Abstract
Human papillomaviruses (HPVs), most commonly the HPV16 genotype, are the principle etiological determinant for cervical cancer, a common cancer worldwide resulting in over 200,000 deaths annually. The oncogenic properties of HPVs are attributable in part to the virally encoded protein E7, best known for its ability to bind to and induce the degradation of the retinoblastoma tumor suppressor, pRb, and related “pocket proteins” p107 and p130. Previously, we defined a role for E7 in the productive stage of the HPV16 life cycle, which takes place in stratified squamous epithelia. HPV perturbs the normal processes of cell growth and differentiation of stratified squamous epithelia. HPVs reprogram cells to support continued DNA synthesis and inhibit their differentiation in the suprabasal compartment of the epithelia, where cells normally have withdrawn from the cell cycle and initiated a well-defined pattern of terminal differentiation. These virus-induced perturbations, which contribute to the production of progeny HPVs, are dependent on E7. In this study, we define the mechanism of action by which E7 contributes to the productive stage of the HPV16 life cycle. We found that the ability of HPV16 to reprogram suprabasal cells to support DNA synthesis correlates with E7's ability to bind pocket proteins but not its ability to induce their degradation. In contrast, the ability of HPV16 to perturb differentiation correlated with both E7's binding to and degradation of pocket proteins. These data indicate that different hallmarks of the productive stage of the HPV16 life cycle rely upon different sets of requirements for E7.
Human papillomaviruses (HPVs) are small DNA tumor viruses that infect stratified squamous epithelial cells. There are over 100 genotypes of HPV that are classified as cutaneous or mucosotropic, based on whether they primarily infect the skin or the anogenital tract/cavity, respectively. The mucosotropic HPVs are further subclassified as low or high risk, depending on their etiological association with human cancers. High-risk HPVs, such as HPV16, are accepted as the main causal factor for cervical cancer, a leading cause of death among women worldwide, and other less common anogenital cancers including cancers of the vagina, vulva, penis, and anus (50, 52). In addition, high-risk HPVs are associated with a subset of oral cancers (17, 52). One of the HPV genes implicated in HPV-associated cancer is E7 (23, 26, 34, 44). E7 also plays a critical role in the viral life cycle by reprogramming cells within the suprabasal compartment of stratified squamous epithelia to support DNA synthesis, a prerequisite for viral DNA amplification and production of progeny virus (12). E7 is a multifunctional protein best known for its ability to bind and inactivate the retinoblastoma tumor suppressor, pRb, and related “pocket proteins” p107 and p130 (10, 28, 36, 42). In this study, we examined the mechanism of action by which E7 contributes to the viral life cycle and, in particular, the importance of E7 interaction with pRb and the other pocket proteins for the viral life cycle.
The life cycle of HPVs is intimately tied to the differentiation of the stratified squamous epithelia these viruses infect (37, 41, 49). HPVs are thought to initiate infection at sites of wounding where they can gain access to the poorly differentiated basal compartment of stratified squamous epithelial cells. There HPV establishes its circular, double-stranded DNA genome as extrachromosomal nuclear plasmids of low number and expresses a subset of viral genes, the early (E) genes, at low levels (6, 48). In this nonproductive stage of the viral life cycle, no late (L) viral structural genes are expressed, and therefore no progeny virions are generated. The productive stage of the life cycle takes place in the suprabasal compartment of the stratified squamous epithelia. Normally, when a basal cell divides, one of the daughter cells loses contact with the basement membrane that separates the epithelium from the underlying stroma. As a consequence, the daughter cell migrates into the suprabasal compartment where it withdraws from the cell cycle and initiates a program of terminal differentiation; these events are hallmarks of normal suprabasal cells. HPV perturbs multiple aspects of this differentiation program including the withdrawal of cells from the cell cycle and the expression of differentiation-specific cellular genes. Most notably, suprabasal cells that harbor HPV genomes undergo DNA synthesis and have delayed expression of differentiation-specific proteins (11, 12, 16, 24). These changes induced by the virus are thought to be critical for the synthesis of viral genomic DNA and its encapsidation to form progeny virus (24). Within the HPV16 life cycle, E7 plays a critical role in this productive stage of the life cycle, as shown by the fact that E7Null HPV16 genomes are deficient in delaying the onset of differentiation, inducing DNA synthesis in the suprabasal cells and supporting viral DNA amplification (12). How E7 contributes to these different processes in the viral life cycle is not known. E7 is a multifunctional protein that can interact with at least 20 different proteins, including cell cycle regulators, transcription factors, and other metabolic factors (33). One of the most-studied partners of E7 is pRb. E7 can bind and induce the degradation of pRb, as well as its family members p107 and p130. The pocket proteins regulate the cell cycle at least in part by their modulation of the E2F family of transcription factors. The E2F family members are DNA-binding proteins that can both inhibit and activate transcription of cellular genes implicated in cell cycle regulation and DNA synthesis (3). The disruption of the pocket proteins by E7 leads to dysregulation of these E2F-responsive genes and thus aberrant cell cycle entry and/or an inability to exit the cell cycle (5, 9, 18, 20, 22). Therefore, the ability of the viral oncogene E7 to bind and induce the degradation of the pocket proteins, especially pRb, has been postulated to contribute to the life cycle of HPV, in particular the ability of the virus to reprogram suprabasal cells to support DNA synthesis.
In this study, we investigated whether the ability of E7 to bind and degrade the pocket proteins is required for the HPV16 life cycle in stratified squamous epithelial cells. For these studies, wild-type HPV16 genomes or E7 mutant HPV16 genomes defective in binding to and/or degradation of the pocket proteins were introduced into a human keratinocyte cell line, NIKS, that supports the viral life cycle (12, 35, 43). These different populations of NIKS were then grown in organotypic raft cultures (rafts) to allow differentiation and completion of the viral life cycle. To determine the effect of the E7 mutations on the life cycle, raft cultures were analyzed for DNA synthesis, cellular proliferation, differentiation, and late viral gene expression. The results of our study indicate that in the context of the entire HPV16 genome, different biochemical properties of E7 are required for it to contribute to these different processes in the viral life cycle.
MATERIALS AND METHODS
Cell lines.
The normal immortalized keratinocyte cell line NIKS (previously termed BC1-Ep/SL cells), was used in this study. NIKS cells are a spontaneously immortalized human keratinocyte cell line that is able to support the entire life cycle of HPV16 (13). NIKS were maintained as subconfluent cultures on mitomycin C-treated m1 3T3 feeder cells, as previously described (16). Early-passage human foreskin keratinocytes (HFKs) were maintained in culture as previously described (12).
Construction of E7 mutant HPV16 genomes.
A recombinant plasmid, pEFHPV-16W12E, derived from W12E cells was used as the source of HPV16 DNA. The E7Null mutant HPV16 genome used in this study was constructed as previously described (12). The four-amino-acid in-frame deletion mutant, E7ΔDLYC, and the five-amino-acid in-frame deletion mutant, E7ΔPTLHE, have been previously described (36). These mutant sequences were constructed in pEFHPV-16W12E in the same fashion as the E7Null mutant HPV16 genome (12). The presence of the desired mutations was confirmed by sequence analysis, and the entire HPV16 genome of representative clones containing the desired mutation was then sequenced to confirm the absence of spurious mutations.
Stable transfectants.
Transfection of NIKS cells was performed as previously described (24). Briefly, to excise the bacterial vector, 5 μg of recombinant plasmid DNA was digested with BamHI for 2 h and then heat inactivated at 80°C for 20 min. DNA was then diluted up to 2 ml in ligase buffer with appropriate amount of ligase and allowed to ligate overnight at 16°C. Religated DNA was then purified with a QIAGEN (Valencia, CA) Spin kit. The purified DNA, along with pEGFP-N1 (Clontech, Palo Alto, CA), was then transfected into NIKS cells using Superfect (QIAGEN), as previously described (12). At 24 h after transfection, populations were subjected to G418 selection for 4 days. Following G418 selection, populations were maintained in culture until harvested for the presence of HPV16 DNA.
Identification of transfected populations that stably harbor HPV16 replicons.
