “NED.”
Samples
DNA samples from 310 individuals were amplified with LMSV2, and DNA samples from 375 individuals were amplified with FMS. The family data for the two studies were very similar; the LMSV2 data consisted of 74 pedigrees with an average of 4.19 people genotyped per pedigree, and the FMS data consisted of 71 pedigrees with an average of 5.28 people genotyped per family. Parents were typed whenever possible, as were some siblings and grandparents.
Genotyping Amplification Protocol
Each amplification used 30 ng of DNA in 6-μl reactions. DNA and reagents were aliquoted, by a Tecan Genesis workstation, into 384-well plates. The microsatellite markers were amplified, by PE Biosystems enzyme AmpliTaq Gold on MJ Research Tetrad thermal cyclers, with the recommended cycling protocol for the enzyme. Each marker was amplified individually and was pooled manually into panels according to the predetermined panels for each panel set.
The pooled PCR products were electrophoresed through polyacrylamide gels on PE Biosystems 377 automated sequencers using the recommended gel conditions and run protocol. PstI-cut lambda-phage DNA labeled with 6-carboxy rhodamine (GS500-ROX; PE Biosystems) was included in each lane, as a size standard.
Genotyping Analysis Protocol
Electrophoresis data were transferred to an offline computer and were tracked as batches by a Quickeys script and GENESCAN 3.1. Tracking of each gel was manually checked before analysis. A standard curve was generated by the “local Southern method” (in the GENESCAN software) for every gel, which thus corrected for any minor gel variations. The size-standard patterns from each lane of any one gel were overlaid to confirm allele assignment, and any allele missasignments were corrected manually. Finally, the microsatellite alleles were sized against the standard curve. A peak was called if it had >10 fluorescent units of peak height.
GENOTYPER 2.1 was used to filter out stutter peaks, A+ peaks, and signals that were generally low in relation to main peaks in a range (peaks <32% of main-peak heights were removed from the call). Templates for this software and bins for allele assignment were constructed on the basis of the sample data. A unique template was prepared for each panel of results. Allele assignments for each panel set of data were used to determine the average size of each allele. This was then used as the size, ±0.5 bp, for the allele and was assigned a bin name to reflect the rounded size. This approach was used for the di-, tri-, and tetranucleotide microsatellite alleles. Several markers have a 1-bp mutation, and, to prevent overlap of bin boundaries, bin assignments were designated as the average size of the allele, with boundaries of ±0.4 bp for these particular markers. We found that, if a marker had a large size range, the alleles did not migrate at exactly 2 bp. Thus, alleles at the beginning of the range may have been rounded to odd whereas the alleles at the end of the range rounded to even (or even to odd, depending on the starting size). To make it clear that these markers did not contain 1-bp alleles, the bins were all changed to reflect the majority—that is, all odd or all even. This process leaves the true allele sizes unchanged; however, the distinction between the 1- and 2-bp alleles was made more clear.
Each genotype generated by use of the template filters and bins was manually checked and corrected as necessary, and the results were saved in an Excel spreadsheet. All manual genotyping was performed without knowledge of the pedigree structure, to ensure unbiased calls. If an allele did not fall within the bin range or if the sample failed to amplify, the sample was repeated once. All repeat samples were run individually, after dilution to reflect the amount of product of the marker when loaded in a panel group. This enabled a more correct fluorescent peak height and, therefore, more-accurate bin assignment. The repeat results were then added to the results table. Identities of samples were checked by a macro written in the software program Excel, to compare the original names to the final results names in the tables; any labeling errors could then be identified and corrected.
Genotyping Errors
Errors were counted for each complete genotype result containing an error. This approach was taken—rather than the counting of specific incorrect alleles—in order to find the total number of affected genotypes that would have been in the data had there been no error checking.
