Abstract
A large percentage of the repetitive elements in mammalian genomes are retroelements, which have been moved primarily by LINE-1 retrotransposons and endogenous retroviruses. Although LINE-1 elements have remained active throughout the mammalian radiation, specific groups of endogenous retroviruses generally remain active for comparatively shorter periods of time. Identification of an unusual extinction of LINE-1 activity in a group of South American rodents has opened a window for examination of the interplay in mammalian genomes between these ubiquitous retroelements. In the course of a search for any type of repetitive sequences whose copy numbers have substantially changed in Oryzomys palustris, a species that has lost LINE-1 activity, versus Sigmodon hispidus, a closely related species retaining LINE-1 activity, we have identified an endogenous retrovirus family differentially amplified in these two species. Analysis of three full-length, recently transposed copies, called mysTR elements, revealed gag, pro, and pol coding regions containing stop codons which may have accumulated either before or after retrotransposition. Isolation of related sequences in S. hispidus and the LINE-1 active outgroup species, Peromyscus maniculatus, by PCR of a pro-pol region has allowed determination of copy numbers in each species. Unusually high copy numbers of approximately 10,000 in O. palustris versus 1,000 in S. hispidus and 4,500 in the more distantly related P.maniculatus leave open the question of whether there is a connection between endogenous retrovirus activity and LINE-1 inactivity. Nevertheless, these independent expansions of mysTR represent recent amplifications of this endogenous retrovirus family to unprecedented levels.
Mammals contain a great array of repetitive sequences in their genomes. These sequences range from tandem repeats to ancient DNA transposons to a vast range of retroelements that have been deposited throughout the mammalian radiation (22, 35, 40). Retroelements alone constitute >43% of the Mus musculus genome (35) and have played significant roles in shaping the evolution of mammalian genomes and controlling gene function. The major autonomous retroelements are the non-long terminal repeat (LTR) elements, consisting of LINEs, and the LTR-containing elements, composed primarily of endogenous retroviruses, respectively, comprising 21% and 8.6% of the Mus genome (35). Table 1 compares some of the relevant similarities and differences between these two types of retroelements. It appears that they coexist in all mammals with the vast majority of these elements being ancient pseudogenes, but it is not known whether there is any interplay between these types of elements in their hosts. For example, do these elements compete directly or indirectly within the host? Does the activity of one group of elements affect the activity of another group, either through burdens placed upon the host, competition for host resources, or through functions satisfied for the host?
TABLE 1.
Comparison of properties of mammalian autonomous retroelements
| Property or characteristic | Non-LTR | LTR |
|---|---|---|
| % of Mammalian genome | ∼20 | ∼8 |
| No. of families | One major (LINE-1); two minor (LINE-2 and 3) | Many |
| Life history | One origin or introduction of LINE-1 into the mammalian genome; few extinctions | Many independent introductions into the genome from extant retroviruses; many extinctions |
| Period when transposition occurs for a group | Long term (>100 Myr) | Short to medium term (usually <20 Myr) |
| Horizontal transfer | None known | Sometimes |
| No. of potentially active members per group | 100 to 1,000s | Usually few |
| Divergence between active members | Usually low; one to a few lineages per host species | Lineage number may increase with age; many distantly related groups in the same host species |
| Ability to move other elements in trans | Yes: SINEs, pseudogenes | Yes: nonautonomous ERVs |
| Modulate gene expression | Yes | Yes |
Endogenous retroviruses arise from infections by exogenous forms. Although there are notable exceptions, the endogenous descendants of a single exogenous infection seem to have a relatively short functional life (less than one million to tens of millions of years) and give rise to a limited number of copies compared to LINE-1 elements, which appear to have resided in mammalian genomes prior to the mammalian radiation (6, 7, 16, 32, 43). Reoccurring infection of mammalian genomes by exogenous retroviruses has given rise to many separate groups, or families, of these endogenous retroviruses (16, 32, 34). On the other hand, LINE-1 elements appear to have been transmitted vertically with no horizontal transmission but with continued activity throughout the entire mammalian radiation of more than 100 million years, giving rise to extremely high numbers of elements phylogenetically related through very few long-term lineages (13, 14). Some of the differences between these two types of elements probably stem from specific aspects unique to retroviruses such as their pathogenicity. However, the reasons for many differences are not obvious.
It is clear that LINE-1 elements have affected their hosts in multiple ways. They are widely considered to be intracellular genomic parasites, but their continued activity throughout the mammalian radiation has led to proposals that they have acquired a function for their hosts. Proposed functions have included a role in double-stranded DNA break repair (21, 37, 49) and in X chromosome inactivation (31). They have recently been shown to play a role in gene regulation through their ability to reduce the rate of transcription elongation upon introduction into transcribed sequences (19). Irrespective of these proposed functions, they have been a major force in shaping mammalian genomes. Insertional mutagenesis can result in inactivation of genes or introduction of new promoters. LINEs provide necessary machinery for movement of SINEs and pseudogenes and are sites for ectopic recombination that leads to genome rearrangements (13). It is estimated that their 3′ transduction of DNA downstream of active elements has moved as much as 1% of the genome (17, 39). Endogenous retroviruses affect their hosts in some of these ways, but the relative contribution of each type of retroelement is unknown (16, 50).