To identify populations that stably carried the HPV16 genome as an extrachromosal plasmid, i.e., replicon, Southern analyses of total genomic DNA were performed as previously described (24) with the following modifications. Briefly, total genomic DNA was extracted from populations maintained subconfluently. A total of 10 μg of DNA extract was digested with BamHI and run on a 0.8% agarose gel to detect linear forms of the HPV16 genome. A total of 20 μg of undigested total genomic extract was also separated by electrophoresis in the same manner to detect supercoiled (SC) and open circular (OC) extrachromosomal forms of the HPV16 genome. Electrophoresed DNAs were transferred from agarose gels onto nitrocellulose membranes (Schleicher & Schuell BioScience, Inc., Keene, NH). The nitrocellulose membranes were then probed witha BamHI-digested pEFHPV-16W12E probe, which was labeled with [α-32P]dCTP with a random primer labeling kit (Amersham, Piscataway, NJ). Following hybridizations and standard washes, 32P-labeled probe was detected with a phosphorimager.
Organotypic raft cultures.
To produce organotypic rafts, populations of NIKS cells were treated as previously described (16), with the following modifications. Transwell inserts (Costar, Acton, MA) were placed into Deep Well plates (BIOCOAT; Becton Dickinson, San Jose, CA). Four days after the collagen dermal equivalent was made in transwell inserts, 1.4 × 105 keratinocytes were added onto the dermal equivalents in keratinocyte plating medium. Two days later, medium was removed from the outer well and fresh keratinocyte plating medium was added. After 2 more days, the collagen dermal equivalent was lifted to the air-liquid interface by placing the transwell inserts on three or four 1- by 1-in. cotton pads (Schleicher & Schuell BioScience, Inc.). At this point, the cultures were fed every other day with cornification medium. Eleven days after the dermal equivalent was lifted to the air-liquid interface, 10 μM bromodeoxyuridine (BrdU) was added to the medium for 8 h prior to harvesting the rafts. After rafts were harvested, they were fixed in 2% agar-1% formalin/phosphate-buffered saline (PBS) overnight at 4°C, embedded in paraffin, and cut into 5 μM sections.
Immunofluorescence.
Immunostaining was performed with 5 μM sections of raft cultures that were formalin fixed and paraffin embedded. All sections were deparaffinized by being submerged in two changes of xylenes for 3 min each and then rehydrated by being submerged in a graded (100%, 95%, 85%, and 70%) series of alcohol:aqueous solutions. For pRb, keratin 10 (K10), BrdU, L1, and involucrin immunostaining, sections were placed in a 10 mM citrate buffer, pH 6.0, and microwaved for 3 min on high power and then at power level 7 for 17 min to unmask the antigens. Raft sections that were analyzed for Ki67 and E4 were also boiled in the same manner but in a 10 mM citrate buffer, pH 9.5, or 10 mM citrate buffer, pH 5.0, respectively. After being unmasked, rafts were cooled to room temperature, rinsed in PBS, and then blocked in 5% normal horse serum (Vector Laboratories, Inc., Burlingame, CA)-PBS at room temperature for 30 min. To detect pRb, sections were incubated with an anti-pRb primary monoclonal antibody (catalog no. 54136; Pharmingen, BD Dickinson, San Jose, CA) at 1:25. All antibodies are diluted in the 5% normal horse serum-PBS blocking solution. For MCM7 staining, MCM7-specific cdc47 monoclonal antibody (NeoMarkers, Fremont, CA) was used at a 1:200 dilution. Ki67-specific staining was achieved with the NA59 monoclonal antibody (clone K-3; Oncogene, San Diego, CA) at 1:100. Clone Ck 8.60 and clone SY-5 (Sigma, St. Louis, MO) monoclonal antibodies were used at 1:200 to detect K10 and involucrin, respectively. H16.D9, an L1-specific antibody, was a gift from Neil Christensen and was incubated with the raft sections at a dilution of 1:20. For BrdU-only immunofluorescence, anti-BrdU monoclonal antibody Ab-2 was used at a 1:50 dilution (Oncogene). In double immunofluorescence involving BrdU, i.e., pRb/BrdU and K10/BrdU staining, BrdU was detected using a biotin-labeled mouse anti-BrdU specific primary antibody (Zymed, San Francisco, CA). In both cases of double immunofluorescence, pRb and K10 were detected as stated above. Incubation times for primary antibodies were either 1 h at 37°C, 3 h at room temperature, or overnight at 4°C in a humid chamber. Primary antibodies were detected with fluorescein or Texas red anti-mouse immunoglobulin G (Vector Laboratories) secondary antibodies at a 1:100 dilution. To detect biotin-BrdU, Texas red-conjugated anti-streptavidin secondary antibody (Vector Laboratories) was used at a 1:50 dilution. Sections were incubated with secondary antibodies in the dark for 30 min at room temperature. E4 was detected as previously described (16).
Quantification of suprabasal DNA synthesis and proliferation.
For all calculations, averages were calculated based on counting 10 fields at a magnification of ×40 of at least three different populations. To determine the average pRb expression for each population of NIKS cells, the total number of cells per field were counted, as well as the total number of pRb-positive cells per field in pRb-specific, immunohistochemically stained raft sections. The percentage of pRb-positive cells was calculated for the basal compartment (pRb-positive basal cells divided by the total number of basal cells) and for the suprabasal compartment (pRb-positive suprabasal cells divided by the total number of suprabasal cells). The percentage of pRb-positive cells in the untransfected NIKS was standardized to 1.0. For each compartment, the percentage of pRb-positive cells in the population harboring HPV16 replicons was divided by the average percentage of pRb-positive in the untransfected NIKS cells, thus giving a relative pRb expression index (e.g., percentage of pRb-positive basal cells in HPV16 rafts/pRb-positive basal cells in NIKS = average basal pRb expression for HPV16). This method was also used to calculate the suprabasal BrdU index. A two-sided Wilcoxon rank sum test was performed with the MStat Program to determine the statistical significance of values. All error bars indicate standard deviations.
MCM7 expression was calculated as the number of MCM7-positive cells in the suprabasal compartment divided by the total number of suprabasal cells in MCM7-specific, immunohistochemically stained raft sections. Ki67 suprabasal expression was calculated in the same fashion. To calculate the percentage of BrdU-positive suprabasal cells in the supraparabasal compartment, the total number of BrdU-positive cells in the suprabasal compartment was determined and the number of BrdU-positive cells in the parabasal layer was subtracted from that value to give the number of BrdU-positive supraparabasal cells. The fractional value of the expression [(number of BrdU-positive supraparabasal cells)/(total number of BrdU-positive suprabasal cells) × 100] gave the percentage of BrdU-positive suprabasal cells in the supraparabasal compartment.
Western analyses.
Analyses of steady-state levels of pRb and E7 were performed with monolayer cultures and raft sections. For monolayer protein analyses, subconfluent cultures were treated with 0.02% EDTA for 2 min at room temperature to remove the mitomycin C-treated m1 3T3 feeders. Subsequently, cells were either directly lysed with the addition of lysis buffer onto the plates at 4°C or treated with 0.05% trypsin-0.53 mM EDTA (Mediatech, Inc., Herndon, VA). Trypsin-treated cells were allowed to detach at 37°C, at which point an equal volume of medium was added to the cells to inhibit further trypsinization. Cell pellets were then isolated by centrifugation at 1,100 rpm for 5 min at 4°C. Cell pellets were washed with cold PBS twice and then resuspended in lysis buffer. The lysis buffer contained 20 mM Tris (pH 8.0), 0.1 M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20, phenylmethylsulfonylfluoride, dithiothreitol, and a general protease inhibitor cocktail that included aprotinin, pepstatin, and leupeptin. For direct lysis of cells on tissue culture plates, cells were washed twice with cold PBS, and then 1 ml lysis buffer was added to 10-cm dishes or 2 ml of lysis buffer was added to 15-cm dishes. The dishes were then placed on a rocker at 4°C for 20 min. After 20 min, cells were collected by being scraped and placed on ice for 20 min, with vortexing every 5 min. After complete lysis, lysates were centrifuged at 14,000 rpm for 10 min at 4°C. The soluble fraction was then subsequently used for protein analyses.
To analyze protein levels from raft sections, rafts were harvested as normal but not formalin fixed or further treated. Instead, rafts were frozen at −80°C or immediately placed into lysis buffer and subjected to low-power homogenization by the PowerGen 125 Homogenizer (Fisher Scientific; Hampton, N.H.) until a single-cell suspension was achieved. The lysate was then treated in the same manner as monolayer lysates.