DNA samples were checked for Mendelian-inheritance errors, by use of the family pedigrees and the Australian Genome Research Facility Data Management Web site that runs PedManager version 0.9 (M. P. Reeve, personal communication). The site manages the PedManager options, allowing for correct ordering of the columns in the pedigree-relationship file, marker ordering of the results file, and allele recoding. Each result file and pedigree-relationship file was saved as a text, tab-delimited Excel file. These files were then imported into the Data Management Web site. The two files were merged, producing a LINKAGE-style pedigree file in the .pre or pre makeped format, which can then be used in many of the linkage-analysis programs. Allele-recoding information and marker-order information were produced and displayed at the site. The merged file was easily cut and pasted back into an Excel spreadsheet. PedManager also was used to check the autosomal genotype results for pedigree-format errors, file errors, and Mendelian-inheritance errors, thereby producing a file that contained information for any errors found in a nuclear family including the alleles for parents and children and a suggested erroneous individual and allele. In this study, results files for each panel’s data were created and then were automatically checked, by PedManager, for pedigree errors. All Mendelian-inheritance errors were manually checked and assigned to the categories described in Appendix A. Of the identified Mendelian-inheritance errors, those determined to be call errors were further categorized, as described in Appendix B.
Duplication Error Analysis
Concordance within gels was checked by use of the FMS data. Sixty-four DNA samples were run in duplicate, for all 60 markers, in adjacent lanes. Concordance between gels was checked for reproducibility by repetition of the sample sets for the first panel, in both the LMSV2 marker set and the FMS marker set. Each duplicate was compared with the original calls, for concordance, and discrepancies were scored.
Results
We measured 124,000 genotypes with the LMSV2 marker set and 22,500 genotypes with the FMS marker set. The failure rate for the LMSV2 study was 3.6% (4,481 repeats), and that for the FMS study was 9.64% (2,169 repeats). The majority of these failures were resolved by repeat genotyping.
PedManager identified 292 and 308 Mendelian-inheritance errors, respectively, in autosomal genotype results generated by use of the LMSV2 marker set and the FMS marker set. There were no cases of nonpaternity in any of the samples, suggesting that all Mendelian-inheritance–errors inconsistencies were due to other causes. Mendelian-inheritance–error types were classified as described in Appendix A, and their proportions are summarized in table 1. Graphic representation of the percentage of each error type is shown in figure 1A, whereas figure 1B represents each error’s percentage of overall genotypes. For FMS, the predominant errors were those classified as missed alleles, with significantly more (χ21=590.12; P=2×10-120) missed alleles in the FMS data than in the LMSV2 data. Missed alleles in the FMS marker set were usually caused by preferential amplification in which the allele of shorter base-pair length amplifies in preference to the longer allele (fig. 2).
Table 1.
Types of Errors Detected
|
Errors |
||||||||
| Mutations |
Other Errors |
|||||||
| Microsatellite | Priming Site | Total | Missed Alleles | Call | Sample Swap | Total | Total No. (%) Detected | |
| Mendelian inheritance: | ||||||||
| LMSV2 (118,420 samples): | ||||||||
| No. of errors | 77 | 64 | 141 | 34 | 76 | 41 | 151 | 292 (.25) |
| Proportion of total errors (%) | 26.37 | 21.92 | 48.29 | 11.64 | 26.03 | 14.04 | 51.71 | |
| Proportion of total genotypes (%) | .065 | .054 | .119 | .029 | .064 | .035 | .128 | |
| FMS (22,500 samples): | ||||||||
| No. of errors | 25 | 16 | 41 | 150 | 53 | 64 | 267 | 308 (1.37) |
| Proportion of total errors (%) | 8.12 | 5.19 | 13.31 | 48.7 | 17.21 | 20.78 | 86.69 | |
| Proportion of total genotypes (%) | .11 | .074 | .184 | .667 | .235 | .284 | 1.186 | |
| Concordance: | ||||||||
| LMSV2 between gels (10,088 samples): | ||||||||
| No. of errors | … | … | … | 5 | 7 | 4 | 16 | 16 (.16) |
| Proportion of total errors (%) | … | … | … | 31.25 | 43.75 | 25 | 100 | |
| Proportion of total genotypes (%) | … | … | … | .05 | .069 | .04 | .159 | |
| FMS: | ||||||||
| Between gels (4,488 samples): | ||||||||
| No. of errors | … | … | … | 79 | 12 | 16 | 107 | 107 (2.38) |
| Proportion of total errors (%) | … | … | … | 73.83 | 11.22 | 14.95 | 100 | |
| Proportion of total genotypes (%) | … | … | … | 1.76 | .267 | .357 | 2.384 | |
| Within gels (3,840 samples): | ||||||||
| No. of errors | … | … | … | 18 | 11 | … | 29 | 29 (.76) |
| Proportion of total errors (%) | … | … | … | 62 | 38 | … | 100 | |
| Proportion of total genotypes (%) | … | … | … | .469 | .29 | … | .759 | |
Figure 1.