LINE-1 elements appear to be active in nearly all mammals examined, but we have previously found one instance of extinction of activity in a group of sigmodontine rodents (9, 18). It is reasonable to assume that loss of LINE-1 activity might have major ramifications for the host species. One predicted outcome from the loss of LINE-1 activity was cessation of activity of SINEs, which depend on functional LINE-1 machinery for their own movement. We have shown that B1 SINE activity has indeed ceased in the sigmodontine species that lost LINE-1 activity, as well as in Sigmodon species that retain active LINE-1s (41). Extinction of LINE-1 activity might also lead to a reduction in the genomic parasite load, loss of genomic plasticity, or loss of functions performed by LINEs. Any of these scenarios could set the stage for the invasion or amplification of an element to fill the genomic niche previously filled by active LINEs.
We initiated a screen to search for repetitive sequences that have been recently amplified in the rice rat, Oryzomys palustris, relative to the cotton rat, Sigmodon hispidus. O. palustris is a member of the group of sigmodontine rodents that lost LINE-1 activity (the “L1-inactive” group), and S. hispidus is in the most closely related genus known to retain active LINE-1s. We used the phylogenetic screening procedure, which is a general method to find any type of rapidly evolving repetitive sequences without prior knowledge of their mode of replication (33, 51). Phylogenetic screening is a differential hybridization method in which labeled genomic DNA from the species of interest (O. palustris) and an outgroup (S. hispidus) are hybridized separately to genomic DNA libraries from each of these species to identify repetitive sequences differentially amplified between those two species. We describe here the isolation and characterization of a family of endogenous retroviruses found as a result of this screen. We have also found that this family is present at unusually high copy numbers in a number of rodent species.
MATERIALS AND METHODS
Specimens examined and genomic DNA extraction.
Phyllotis xanthopygus AK13012 tissue came from the Texas Cooperative Wildlife Collection at Texas A&M University. Neacomys spinosus NK25265 and Thomosomys baeops NK27679 were provided by the Museum of Natural History collection at New Mexico. Sigmodon hispidus TK72547 and Peromyscus maniculatus TK25418 were from The Museum at Texas Tech University. O. palustris KE02 was obtained from Kent Edmonds (Indiana University, New Albany, IN). M. musculus is the Swiss Webster strain. S. mascotensis JS2014 was obtained from Jack Sullivan at the University of Idaho. Genomic DNA was extracted as previously described (29).
Construction of libraries.
Genomic DNA libraries were constructed by standard techniques (1). Libraries containing small inserts were produced for O. palustris, S. hispidus, and P. maniculatus using DNA sheared to an average size of 1 to 2 kb. The O. palustris cosmid library was constructed by shearing genomic DNA to an average insert size of 30 to 50 kb with ligation into the cosmid vector SuperCosI (28) (Stratagene, La Jolla, CA).
Repetitive sequence screens.
Screening was carried out by a modification of the phylogenetic screen originally described by Wichman and coworkers. (33, 51). Replicate clones from the species of interest were probed with labeled DNA from that species and from an outgroup. Single-copy and lowly repetitive sequences in labeled genomic DNA are at such low concentrations that the only clones visibly hybridizing should be those containing middle to highly repetitive DNA. Colony hybridizations were designed to identify clones from the O. palustris library that gave a positive hybridization signal when probed with O. palustris genomic DNA, but either no signal or a much lower signal when probed with S. hispidus genomic DNA. The same types of hybridizations were also done on the S. hispidus library.
Clones from each library were arrayed onto Magna nitrocellulose membranes (Fisher Corp., Pittsburgh, PA) and probed with 20 ng of sheared, random-primed 32P-labeled genomic DNA at 106 cpm/ml. These colony hybridizations were done for ca. 40 h at 55°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.3% sodium dodecyl sulfate (SDS), 40 μg of salmon sperm DNA/ml, and 10× Denhardt solution, followed by washing to 58°C in 6× SSCP-0.1% SDS. Differentially hybridizing clones were confirmed by Southern hybridizations under the same conditions.
Colony hybridization to small insert libraries for determination of copy numbers was done as described above.
Dot blots and Southern hybridization to genomic DNA.