To detect E7, protein lysates were subjected to electrophoretic separation on a sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis gel or a gradient gel (4.5 to 15%; Bio-Rad, Hercules, CA) and then transferred to a polyvinylidene difluoride Immobilon-P membrane (Millipore, Billerica, MA). Subsequently, the membrane was blocked for 1 h at room temperature in 5% skim milk-PBST (PBST is 0.1% Tween 20-PBS). A primary antibody mixture containing a 1:150 dilution of 8C9 (Zymed, Burlingame, CA) and a 1:200 dilution of ED17 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) in blocking solution was then incubated with the membrane overnight with shaking at 4°C. After blots were washed with PBST, they were then treated with a goat anti-mouse peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) at a 1:5,000 dilution in blocking solution at room temperature for 1 h. Blots were subsequently washed and developed with ECL Plus (Amersham, Piscataway, NJ).
Detection of pRb, actin, and filaggrin was similar to the detection methods used for E7, with the following exceptions. pRb lysates were electrophoretically separated on a 4.5 to 15% gradient gel (Bio-Rad), and pRb was detected using a mouse anti-pRb primary antibody (SC-102; Santa Cruz) at a 1:500 dilution in blocking solution. β-Actin was detected with the monoclonal antibody clone SY-5 (Sigma) at a dilution of 1:5,000 to 1:10,000. Filaggrin-specific primary antibody (Covance, Richmond, CA) was used at a 1:200 dilution.
RESULTS
Binding of E7 to pocket proteins and induction of their degradation are not required for the nonproductive stage of the life cycle of HPV16.
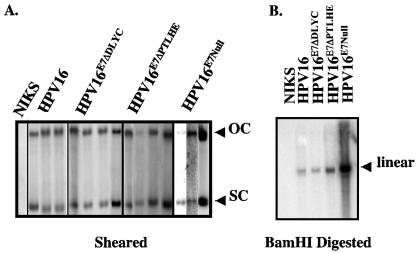
To examine the role of E7 interacting with pocket proteins in the nonproductive stage of the life cycle, we examined the ability of wild-type and E7 mutant HPV genomes to be maintained as an extrachromosomal nuclear plasmid (defined here as a replicon) within NIKS cells. NIKS cells are a spontaneously immortalized human keratinocyte cell line that can fully support the HPV replicative life cycle (13). E7 mutants were made in the context of the entire HPV16 genome. The mutants included a five-amino-acid in-frame deletion mutant, ΔPTLHE, which is previously described and is competent to bind the pocket proteins, but defective in the degradation of the pocket proteins (18, 22, 36). Another mutant, ΔDLYC, contains a four-amino-acid in-frame deletion previously shown to be defective in binding and in degradation of the pocket proteins (18, 22, 35). The properties of these E7 mutant genomes were compared to the previously characterized, E7Null HPV16 genome that contains a translation termination linker in the E7 open reading frame (12). Populations of NIKS were cotransfected with plasmid pEGFPN1 (a plasmid conferring neomycin resistance) and either the wild-type HPV16 genome or the E7 mutant HPV16 genomes. Populations were selected for resistance to G418. After expansion, total genomic DNA was extracted, sheared and/or digested with BamHI, and subjected to HPV16-specific Southern analysis to determine the status of the HPV16 genomes. Southern analyses of representative populations are shown in Fig. 1. NIKS populations transfected with either wild-type or E7 mutant HPV16 genomes (ΔDLYC, ΔPTLHE and E7Null), harbored the HPV16 genome as an extrachromosomal nuclear plasmid, as illustrated by the presence of SC and OC HPV16 DNA in samples that were only sheared (Fig. 1A). Digestion with BamHI, which cleaves the HPV16 genome once, indicated the presence of unit-length viral genomes (Fig. 1, right). The absence of additional HPV16-specific bands in the BamHI digest was consistent with an absence of viral DNA integration. These data confirm an earlier study that demonstrated HPV16 E7Null genomes replicate as nuclear plasmids in NIKS cells (12) and furthermore demonstrate that the ability of E7 to bind and/or degrade pocket proteins is dispensable for the viral plasmid maintenance.
FIG. 1.
Establishment of wild-type and E7 mutant HPV16 populations that stably harbor the HPV16 genome as an extrachromosomal plasmid (replicon). NIKS cells were cotransfected with a neomycin-resistant plasmid and wild-type or E7 mutant HPV16 genomes. After G418 selection, total genomic DNA was extracted from the transfected populations to determine if the HPV16 genomes were maintained as an extrachromosomal nuclear plasmid (replicon) within NIKS cells. (A) Total genomic DNA extracted from NIKS cells and NIKS cells transfected with wild-type or E7 mutant HPV16 genomes was sheared and subsequently analyzed by Southern blotting. Blots were probed with a full-length HPV16 probe labeled with 32P. The presence of OC and SC forms of the HPV16 genome confirms that the populations harbor HPV16 as a replicon. (B) Total genomic extracts of populations were sheared, digested with BamHI, which cuts the HPV16 genome only once, and then analyzed by Southern analysis as in panel A. As evidenced by the singular band representing the linear form of the HPV16 genome, the NIKS populations transfected with wild-type or E7 mutant genomes contained wild-type genomes with no gross aberrations and/or integrations.
HPV16 causes a reduction in the abundance of pRb selectively in the basal compartment of stratified squamous epithelia, and this reduction correlates with E7's ability to induce degradation of pRb.
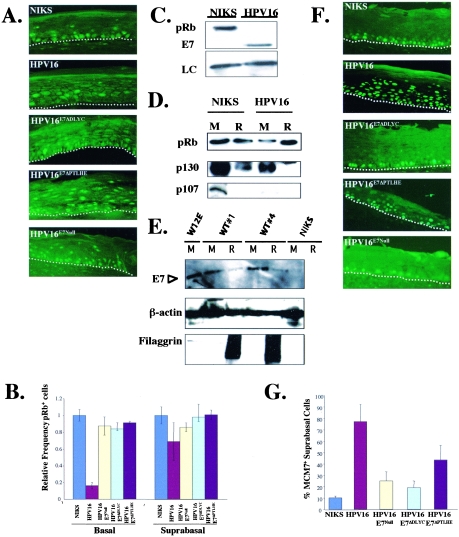
To study the contribution of binding and degradation of the pocket proteins by E7 in the productive stage of the HPV16 life cycle, we placed our keratinocyte populations into organotypic (raft) cultures to allow their stratification and terminal differentiation. Prior studies carried out exclusively with monolayer cultures of human epithelial cells indicated that E7 can destabilize pRb and that both ΔDLYC and ΔPTLHE E7 mutants are defective for inducing the degradation of the pocket proteins (5). We therefore predicted that in raft cultures of human keratinocytes harboring wild-type HPV16 replicons, but not in those harboring E7 mutant HPV16 replicons, pRb levels would be decreased compared to those levels observed with raft cultures of cells not harboring any HPV16 genomes. To test this prediction, we carried out pRb-specific immunofluorescence analyses (Fig. 2A and B). In the basal compartment of rafts harboring wild-type HPV16 replicons, there was a significant (P = 0.02) loss of pRb-positive cells, down to 17% of that seen with HPV16-negative rafts (Fig. 2B). However, the reduction of pRb levels in the suprabasal compartment was less. Seventy percent of cells expressed pRb detectably compared to rafts not harboring HPV16 replicons (Fig. 2B). This reduction was still statistically significant (P = 0.04), indicating that HPV16 retains some capacity to destabilize pRb in suprabasal cells, albeit less so than in basal cells. pRb-specific immunofluorescence was also performed with rafts of HFKs in the absence and presence of HPV16 DNA (data not shown). Results were similar to those observed with NIKS cells, i.e., pRb expression in the presence of HPV16 led to a striking decrease in pRb expression in the basal compartment of the rafts, but pRb was largely retained in the suprabasal compartment (data not shown). These data further emphasize the usage of NIKS as a valid keratinocyte cell line, which can be used to study the life cycle of HPV16.
FIG. 2.