Percentage of total errors (A) and total genotypes (B), for each error type.
Figure 2.
Preferential amplification. Paternal allele 168 (children 2 and 3) and 172 (child 1) did not amplify as well as did the allele 148.
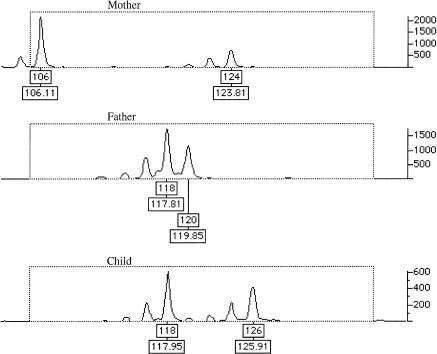
Mutations in which there was a gain or loss of a repeat unit were much more easily identified, in both marker sets. Figure 3 illustrates an example of a 2-bp mutation event. For this example, maternal alleles were 106 and 124, whereas paternal alleles were 118 and 122. The mutation has led to an expansion of the maternal allele in the offspring, resulting in a genotype of 118 (paternal allele) and 126 (new maternal allele). Mutation rates were calculated as being 3×10-4 (for LMSV2) and 5.6×10-4 (for FMS) per meiosis, by use of the number of observed microsatellite mutations in both study sets (table 2).
Figure 3.
Microsatellite mutation to new allele. The child’s maternal allele (124) has mutated to a new allele (126), whereas the paternal allele (118) is normal.
Table 2.
Mutation/Error Rates
| Marker Set | No. of Genotypes | No. of Errors | Error Rate(%) | No. of Mutations | Mutation Rate(%) |
| LMSV2 | 118,420 | 151 | .13 | 77 | 3×10-4 |
| FMS | 22,500 | 267 | 1.19 | 25 | 5.6×10-4 |
Mendelian-inheritance errors designated as “call errors” were further classified into the groups described in Appendix B and are summarized in table 3. Using the results from the LMSV2 marker set, we found that incorrect calls were less than half (30.26%) of the human-error calls, whereas 28.94% were GENOTYPER handling errors (i.e., incorrect updating bin assignment), 17.11% were sample-loading errors (i.e., due to lane leakage), and 7.89% were due to low fluorescence of a sample. In total, 151 human-related errors were made with the LMSV2 marker set and 267 were made with the FMS marker set. Error rates for the LMSV2 and FMS marker sets were thus calculated as being 0.13% and 1.19%, respectively (table 2). These error rates were calculated by subtraction of the observed mutations from the total error counts.
Table 3.
Call Errors
|
Call Error |
|||||||
| Genotype Not Updated | Incorrect Call | Leak | Sample | Binning | Other | Total | |
| LMSV2 (118,420 samples): | |||||||
| No. of errors | 6 | 23 | 13 | 6 | 16 | 12 | 76 |
| Proportion of total errors (%) | 7.89 | 30.26 | 17.11 | 7.89 | 21.05 | 15.8 | 100 |
| Proportion of total genotypes (%) | .005 | .019 | .011 | .005 | .014 | .01 | .064 |
| FMS (22,500 samples): | |||||||
| No. of errors | 1 | 24 | … | 5 | 21 | 2 | 53 |
| Proportion of total errors (%) | 1.9 | 45.28 | … | 9.43 | 39.62 | 3.77 | 100 |
| Proportion of total genotypes (%) | .004 | .1067 | … | .022 | .093 | .009 | .2347 |
Finally, we tested the sample-duplication processes, for their ability to detect errors and to assess our process for reproducibility. We first ran the same panel of PCR products in consecutive lanes of the same gel and then on gels run on different days. We found a discordance of 0.76% when testing within a gel (3,840 genotypes, in FMS) and found a discordance of 2.38% (4,488 genotypes, in FMS) and 0.16% (10,088 genotypes, in LMSV2) between gels. As in the case of the Mendelian-inheritance errors, concordance-error types were assigned to the categories described in Appendix A, and the results are summarized in table 1.