HPV16 significantly degrades pRb in the basal compartment versus the suprabasal compartment yet still can inactivate pRb in the suprabasal compartment, as judged by the induction of an E2F-responsive gene. (A) pRb levels in raft cultures of NIKS cells harboring no (NIKS), wild-type (HPV16), or E7 mutant (HPV16ΔDLYC, HPV16ΔPTLHE, and HPV16E7−Null) HPV16 replicons. Formalin-fixed, paraffin-embedded sections of raft cultures were subjected to pRb-specific immunofluorescence. The white dotted lines indicate the bottom of the basal compartment of the stratified squamous epithelium. (B) Quantification of pRb-positive cells in the basal and suprabasal compartments. For quantification, pRb-positive cells in the basal and the suprabasal compartments of raft sections were counted in 10 random fields (magnification, ×40) of sections from raft cultures of at least three different cell populations for each HPV replicon, as described in Materials and Methods. For each compartment, the average number of pRb-positive cells in the HPV-negative populations was set at a value of 1.0, and the number of pRb-positive cells in the HPV-positive populations is shown relative to that of the HPV-negative populations. A significant decrease in pRb-positive cells in rafts harboring wild-type HPV16 replicons versus NIKS cells was observed with the basal (P = 0.02092) and the suprabasal (P = 0.04331) compartments. (C) Immunoblot analyses of pRb levels in monolayer cultures of keratinocytes. Western blot analyses of cell lysates of monolayer cultures of NIKS cells and NIKS cells harboring wild-type HPV16 replicons were performed. Loading control (LC) levels of a nonrelevant protein were used to confirm that equal amounts of cellular protein were loaded on the gel. (D) Comparison of levels of pRb, p130, and p107 in raft (R) cultures versus monolayer (M) cultures. Western blot analyses were performed with 20 μg (each) of protein-isolated monolayer (M) and raft (R) cultures of NIKS cells and wild-type HPV16 replicons. (E) Comparison of levels of E7 protein in raft cultures versus monolayer cultures. E7-specific Western analysis was performed on 100 μg of protein lysate from monolayer and raft cultures as described in Materials and Methods. The intensity of the E7-specific signal in the two raft cultures was one-third that in the monolayer cultures for the same HPV16-positive cell cultures. The same blot was probed with antibodies either to β-actin to confirm that equivalent amount of cellular protein were loaded or to filaggrin to assess the differentiation state of the cells being analyzed. (F) Expression of the E2F-responsive gene MCM-7 in raft cultures of NIKS cells harboring no HPV16 replicons (NIKS), wild-type HPV16 (HPV16) replicons, or E7 mutant HPV16 (HPV16ΔDLYC, HPV16ΔPTLHE, and HPV16E7−Null) HPV16 replicons. MCM7-specific immunofluorescence was performed on sections of raft cultures. The white dotted lines indicate the bottom of the basal compartments of the stratified squamous epithelium. (G) The number of MCM7-positive cells in the suprabasal compartment versus the total amount of MCM7-positive cells (percentage of MCM7 suprabasal cells) was graphed. To calculate the averages, 10 random fields (magnification, ×40) of three different populations of each replicon were counted.
To further analyze the expression patterns of the pocket proteins in the presence of HPV16, monolayer and raft culture lysates were examined by Western blotting. Monolayer cultures were grown subconfluently to maintain cells in the poorly differentiated, basal-like state. Whereas raft cultures are highly stratified and thus are primarily composed of suprabasal, differentiated cells, Western blot analyses (Fig. 2C) indicated that the steady-state levels of pRb, p130, and p107 were reduced in the monolayer cultures of NIKS cells harboring wild-type HPV16 replicons compared to NIKS cells not harboring any HPV16 DNA. This reduction confirms previous reports that E7 HPV16 can destabilize the pocket proteins; these data further demonstrate that E7 is able to destabilize the pocket proteins in the context of the HPV16 genome in stratified keratinocytes. In contrast to the monolayer data, the expression pattern of pocket proteins in the suprabasal compartment was not uniform. pRb expression did not significantly change in the presence of absence of HPV16 replicons, providing further evidence that HPV16 reduces pRb most strongly in the basal compartment of stratified keratinocytes and that pRb is largely retained within the suprabasal compartment. However, in strong contrast to pRb, p130 expression in raft cultures was significantly diminished in the presence of the HPV16 replicons. The suprabasal expression pattern of p107 in the presence or absence of HPV16 replicons was not clear, as p107 could not be detected in either HPV16-positive or -negative raft lysates.
To better understand the different expression patterns of the pocket proteins and their regulation, we decided next to analyze the expression of E7 in the basal and suprabasal compartments. In an earlier, collaborative study, E7 expression was observed with both the basal, as well as suprabasal, compartment of rafts harboring wild-type HPV16 based upon E7-specific immunofluorescence (31). This technique does not allow one to quantify levels of E7. We therefore extended these findings by quantifying the level of E7 in protein lysates extracted from monolayer and raft cultures by Western analysis (Fig. 2C and E). We observed E7 protein in both the monolayer and the raft cultures; however, the level of E7 protein in the raft culture was approximately 30% of that seen in the monolayer culture. These data indicate that the steady-state levels of E7 are lower in the suprabasal, differentiated compartment than in the basal compartment.
To assess whether these lower levels of E7 can inactivate pRb and the other pocket proteins p107 and p130 in the suprabasal compartment of rafts, we monitored levels of MCM7 protein by immunofluorescence (Fig. 2F). The MCM7 gene is highly induced by active E2F, and therefore provides a readout for the inactivation of the pocket proteins pRb, p107, and p130, all of which modulate the transcriptional activity of E2F family members. We found that MCM7 was highly induced in the suprabasal compartment of rafts harboring the wild-type HPV16 genome, but not in the rafts harboring either the E7ΔDLYC or E7Null HPV16 replicons (Fig. 2F and G). These data are consistent with E7 inactivating pocket protein function in the suprabasal compartment and that this inactivation depends on E7's ability to bind these pocket proteins. We also observed an induction of MCM7 (P = 0.05) in the suprabasal compartment of rafts harboring the E7ΔPTLHE mutant HPV16 replicon, but this induction was significantly reduced compared to that seen in rafts harboring wild-type HPV16 replicons (Fig. 2G). These data support the hypothesis that the ability of E7 to degrade the pocket proteins contributes to the functional inactivation of the pocket proteins, even though the degradation of the pocket proteins in the suprabasal compartment was not as uniform as in the basal compartment.
The ability of E7 to inactivate pRb and the other pocket proteins correlates with its capacity to reprogram the suprabasal compartment of keratinocytes to support DNA synthesis.
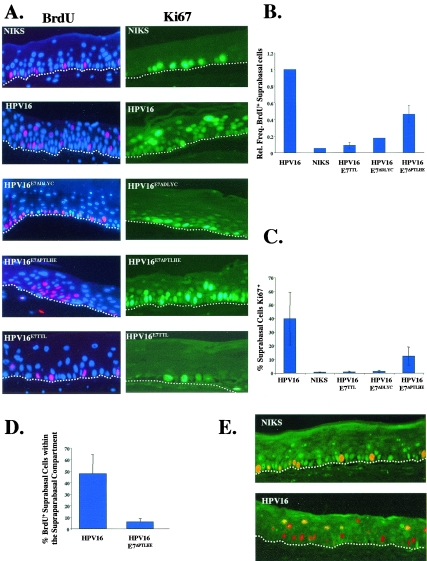
Cells within the suprabasal compartment of stratified squamous epithelia are normally quiescent and cannot initiate new rounds of DNA synthesis. A hallmark of the papillomavirus life cycle is the reprogramming of suprabasal cells to support DNA synthesis. We previously discovered that E7 is required for conferring this property in the context of the viral life cycle (12). To assess whether E7's ability to inactivate the pocket proteins contributes to suprabasal DNA synthesis, we analyzed BrdU incorporation into cells in raft cultures harboring wild-type or E7 mutant HPV16 genomes. The raft cultures were incubated with BrdU for 8 h prior to harvesting. After the cultures were harvested, formalin fixed, and paraffin embedded, the histological sections were subjected to BrdU-specific immunofluorescence (Fig. 3A, left). Raft cultures of keratinocytes harboring no HPV DNA exhibited BrdU-positive cells only in the basal compartment, as expected; raft cultures of keratinocyte populations harboring the wild-type HPV16 replicons exhibited BrdU-positive cells in both the basal and suprabasal compartments, consistent with prior studies (12, 13, 16). Raft cultures of keratinocytes harboring the E7Null HPV16 genomes did not exhibit any BrdU-positive cells in the suprabasal compartment, also consistent with prior studies (12). The same was true for raft cultures of keratinocytes harboring the E7ΔDLYC mutant HPV16 replicon (Fig. 3A), indicating that the ability of E7 to bind pocket proteins correlates with its ability to reprogram suprabasal cells to support DNA synthesis. In contrast, BrdU-positive cells were observed with the suprabasal compartment of raft cultures harboring E7ΔPTLHE mutant HPV16 replicons, indicating that while binding of the pocket proteins by E7 may be required to reprogram the suprabasal compartment to support DNA synthesis, degradation of the pocket proteins by E7 is not essential. To further clarify the role of E7's interactions with pocket proteins in the reprogramming of suprabasal cells to support DNA synthesis, the frequency of BrdU-positive cells within the suprabasal compartment was quantified for the various raft cultures analyzed (Fig. 3B). The average frequency of BrdU-positive cells in the suprabasal compartment of raft cultures harboring wild-type HPV16 replicons was set to a value of 1.0, and the frequencies obtained for the other cultures were graphed relative to that value. The frequency of BrdU-positive suprabasal cells in raft cultures harboring the E7Null and E7ΔDLYC mutant HPV16 replicons was not significantly different from that obtained with raft cultures of cells harboring no HPV16 DNA (Fig. 3B). In contrast, the frequency of BrdU-positive suprabasal cells with raft cultures of cells harboring the E7ΔPTLHE mutant HPV16 replicon was significantly greater than that obtained with raft cultures harboring no HPV genome (P = 0.04) (Fig. 3B), but that frequency was significantly lower (P = 0.03) than that observed with raft cultures harboring wild-type HPV16 replicons. These quantitative differences indicate that the ability of HPV16 to induce the degradation of the pocket proteins is not absolutely necessary, yet does contribute quantitatively to the efficiency to which HPV16 induces suprabasal DNA synthesis.