Discussion
One current approach to the mapping of loci involved in complex diseases is a genomewide scan using microsatellite genetic markers. A complete understanding of the types of errors associated with the measurement of microsatellite marker size used for genome screens can help to maximize the information obtained from the final data. Commercial marker sets are made from primers chosen for both their location and ability to amplify DNA under common PCR conditions. Any primers that display amplification problems are substituted by those which perform optimally. In contrast, when fine-mapping sets are constructed, markers are chosen according to their chromosomal location, and so their amplification performance may not always be optimal. This difference in primer-design strategy was reflected in the initial PCR failure rates found in this study. At the outset, this observation prompted us to repeat all failed samples once, immediately increasing the amount of usable data for any linkage study. We also found that the total (detectable) human-error rate (as defined by Mendelian-inheritance errors) was significantly higher (χ21=561.49; P=4×10-124) for the FMS marker set (1.19%) than for the LMSV2 marker set (0.13%), mainly because of differences in amplification of fine-mapping markers and, thereby, difficulty in the scoring of some alleles. It is important to note that this study was genotyped independently of knowledge of the pedigree structure, to prevent introduction of bias. Pedigrees were referred to only for error analysis.
Error checking using concordance methods found 0.16% error between gels of the LMSV2 marker set and 2.38% between gels of the FMS marker set; in comparison, use of Mendelian-inheritance–error checking to identify errors found more errors in the LMSV2 marker set (0.25%) but fewer in the FMS marker set (1.37%). There were only three types of errors (i.e., missed alleles, call errors, or sample swaps) identified through concordance checking, with all microsatellite or priming-site mutation–related errors missed. Mendelian-inheritance–error checking found similar numbers for the error types listed above (0.13% for LMSV2 and 1.19% for FMS). This indicates that Mendelian-inheritance–error checking is a valuable method for the detection of these error types. Concordance checking, however, will miss the mutational error types, which can be a large proportion of the errors (nearly 50% of errors in LMSV2 were missed).
Overall, 26.37% of all errors found in the LMSV2 marker set were due to mutation of the microsatellite sequence, and 21.92% were due to assumed mutations in the priming sites (i.e., to null alleles). This represents nearly half (48.29%) of all errors; and no improvement in genotyping methods will change the frequency of these errors. The remaining 51.71% of errors are correctable errors; half of these (26.03% of total errors) are caused by call errors, whereas 11.64% are errors caused by alleles that have been missed and 14.04% are errors caused by mislabeling of samples. The proportion of errors is different for the FMS marker set. Only 13.31% of all errors were due to mutation events or null alleles. However, if the overall percentage of affected genotypes is compared with that in the LMSV2 study (0.184% in FMS, vs. 0.119% in LMSV2), and if the mutation rates (3×10-4 and 5.6×10-4, respectively) are compared, they are not significantly different (after multiple-comparison Bonferroni corrections of P values). Of the correctable errors, 20.78% of errors were due to sample swap, whereas 17.21% of errors were due to call errors.
Between gels, concordance checking found more errors in the FMS data than did Mendelian-inheritance–error checking. The predominant reason for errors in the FMS data was missed alleles. Approximately half (48.7%) of all FMS errors found by Mendelian-inheritance–error checking and nearly three-quarters (73.83%) of all FMS errors found by concordance checking were due to missed alleles, whereas only 11.64% of LMSV2 errors were missed alleles (Mendelian-inheritance–error checking). Even though concordance checking of the FMS marker set found more errors overall, only the number of missed alleles identified was statistically different (χ21=25.47; P=4.5×10-7). It should be noted that the 2.38% discordancy rate for duplicate genotypes in the FMS data reflects a large contribution from three markers with high rates of preferential amplification, as well as a sample swap, errors that were easily eliminated prior to linkage analyses. Thus, the rate of errors that might have gone undetected in the absence of duplicate genotyping was substantially smaller—and certainly <1%. Since the focus of the present study is on the types of errors detected by different methods, rather than on error rates of particular studies, more-detailed analysis of the error rate for the FMS data will be presented elsewhere.
Call errors in both the LMSV2 data and the FMS data were further analyzed to determine where improvement could be made. More than 50% of errors were due to software handling, gel handling, fail criteria, and allele binning; 7.89% of call errors in the LMSV2 data were due to the genotyping software not being updated to capture the change after a manual correction, and 17.11% were due to gel handling (i.e., lane leakage), which should be reduced if more care is given by the operator. An increase (from 10 fluorescent units to 15 fluorescent units) of the minimum peak height as the threshold for failure of a sample may also help to reduce errors. Binning accounted for 21.05% (in LMSV2) and 39.62% (in FMS) of all call errors. LMSV2 markers were generally easier to bin, because of preselection and PIG-tailing; however, markers displaying 1-bp alleles or amplification problems still caused a number of bin-assignment errors. To reduce these errors, we suggest that bin boundaries be made smaller, which may increase the failure rate of the sample but should reduce incorrect bin assignments and, therefore, the overall number of errors.
Null alleles are likely to be due to a mutation in one of the priming sites of the amplifying primers. This contention would need to be verified by sequencing the primer regions. There was a large percentage (>12% of errors) of priming-site mutations, in both the LMSV2 data and the FMS data. At least in the LMSV2 data, some of these findings are explained by null alleles, since, for three of the markers, these alleles segregate in many of the families (data not shown). Other studies also have found that these three microsatellites have null alleles (data not shown), suggesting that the priming-site mutations are possibly quite old and prevalent in Australian populations.
New mutations arising within the microsatellite repeat were used to calculate mutation rates of 3×10-4 and 5.6×10-4 for LMSV2 and FMS, respectively. These calculated rates are in agreement with the rate of 4.53×10-4 that was found by Ghosh et al. (1997) and with rates measured in other studies (Tautz 1989; Henderson and Petes 1992). Errors arising from mutations were left in the data, since they should not necessarily be removed from data sets during the initial analysis; rather, their identity should be made known to the linkage analysts, for their assessment.
Importantly, this study has suggested strategies for reduction of laboratory error. Genotyping laboratories are required to process a large number of genotypes within a reasonable time frame. Suggestions for decreasing the error rates have included repetition of all assays, repetition of the running of gels on common PCR material, or having duplicate allele calls. Our data would indicate that none of these approaches would decrease the error rate sufficiently to warrant application to complete marker sets, especially commercial sets. This has been conclusively demonstrated by our showing that the rate of discordance between repeat samples (i.e., 0.16% error detected) is much less than the overall detected error rate of 0.25% in the LMSV2 data. By relying on duplication to identify all errors, our results have shown that a whole group of errors caused by mutations and null alleles will be missed. It is worth noting that concordance checking using duplicate samples between gels can be of benefit for further error reduction in markers identified, by Mendelian-inheritance–error analysis, as displaying preferential amplification. This observation is of greater importance in fine-mapping data sets, in which the marker performance is not as robust as that in the commercial data set.
The single greatest source of preventable errors in our laboratory is incorrect labeling of samples. This can be ameliorated by routinely confirming the identity of samples by double typing the sample sheets and checking the label concordance both before and after the run, by means of simple macros written in Excel; any incorrect sample sheets are then easily replaced. These simple procedures should remove all sample-file errors.
The use of PedManager (or other pedigree-checking software—such as PedCheck, from the Division of Statistical Genetics, Department of Human Genetics, University of Pittsburgh) (O'Connell and Weeks 1998) enables any marker-related problems or mutation events to be easily identified, provided that genotypes of relatives are available and that the pedigree information is correct. As a result of our studies, we routinely repeat any marker showing >5% Mendelian-inheritance errors that cannot be explained as either a laboratory error or a null mutation. This ensures that, if a marker has a 1-bp allele or preferential amplification, then two calls are made from different amplifications, ensuring more accurate results.