FIG. 3.
Degradation of the pocket proteins by HPV16E7 does not correlate with E7's ability to induce suprabasal DNA synthesis. (A, left) Eight hours prior to harvesting rafts, rafts of NIKS cells harboring no HPV replicons (NIKS; top row) or NIKS harboring wild-type or E7 mutant HPV16 replicons (second through fifth rows) were incubated with BrdU. BrdU was detected by BrdU-specific immunodetection and visualized by Texas red-conjugated secondary antibody. Raft sections were also costained with DAPI (4′,6′-diamidino-2-phenylindole) nuclear counterstain; DAPI stains nuclei blue. Shown are representative images of the populations studied. The white dotted lines indicate the bottom of the basal compartment of the stratified squamous epithelium in each panel. (Right) Ki67 expression in the same populations was determined by Ki67-specific immunohistochemistry of rafts. Ki67-positive cells are brown. Hematoxylin was used as a counterstain to detect nuclei; hematoxylin-positive cells are blue. Shown are representative images. (B) Relative suprabasal DNA synthesis was determined as described in Materials and Methods for at least three populations of each mutant and wild-type HPV16 replicon. (C) Ki67 expression was quantified as the percentage of suprabasal Ki67 expression as described in Materials and Methods. Ten random fields (magnification, ×40) of three different populations of each replicon were counted to calculate the percentage of suprabasal Ki67 expression. (D) BrdU incorporation in raft cultures that harbor E7ΔPTLHE mutant HPV16 replicons is spatially restricted to the lower levels of the suprabasal compartment. Supraparabasal BrdU incorporation in sections of rafts of NIKS cells harboring wild-type and E7ΔPTLHE mutant HPV16 replicons was determined. The number of BrdU-positive supraparabasal cells was divided by the total number of BrdU-positive suprabasal cells for each set of rafts. To calculate the averages, 10 random fields (magnification, ×40) of three different populations of each replicon were counted. (E) pRb degradation is not required for HPV to reprogram the suprabasal compartment. Sections of raft cultures of NIKS cells harboring no viral DNA (top) or harboring wild-type HPV16 replicons (bottom) were analyzed for pRb expression and BrdU incorporation by coimmunofluorescence. pRb-specific antibodies were used in conjunction with a fluorescein-conjugated secondary antibody to visualize pRb-positive cells. BrdU was detected using a biotin-labeled BrdU-specific primary antibody, followed by incubation with a Texas red-conjugated anti-streptavidin secondary. Cells that contain both pRb and BrdU are yellow.
To examine how interactions of E7 with pocket proteins affect the regulation of cellular factors that may contribute to and/or reflect the reprogramming of the suprabasal compartment to support DNA synthesis, we analyzed the expression of Ki67. Ki67 is a cell cycle-specific nuclear antigen that is not expressed in quiescent cells and is often used as a marker for cells that are proliferating (2). The expression of Ki67 was monitored by immunofluorescent staining. In raft cultures that did not contain any HPV DNA, Ki67 expression was restricted to the basal compartment of the epithelia (Fig. 3A, right). However, in rafts harboring wild-type HPV16 replicons, Ki67 expression was seen in the basal, as well as the suprabasal, compartment. In contrast, the raft cultures harboring E7Null or E7ΔDLYC mutant HPV16 replicons did not contain suprabasal Ki67 expression. This finding indicates that the ability of HPV16 to induce suprabasal proliferation and DNA synthesis was dependent on E7 and the ability of E7 to bind the pocket proteins. Raft cultures harboring the E7ΔPTLHE mutant HPV16 replicons exhibited Ki67 expression in the basal and the suprabasal compartments. Quantification of Ki67 staining in the suprabasal compartment confirmed that the frequency of Ki67-positive, suprabasal cells in the raft cultures harboring the E7ΔPTLHE mutant HPV16 replicons was lower than that observed with rafts harboring the wild-type HPV16 replicon (Fig. 3C). These findings parallel the results obtained with BrdU-specific immunofluorescence and indicate that the ability of E7 to bind the pocket proteins is necessary to establish a proliferative-like environment in the suprabasal compartment and that the ability of E7 to degrade the pocket proteins contributes quantitatively to this property as well.
While Ki67 expression and BrdU incorporation was observed with the raft sections of cells harboring E7ΔPTLHE mutant HPV16 replicons, it was noted that these markers of proliferation and DNA synthesis were restricted more to the lower levels of the suprabasal compartment than seen in rafts of cells harboring wild-type HPV16 replicons. To determine if there was a quantitative difference in this regard, the distribution of BrdU-positive cells in the parabasal (the first layer of cells above the basal layer) versus supraparabasal (all cells above the parabasal layer) compartments was determined for rafts of cells harboring wild-type versus E7ΔPTLHE mutant HPV16 replicons. There indeed was a significant decrease (P = 0.05) in the relative proportion of the BrdU-positive cells in the supraparabasal compartment of the rafts harboring the E7ΔPTLHE mutant HPV16 replicons compared to the rafts harboring wild-type HPV16 replicons (Fig. 3D). These data indicate that while HPV16 does not need to degrade the pocket proteins to induce suprabasal DNA synthesis, the ability to degrade the pocket proteins yields a more efficacious reprogramming of the suprabasal compartment to support DNA synthesis throughout a greater portion of the stratified epithelia.
Suprabasal cells that support DNA synthesis retain pRb.
E7's ability to degrade the pocket proteins contributed quantitatively to a reduction in the frequency of suprabasal cells either retaining pRb (Fig. 2) or supporting DNA synthesis (Fig. 3), compared to that seen with HPV16 genomes expressing wild-type E7. These findings led us to hypothesize that only in those suprabasal cells in which E7 HPV16 has depleted pRb levels would we find DNA synthesis to be induced. We therefore carried out double-immunofluorescence studies to identify the pRb status of suprabasal cells supporting DNA synthesis (Fig. 3E). In the HPV-negative NIKS rafts, BrdU incorporation was restricted to the basal compartment. Nearly all of these BrdU-positive cells were also pRb positive (Fig. 3E, top). In the raft cultures of cells harboring wild-type HPV DNA, BrdU-positive cells were found in the basal and the suprabasal compartments. In the basal compartment, these cells supporting DNA synthesis were largely pRb negative (Fig. 3E, bottom), consistent with our earlier analyses (Fig. 2) demonstrating that HPV can lower the levels of pRb in virtually all basal cells. However, in the suprabasal compartment, cells supporting DNA synthesis often expressed pRb (Fig. 3E, bottom). Thus, the depletion of pRb is not essential for HPV16 E7 to reprogram suprabasal cells to support DNA synthesis in the context of the HPV16 life cycle. These results were also observed with HFK raft cultures that harbored wild-type HPV16 replicons, as well as NIKS raft cultures that harbored HPV18 genomes (data not shown).
The ability of E7 to bind and degrade the pocket proteins correlates with the ability of HPV16 to disrupt differentiation.