It is impossible to detect all errors; however, recognition of the type and cause of allele-calling errors will reduce their incidence. In large-scale genotyping laboratories using either commercial marker sets or custom-designed fine-mapping sets, the reduction of error incidence is crucial. Finally, the inclusion of family data to identify Mendelian-inheritance errors is imperative for the success of projects, since errors can seriously interfere with data interpretation. This study has demonstrated the value of family genotyping data, since they allow Mendelian-inheritance–error checking. It also has highlighted the value of close collaboration between the client and the genotyping laboratory, since such communication ultimately provides the highest-quality data. The genotyping laboratory should have access to the pedigree data, to detect poorly performing markers; however, the genotyping analyst should not attempt to correct errors that are not due to human error, since this may lead to bias. Such a procedure also allows the detection of pedigree errors and sample swaps at the earliest point in time. This enables early decisions to be made regarding the potential need to collect further samples.
The remaining genotyping errors are easily identified by the statistical geneticist who may be able to recover some data. Alternatively, the prudent strategy is to declare that the genotypes found to be in error are missing data.
Concordance checking alone will not identify some errors, such as those caused by mutations, since these will appear consistently. Furthermore, even after concordance checking, some human errors may remain hidden. In contrast, Mendelian-inheritance–error checking is able to detect a broader range of errors, including mutations. The ability to detect errors by means of Mendelian-inheritance–error checking is highly dependent on the amount of genotyping data available. In general, the availability of parental genotyping data determines the efficacy of Mendelian-inheritance–error checking. For example, in sibling-pair data for which no parental genotyping data are available, it is not possible to carry out Mendelian-inheritance–error checking on autosomal data. In late-onset diseases, such as glaucoma, this deficiency is a common occurrence and has considerably hampered linkage analysis investigating such diseases. An alternative is to examine such sib-pair data for the presence of double recombinants, effectively using a multipoint approach (Douglas et al. 2000). This is a successful strategy in experimental crosses but has proved both to have a high false-negative rate with markers of density <5 cM and to be prone to bias (Bahlo and Broman 1999).
This study has shown that the optimal strategy for genotyping-error checking is dependent on the type of study. Mendelian-inheritance–error checking should be used for all types of genotyping studies, since it identifies error types additional to those detected by concordance checking. For fine-mapping studies, where the markers perform less consistently, there is an advantage to concordance checking, and it should be employed at least for markers found to be problematic. When markers were more robust, as in the commercial sets, we did not find any increase in the errors detected by concordance checking, and so we would suggest that Mendelian-inheritance–error checking should identify most of the initial errors. These precautions will optimize the amount of usable data derived from a project and so will increase the chance of discovering linkage.
Acknowledgments
We thank Nancy De Filippis, Margareta Go, Tim Barlow, and Paige Stevenson for their technical assistance; and Wayne Ward, Dr. Garry Myers, and Dianne Arnold for genotyping. This study was supported in part by the CRC for Discovery of Genes for Complex Human Diseases, which is established and supported by the Australian Government’s Cooperative Research Centre’s Program and by National Institute of Mental Health grants MH45097 and KO2-01207.
Appendix A: Mendelian-Inheritance–Error Categories
| Microsatellite mutation | Alleles are called correctly but there is a size shift of 1–2 bp (in the LMSV2 data) or 1–4 bp (in the FMS data), depending on repeat type |
| Priming-site mutation | An individual is missing an allele from one of the parents or has inherited a “null” (nonamplifying) allele from a parent |
| Missed allele | An allele in the genotype is missed during calling |
| Sample swap | Sample is incorrectly labeled |
| Call error | Allele is incorrectly assigned |
Appendix B: Call-Error Subcategories
| Binning | Bins are incorrectly assigned, so that samples are incorrectly binned or, because of inconsistent A+ preferential amplification, fall out of the bin |
| Sample | Alleles are sized incorrectly, because of low fluorescence |
| Incorrect call | Alleles are incorrectly assigned |
| GENOTYPER not updated | Manual genotype change is performed, but software is not updated with the correction |
| Leak | The adjacent lane’s sample bleeds through to the lane during loading and is called when genotyped |
| Other | Pull-up peaks caused by spectral overlap or overfluorescence |
Electronic-Database Information
The URLs for data in this article are as follows:
- Australian Genome Research Facility, http://www.agrf.org.au/ (for PedManager version 0.9)
- Division of Statistical Genetics, Department of Human Genetics, University of Pittsburgh, http://watson.hgen.pitt.edu/register/soft_doc.html (for PedCheck)
References
- Bahlo M, Broman KW (1999) Identification of and adjustment for genotyping errors in data on sibpairs when parental genotypes are unavailable. Am J Hum Genet Suppl 65:A241 [Google Scholar]
- Bowcock AM, Ruix-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457 [DOI] [PubMed] [Google Scholar]
- Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20:1004–1010 [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Dixon J, Raskova D, Le Beau MM, Williamson R, Klinger C, Landes GM (1992) Genetic and physical mapping of the Treacher Collins syndrome locus: refinement of the localisation to chromosome 5q 32-33.2.1. Hum Mol Genet 1:249–253 [DOI] [PubMed] [Google Scholar]
- Douglas JA, Boehnke M, Lange K (2000) A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data. Am J Hum Genet 66:1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges M, Dietz AB, Mishra A, Nielsen D, Sargeant TLS, Sorensen A, Steele MR, Zhao X, Leipolo H, Womack JE, Lathrop M (1993) Microsatellite mapping of the causing weaver disease in cattle will allow the study of an associated quantitative trait locus. Proc Natl Acad Sci USA 90:1058–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Karanjawala ZE, Hauser ER, Ally D, Knapp JI, Rayman JB, Musick A, Tannenbaum J, Te C, Shapiro S, Eldridge W, Musick T, Martin C, Smith JR, Carpten JD, Brownstein MJ, Powell JI, Whiten R, Chines P, Nyland SJ, Magnuson VL, Boehnke M, Collins FS, FUSION Study Group (1997) Methods for precise sizing, automated binning of alleles, and reduction in large-scale genotyping using fluorescently labelled dinucleotide markers. Genome Res 7:165–178 [DOI] [PubMed] [Google Scholar]
- Goldstein DR, Zhao H, Speed TP (1997) The effects of genotyping errors and interference on estimation of genetic distance. Hum Hered 47:86–100 [DOI] [PubMed] [Google Scholar]
- Henderson ST, Petes TD (1992) Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol 12:2749–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herber B, Herold K (1998) DNA typing of human dandruff. J Forensic Sci 43:648–656 [PubMed] [Google Scholar]
- Huges CR, Queller DC (1993) Detection of highly polymorphic microsatellite loci in a species with little allozyme polymorphism. Mol Ecol 2:131–137 [DOI] [PubMed] [Google Scholar]
- Levinson DF, Mahtani MM, Nancarrow DJ, Brown DM, Kruglyak L, Kirby A, Hayward NK, Crowe RR, Andreasen NC, Black DW, Silverman JM, Endicott J, Sharpe L, Mohs RC, Siever LJ, Walters MK, Lenon DP, Jones HL, Nertney DA, Daly KJ, Gladis M, Mowry BJ (1998) Genome scan of schizophrenia. Am J Psychiatry 155:741–750 [DOI] [PubMed] [Google Scholar]
- Lincoln SE, Lander ES (1992) Systematic detection of errors in genetic linkage data. Genomics 14:604–610 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pálsson B, Pásson F, Perlin M, Gudbjartsson H, Stefánsson K, Gulcher J (1999) Using quality measures to facilitate allele calling in high-throughput genotyping. Genome Res 9:1002–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LJ, Baldwin TM, Speed TP, Handman E, Foote SJ (1999) Chromosomes X, 9 and the H2 locus interact epistatically to control Leishmania major infection. Eur J Immunol 29:3047–3050 [DOI] [PubMed] [Google Scholar]
- Sacchetti L, Calcagno G, Coto I, Tinto N, Vuttariello E, Salvatore F (1999) Efficiency of two different nine-loci short tandem repeat systems for DNA typing purposes. Clin Chem 45:178–183 [PubMed] [Google Scholar]
- Tautz D (1989) Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res 17:6463–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AC, Sherwin WB, Wayne RK (1994) Genetic variation of microsatellite loci in a bottlenecked species: the northern hairy-nosed wombat Lasiorhinus krefftii. Mol Ecol 3:277–290 [DOI] [PubMed] [Google Scholar]