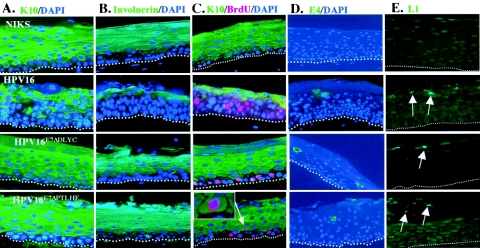
Early in the productive stage of the viral life cycle within the suprabasal compartment, the cellular differentiation program is disturbed, as evidenced by a delay in the onset of expression of the differentiation markers, K10 and involucrin. These markers normally are induced immediately upon cells migrating into the suprabasal compartment. However, in raft cultures of cells harboring wild-type HPV16 replicons, this induction is delayed, leading to the presence of multiple layers of K10-negative, involucrin-negative cells within the suprabasal compartment (13). A prior study demonstrated that HPV16 E7 is necessary for this disturbance of normal differentiation in the context of the viral life cycle (12) and sufficient to induce this delay in differentiation in mouse epidermis (1). To determine if the ability of E7 to bind and/or degrade the pocket proteins correlates with this delay in keratinocyte differentiation in the context of the viral life cycle, we analyzed the expression of K10 (Fig. 4A) and involucrin (Fig. 4B) in our raft cultures of cells harboring wild-type or E7 mutant HPV16 replicons. In the HPV-negative NIKS rafts, K10 and involucrin were expressed throughout the spinous layer of the rafts, as expected. The raft cultures of cells harboring wild-type HPV16 genomes displayed a delayed onset of K10 and involucrin expression within the spinous layer. In addition, expression of these markers was patchy in the more terminally differentiated granular layer. These defects in expression of K10 and involucrin are consistent with that observed with prior studies (12, 16, 24). In contrast to what was observed with the raft cultures of cells harboring wild-type HPV16, the rafts of cell populations harboring the E7ΔDLYC or the E7ΔPTLHE mutant HPV replicons displayed differentiation staining patterns similar to that seen with HPV-negative rafts (Fig. 4A and B) and rafts harboring E7Null HPV16 replicons (reference 12 and data not shown). These data indicate that the ability of HPV16 to delay differentiation correlates with both the ability of E7 to bind and to degrade the pocket proteins.
FIG. 4.
The ability of E7 to bind and degrade the pocket proteins correlates with the ability of HPV16 to disrupt differentiation in raft cultures. Immunofluorescence was performed on raft sections from raft cultures of NIKS (first row), HIKS cells harboring wild-type HPV16 replicons (second row), NIKS cells harboring E7ΔDLYC mutant HPV16 replicons (third row), and NIKS cells harboring E7ΔPTLHE mutant HPV16 replicons (fourth row). (A) Early differentiation is not perturbed in the absence of binding or degradation of the pocket proteins by E7. K10 expression was detected by K10-specific primary monoclonal antibody, followed by anti-mouse fluorescein-conjugated secondary antibody. DAPI nuclear counterstaining was also used. (B) Later steps in the differentiation program are not disrupted when E7 cannot disrupt pocket proteins. Raft sections were incubated with monoclonal antibody raised against involucrin, followed by detection with a fluorescein-conjugated anti-mouse secondary antibody. (C) Suprabasal DNA synthesis is not precluded in cells undergoing a normal differentiation program. K10- and BrdU-specific coimmunofluorescence was performed on raft sections; K10 is detected with a fluorescein-conjugated secondary antibody; BrdU is detected with a Texas red secondary antibody. DAPI counterstaining was also applied to sections. The white arrow highlights a cell undergoing suprabasal DNA synthesis and expressing the early differentiation marker K10. (D) Late viral gene expression does not require E7-pocket protein interactions. To determine late viral gene expression in E7 mutant HPV16 replicons, E4-specific immunofluorescence was performed using a fluorescein-conjugated anti-E4 antibody in conjunction with DAPI nuclear counterstaining. (E) L1-specific immunofluorescence was performed on raft sections using the L1-specific monoclonal antibody H16D9.
In contrast to the above-observed correlations between E7 ability to bind and degrade the pocket proteins and the ability of HPV16 to delay the onset of differentiation (Fig. 4A and B), E7 did not need to degrade the pocket proteins to induce suprabasal DNA synthesis (Fig. 3A and B). This led to the prediction that suprabasal DNA synthesis that arises in the raft cultures of cells harboring the E7ΔPTLHE mutant HPV16 replicons will have already initiated their normal differentiation program. To address this, we performed BrdU and K10 double immunofluorescence (Fig. 4C). As predicted, K10 and BrdU double-positive cells were selectively observed with the suprabasal cells of rafts of cells harboring the E7ΔPTLHE mutant HPV16 replicons. This result demonstrates that E7's disruption of differentiation is not required for the reprogramming suprabasal cells to support DNA synthesis.
Late viral gene expression is not affected by loss of E7-pocket protein interactions.
E1∧E4 is a papillomaviral protein that is highly expressed late in the viral life cycle. Expression of E4 has been shown to correlate with viral DNA amplification and to precede onset of expression of the structural viral genes L1 and L2. Therefore, we examined the expression of E4 in the presence and absence of E7 pocket protein interactions, using the raft cultures harboring wild-type versus E7 mutant HPV16 replicons (Fig. 4D). Whereas no expression of E1∧E4 was observed in raft cultures not harboring HPV16 DNA, E4 expression was seen in the more superficial suprabasal layers in rafts harboring wild-type HPV16. E1∧E4 expression was not dependent upon E7's ability to bind and/or degrade the pocket proteins, as E1∧E4-positive cells were also seen in rafts harboring either the E7ΔDLYC or E7ΔPTLHE mutant HPV16 replicons. In addition, L1, a viral capsid late gene, exhibited a similar expression pattern (Fig. 4E). Thus, late gene expression is separable from requirements for induction of suprabasal DNA synthesis and the delay in onset of differentiation.
DISCUSSION
This study is the first to examine the contribution of E7-pocket protein interactions to the HPV16 life cycle. The HPV16 life cycle does not require E7 for the nonproductive stage of its life cycle but does require E7 for the productive stage (12). This is also the case for HPV18, another high-risk HPV (30). In contrast, the mid-risk HPV31 seems to require E7 for the nonproductive, as well as the productive, stage of the life cycle (29). These differences may be due to unique technical aspects of analyses performed by different groups but is most likely the result of genotypic differences between the high-risk HPVs and the lower-risk viruses. Since HPV16 does require E7 for its productive stage, we examined how the E7-pocket protein interactions contributed to the different facets of this stage of the life cycle in the context of the HPV16 genome.
We found different facets of the productive stage of the HPV16 life cycle differentially depend on E7's ability to bind and degrade the pocket proteins. In particular, HPV16's delay of the onset of terminal differentiation depended on the ability of E7 to both bind and degrade the pocket proteins. In contrast, the ability of HPV16 to reprogram cells to support DNA synthesis required E7's binding the pocket proteins but was not wholly dependent upon E7's inducing their degradation. Consistent with these findings, we found that DNA synthesis occurred in suprabasal cells even though they retained Rb protein and had initiated differentiation, as indicated by their expression of K10. Lastly, we found that neither E7's binding nor degradation of pocket proteins was required for viral late gene expression.
One of the most striking implications of our work comes from the observation that HPV16 can reprogram cells to support DNA synthesis under conditions in which it fails to delay the onset of differentiation. This finding is clearly demonstrated by the properties of rafts harboring the E7ΔPTLHE mutant HPV16 replicon, in which the differentiation program is unaltered, yet DNA synthesis is induced in the suprabasal compartment (Fig. 4C). These findings refute the hypothesis that suprabasal DNA synthesis is merely a consequence of cells being maintained in a basal compartment-like differentiation state. Rather, it supports the alternative hypothesis that E7 directly alters the regulation of the cell cycle in a manner that is separable from the disruption of the differentiation program. A second set of implications comes from the finding that pRb expression is significantly diminished in the basal compartment of rafts harboring wild-type HPV16 replicons, yet pRb is largely retained in the suprabasal compartment of these rafts (Fig. 2A to D). This is in stark contrast to the expression pattern of p130, which has the opposite suprabasal expression pattern of pRb in raft cultures of cells harboring HPV16 replicons. Relative to pRb, p130 is greatly diminished in raft cultures harboring HPV16 replicons (Fig. 2D). This difference in expression patterns of p130 versus pRb may reflect the unique roles of E7-pocket protein interactions in reprogramming keratinocytes to support the HPV16 life cycle. The lack of substantive pRb expression in monolayer HPV16 cultures, combined with the lack of p130 expression in HPV16 raft cultures, may illustrate a switch in E7 targeting as necessary for the different steps of the HPV life cycle. Previous studies have demonstrated that while both pRb and p130 can contribute to cell cycle regulation and differentiation, pRb is required for the switch from proliferation to early differentiation while p130 may largely regulate later stages of differentiation, especially the maintenance of terminal differentiation (14, 25, 40). These data indicate that early in the life cycle, as seen in the basal compartment, it is most important for the virus to dysregulate the cell cycle and impair the initial differentiation pathway; yet when the virus moves to the suprabasal compartment later in the life cycle, the emphasis switches to disruption of the terminal differentiation program. The dysregulation of the cell cycle and initial differentiation program in the life cycle may be accomplished by degradation of pRb in the basal compartment, while the disruption of the terminal differentiation pathway may be gained by degradation of p130 (in conjunction with or independent of pRb inactivation) in the suprabasal compartment. If such mechanisms are in place to independently disrupt the cell cycle or the differentiation program by HPV16, it would also explain how suprabasal cells undergoing DNA synthesis are also able to express differentiation markers, as seen in Fig. 4C. In any case, it will be interesting to determine how E7 is able to modulate its interactions with the different pocket proteins in the basal versus the suprabasal compartment.
That E7 can inactivate pRb function without degrading pRb, as evidenced by the efficient induction of MCM7 in rafts of cells harboring wild-type or E7ΔPTLHE mutant HPV16 replicons, as well as the presence of pRb-BrdU double-positive cells in the suprabasal compartment of rafts harboring wild-type HPV16 replicons, has additional implications. One is that E7 is present at sufficient stoichiometric levels to bind and sequester sufficient amounts or an appropriate subpopulation of pRb or other pocket protein molecules to deregulate the cell cycle, thereby circumventing the need to degrade the pocket protein. Alternatively, pRb may not be a critical target to degrade in this phase of the life cycle. The data may also indicate that the ability of E7 to disrupt the function of other cellular factors, in addition to the pocket proteins, contributes to the deregulation of the cell cycle. For example, E7 can bind and inactivate the cyclin-dependent kinase inhibitor p21 (15, 27), which correlates with the ability of E7 to inhibit the DNA damage response, a property of E7 that likely reflects its ability to override normal cell cycle control (21). In another study, investigators have argued that E7's ability to be phosphorylated by casein kinase 2 is critical for E7 to reprogram cells to reenter S phase (5). How the phosphorylation status of E7 affects its function is not clear. Note that both E7ΔDLYC and E7ΔPTLHE when expressed ectopically or in vitro are competent for casein kinase 2 phosphorylation, as well as p21 inactivation. Therefore, this study does not address the contribution of these pocket protein-independent factors to the life cycle of HPV16. Whether such properties of HPV16 E7 contribute to its induction of suprabasal DNA synthesis in the context of the viral life cycle remains unknown. Studies with mouse skin using genetically engineered mouse strains clearly demonstrate that complete inactivation of the pRb pathway is sufficient to induce suprabasal DNA synthesis (1); when placed on a genetic background encoding a pRb that does not interact with E7, E7 is no longer able to induce suprabasal DNA synthesis (S. J. Balsitis and P. F. Lambert, manuscript in preparation). Thus, inactivation of pRb is both necessary and sufficient to induce suprabasal DNA synthesis in the mouse epidermis. However, it remains unclear whether, in the context of the viral life cycle, E7 achieves a complete deregulation of the cell cycle solely through its binding to pRb. It is in this regard that properties like E7's inactivation of p21 are clearly relevant and warrant further investigation.
Where the induction of DNA synthesis and activation of MCM7- and Ki67-positive staining in the suprabasal compartment of raft cultures harboring HPV16 replicons were dependent on the ability of E7 to bind but not degrade the pocket proteins, the delayed onset of differentiation within the spinous layer relied on both properties of E7. These data suggest that the delay in differentiation requires the efficient depletion of one or more pocket proteins by E7. pRb, p107, and p130 are each implicated in differentiation processes (40, 45, 46). The complete inactivation of pRb is sufficient to delay the onset of differentiation in the spinous layer in the mouse (1). Furthermore, E7's ability to delay the onset of differentiation in the mouse epidermis is mostly lost on a genetic background encoding a pRb that cannot bind to E7 (Balsitis and Lambert, manuscript in preparation). These data demonstrate that pRb is a critical target for HPV16 to disrupt normal keratinocyte differentiation. How pRb promotes differentiation is not clearly understood. It is known that the differentiation-promoting activity of pRb is distinct from its ability to interact with E2F and promote G1/S arrest (47). Generally, epithelial cells, muscle cells, and adipocytes require pRb for differentiation (4, 7, 19, 38, 46, 51). For muscle cells and adipocyte differentiation, pRb functions as a required transcriptional coactivator (4, 32, 39, 47). In addition, posttranslational modifications of pRb are also postulated to promote permanent cell cycle arrest and/or an environment more responsive to differentiation factors (37). Whatever the mechanism(s), our data correspond well with previous findings that indicate that the ability of pRb to inactivate E2F activity is separable from its role in differentiation. Furthermore, our data indicate that E7's effect on differentiation cannot be driven solely by its binding to pRb but must also require its ability to induce pRb's degradation.
One of the last steps of the viral life cycle is the expression of late viral genes. While interactions of E7 with pocket proteins have a significant effect on the induction of suprabasal DNA synthesis, the proliferative capacity of the suprabasal compartment and differentiation, they do not affect late viral gene expression. A possible explanation for this lack of dependence on E7 for late gene expression is that late viral gene expression is regulated by differentiation-specific events arising in the more superficial compartments. E7 is reduced in its expression in the raft cultures compared to the monolayer cultures, consistent with a model in which the level of E7 wanes as cells migrate more superficially within stratified epithelial tissue. A reduction in the levels of E7 protein in the more superficial cells may reduce the efficiency with which E7 modulates differentiation, thus allowing normal differentiation and late viral gene expression.
From these data, we put forth the following working model to explain mechanistically E7's role in the HPV16 life cycle. In the basal compartment, E7, while not required for viral DNA replication, nevertheless efficiently degrades pRb. The consequence of this property in the nonproductive stage of the life cycle remains obscure from our raft culture studies, but in vivo studies demonstrate that the inactivation of pRb enhances the proliferative index of the basal compartment (1). The absence of such a difference in raft culture could simply reflect a limitation of this dynamic in vitro model system. As basal cells divide and daughter cells migrate into the suprabasal compartment, E7 delays the onset of differentiation and reprograms these cells to support DNA synthesis. Both properties correlate with E7's ability to bind the pocket proteins, consistent with the pocket proteins being relevant targets of E7. However, the delay in differentiation relies more on E7's ability to degrade the pocket proteins, perhaps specifically p130. We hypothesize that the different facets of HPV16's life cycle requires different modes of pocket protein inactivation, due to the ability of E7 to inhibit pocket protein function in the context of cell cycle by pocket protein-independent mechanisms, including its inactivation of p21. In contrast, the inhibition of pocket protein function in differentiation relies upon a more complete inactivation of the pocket proteins directly by E7. What remains unclear is an understanding of which pocket proteins are the relevant targets for E7 in mediating its different effects in the viral life cycle and identification of other biologically significant cellular targets.
Acknowledgments
We acknowledge Elsa Flores for constructing the E7 mutant HPV16 genomes; Sybil Genther and Elsa Flores for performing some of the raft cultures; and Neil Christensen and John Doorbar for antibodies. We thank all members of the Lambert laboratory for helpful discussions and comments and Bill Sugden for critically reading the manuscript.
This work was supported by NIH grants CA22443, CA09135, and CA07175.
REFERENCES
- 1.Balsitis, S. J., J. Sage, S. Duensing, K. Munger, T. Jacks, and P. F. Lambert. 2003. Recapitulation of the effects of the human papillomavirus type 16 E7 oncogene on mouse epithelium by somatic Rb deletion and detection of pRb-independent effects of E7 in vivo. Mol. Cell. Biol. 23:9094-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, D. C., and K. C. Gatter. 2002. Ki67 protein: the immaculate deception? Histopathology 40:2-11. [DOI] [PubMed] [Google Scholar]
- 3.Cam, H., and B. D. Dynlacht. 2003. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell 3:311-316. [DOI] [PubMed] [Google Scholar]
- 4.Chen, P. L., D. J. Riley, Y. Chen, and W. H. Lee. 1996. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 10:2794-2804. [DOI] [PubMed] [Google Scholar]
- 5.Chien, W. M., J. N. Parker, D. C. Schmidt-Grimminger, T. R. Broker, and L. T. Chow. 2000. Casein kinase II phosphorylation of the human papillomavirus-18 E7 protein is critical for promoting S-phase entry. Cell Growth Differ. 11:425-435. [PubMed] [Google Scholar]
- 6.Chow, L. T., and T. R. Broker. 1994. Papillomavirus DNA replication. Intervirology 37:150-158. [DOI] [PubMed] [Google Scholar]
- 7.Classon, M., B. K. Kennedy, R. Mulloy, and E. Harlow. 2000. Opposing roles of pRB and p107 in adipocyte differentiation. Proc. Natl. Acad. Sci. USA 97:10826-10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Dick, F. A., E. Sailhamer, and N. J. Dyson. 2000. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol. Cell. Biol. 20:3715-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyson, N., P. Guida, K. Munger, and E. Harlow. 1992. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J. Virol. 66:6893-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehrmann, F., D. J. Klumpp, and L. A. Laimins. 2003. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J. Virol. 77:2819-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, C. A. Sattler, and P. F. Lambert. 1999. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology 262:344-354. [DOI] [PubMed] [Google Scholar]
- 14.Francesconi, C. M., A. E. Hutcheon, E. H. Chung, A. C. Dalbone, N. C. Joyce, and J. D. Zieske. 2000. Expression patterns of retinoblastoma and E2F family proteins during corneal development. Investig. Ophthalmol. Vis. Sci. 41:1054-1062. [PubMed] [Google Scholar]
- 15.Funk, J. O., S. Waga, J. B. Harry, E. Espling, B. Stillman, and D. A. Galloway. 1997. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 11:2090-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genther, S. M., S. Sterling, S. Duensing, K. Munger, C. Sattler, and P. F. Lambert. 2003. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J. Virol. 77:2832-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillison, M. L., W. M. Koch, R. B. Capone, M. Spafford, W. H. Westra, L. Wu, M. L. Zahurak, R. W. Daniel, M. Viglione, D. E. Symer, K. V. Shah, and D. Sidransky. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92:709-720. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu, W., J. W. Schneider, G. Condorelli, S. Kaushal, V. Mahdavi, and B. Nadal-Ginard. 1993. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell 72:309-324. [DOI] [PubMed] [Google Scholar]
- 20.Gulliver, G. A., R. L. Herber, A. Liem, and P. F. Lambert. 1997. Both conserved region 1 (CR1) and CR2 of the human papillomavirus type 16 E7 oncogene are required for induction of epidermal hyperplasia and tumor formation in transgenic mice. J. Virol. 71:5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helt, A. M., J. O. Funk, and D. A. Galloway. 2002. Inactivation of both the retinoblastoma tumor suppressor and p21 by the human papillomavirus type 16 E7 oncoprotein is necessary to inhibit cell cycle arrest in human epithelial cells. J. Virol. 76:10559-10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helt, A. M., and D. A. Galloway. 2001. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 75:6737-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herber, R., A. Liem, H. Pitot, and P. F. Lambert. 1996. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J. Virol. 70:1873-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmgren, S. C., N. A. Patterson, M. A. Ozbun, and P. F. Lambert. 2005. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J. Virol. 79:3938-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huh, M. S., M. H. Parker, A. Scime, R. Parks, and M. A. Rudnicki. 2004. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol. 166:865-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeon, S., B. L. Allen-Hoffmann, and P. F. Lambert. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 69:2989-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, D. L., R. M. Alani, and K. Munger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, D. L., and K. Munger. 1997. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J. Virol. 71:2905-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longworth, M. S., and L. A. Laimins. 2004. The binding of histone deacetylases and the integrity of zinc finger-like motifs of the E7 protein are essential for the life cycle of human papillomavirus type 31. J. Virol. 78:3533-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLaughlin-Drubin, M. E., J. L. Bromberg-White, and C. Meyers. 2005. The role of the human papillomavirus type 18 E7 oncoprotein during the complete viral life cycle. Virology 338:61-68. [DOI] [PubMed] [Google Scholar]
- 31.Middleton, K., W. Peh, S. Southern, H. Griffin, K. Sotlar, T. Nakahara, A. El-Sherif, L. Morris, R. Seth, M. Hibma, D. Jenkins, P. Lambert, N. Coleman, and J. Doorbar. 2003. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J. Virol. 77:10186-10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris, E. J., and N. J. Dyson. 2001. Retinoblastoma protein partners. Adv. Cancer Res. 82:1-54. [DOI] [PubMed] [Google Scholar]
- 33.Munger, K., J. R. Basile, S. Duensing, A. Eichten, S. L. Gonzalez, M. Grace, and V. L. Zacny. 2001. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20:7888-7898. [DOI] [PubMed] [Google Scholar]
- 34.Munger, K., and W. C. Phelps. 1993. The human papillomavirus E7 protein as a transforming and transactivating factor. Biochim. Biophys. Acta 1155:111-123. [DOI] [PubMed] [Google Scholar]
- 35.Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen, D. X., L. A. Baglia, S. M. Huang, C. M. Baker, and D. J. McCance. 2004. Acetylation regulates the differentiation-specific functions of the retinoblastoma protein. EMBO J. 23:1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novitch, B. G., G. J. Mulligan, T. Jacks, and A. B. Lassar. 1996. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J. Cell Biol. 135:441-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novitch, B. G., D. B. Spicer, P. S. Kim, W. L. Cheung, and A. B. Lassar. 1999. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr. Biol. 9:449-459. [DOI] [PubMed] [Google Scholar]
- 40.Paramio, J. M., S. Lain, C. Segrelles, E. B. Lane, and J. L. Jorcano. 1998. Differential expression and functionally co-operative roles for the retinoblastoma family of proteins in epidermal differentiation. Oncogene 17:949-957. [DOI] [PubMed] [Google Scholar]
- 41.Peh, W. L., K. Middleton, N. Christensen, P. Nicholls, K. Egawa, K. Sotlar, J. Brandsma, A. Percival, J. Lewis, W. J. Liu, and J. Doorbar. 2002. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 76:10401-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phelps, W. C., S. Bagchi, J. A. Barnes, P. Raychaudhuri, V. Kraus, K. Munger, P. M. Howley, and J. R. Nevins. 1991. Analysis of trans activation by human papillomavirus type 16 E7 and adenovirus 12S E1A suggests a common mechanism. J. Virol. 65:6922-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phelps, W. C., K. Munger, C. L. Yee, J. A. Barnes, and P. M. Howley. 1992. Structure-function analysis of the human papillomavirus type 16 E7 oncoprotein. J. Virol. 66:2418-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phelps, W. C., C. L. Yee, K. Munger, and P. M. Howley. 1988. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 53:539-547. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz, S., C. Segrelles, A. Bravo, M. Santos, P. Perez, H. Leis, J. L. Jorcano, and J. M. Paramio. 2003. Abnormal epidermal differentiation and impaired epithelial-mesenchymal tissue interactions in mice lacking the retinoblastoma relatives p107 and p130. Development 130:2341-2353. [DOI] [PubMed] [Google Scholar]
- 46.Schneider, J. W., W. Gu, L. Zhu, V. Mahdavi, and B. Nadal-Ginard. 1994. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science 264:1467-1471. [DOI] [PubMed] [Google Scholar]
- 47.Sellers, W. R., B. G. Novitch, S. Miyake, A. Heith, G. A. Otterson, F. J. Kaye, A. B. Lassar, and W. G. Kaelin, Jr. 1998. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 12:95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoler, M. H., and T. R. Broker. 1986. In situ hybridization detection of human papillomavirus DNAs and messenger RNAs in genital condylomas and a cervical carcinoma. Hum. Pathol. 17:1250-1258. [DOI] [PubMed] [Google Scholar]
- 49.Stubenrauch, F., and L. A. Laimins. 1999. Human papillomavirus life cycle: active and latent phases. Semin. Cancer Biol. 9:379-386. [DOI] [PubMed] [Google Scholar]
- 50.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 51.Zacksenhaus, E., Z. Jiang, D. Chung, J. D. Marth, R. A. Phillips, and B. L. Gallie. 1996. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 10:3051-3064. [DOI] [PubMed] [Google Scholar]
- 52.zur Hausen, H. 2001. Oncogenic DNA viruses. Oncogene 20:7820-7823. [DOI] [PubMed] [Google Scholar]