Genomic DNAs were quantified as described previously (41). For dot blots, 500 ng of genomic DNA from each species was blotted onto a charged nylon membrane by standard procedures (1) (Amersham Biosciences, Piscataway, NJ). The following clade specific oligonucleotide probes were 32P labeled with polynucleotide kinase (1): O419, 5′-ATATGGATTCCTCAG-3′; S205, 5′-ATCTCCTACGACAAT-3′;and P805, 5′-CCTCCCACAGGGAAT-3′. Tetramethylammonium chloride (TMAC) hybridizations and washes were done as previously described (1) with the addition of each of the nonlabeled oligonucleotides to the hybridizations at 50 pmol/ml, which is 50 times the molar concentration of the labeled oligonucleotide. The TMAC hybridization conditions required 100% sequence identity in order to give a positive signal. Dot blot hybridizations with species-specific DNA probes of approximately 930 bp were carried out under the hybridization conditions described for the colony hybridizations but with washes in 2× SSCP-0.1% SDS at 60°C. Southern hybridization to genomic DNA was performed as in the colony hybridizations but with the 1.2-kb OpalH6 insert labeled by random prime labeling.
Sequencing and sequence analysis.
Sequencing was done with a 3730 DNA analyzer (Applied Biosystems, Foster City, CA). Unless otherwise specified, contig analyses and sequence analyses were done by using the DNASTAR (Madison, WI) and Vector NTI (Informax, Bethesda, MD) analysis packages. Additional BLAST searches and analyses of open reading frames (ORFs) were done by using blastn, blastp, and ORF Finder (National Center for Biotechnology Information, Bethesda, MD). Repeat searches were performed on the Repbase Censor Server (http://www.girinst.org). Stop codon maps were generated by using a PerlScript written by Gregory Baillie (Terry Fox Laboratory, British Columbia Cancer Agency). Probable tRNA specificity for primer binding sites was determined as previously described (2) but using two databases (30, 45) for stand-alone BLAST searches.
Sequences were aligned by using the CLUSTAL W algorithm as implemented in DNASTAR (Madison, WI) and then adjusted manually. Appropriate models of sequence evolution were determined by using DT-ModSel (36). Maximum-likelihood trees were determined by using PAUP* 4.0b10 (47) with stepwise addition (10 random sequence additions) and tree bisection-reconnection branch swapping. Nodal support for the likelihood trees was estimated by using bootstrap analysis (100 replicates) under the appropriate model.
Fluorescent in situ hybridization.
Karyotypes were prepared from O. palustris (TK110999 and TK111000) and S. hispidus (TK93765 and TK93768; The Museum at Texas Tech University) using the in vivo bone marrow/yeast stress method (3). The mysTR plasmid MP1, which contains Opalc65 sequence in the region indicated in Fig. 4, was used as a probe for in situ hybridization to O. palustris and S. hispidus. Probes were labeled by standard nick translation with biotinylated dATP following the BioNick labeling kit instructions (Gibco-BRL, Gaithersburg, MD). Hybridization procedures have been previously described (4, 38). Patterns of hybridization were examined by using an Olympus epi-fluorescence microscope BX51 with a dual-band-pass filter allowing the simultaneous viewing of propidium iodide and fluorescein. Images were photographed by using an Applied Imaging camera and captured using the Genus System 3.1 from Applied Imaging Systems (San Jose, CA).
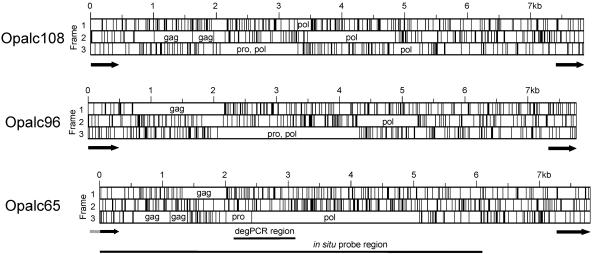
FIG. 4.
Maps of mysTR elements Opalc108, Opalc96, and Opalc65. Vertical lines show the position of stop codons in each reading frame. ORFs for gag, pro (protease), and pol (polymerase) coding regions are indicated. Black arrows show the regions covered by each LTR, and the gray tail next to the left LTR of c65 indicates the part of that LTR not present because the border of the clone containing c65 was within that LTR. The pro/pol region amplified by degenerate PCR is indicated by a horizontal line as is the region of Opalc65 used as a probe for in situ hybridizations.
PCR primer design and amplification.
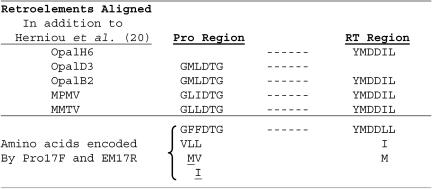
PCR primers for amplification of mysTR-related elements were designed to the same conserved regions extending from the 3′ portion of the protease gene through the conserved reverse transcriptase domains used by Herniou et al. (20) for amplification of betaretroviruses but contained 5′ clamps and modifications as shown in Fig. 1. The protease primer, PRO17F, is 5′-ACGAATTGCTCGAGA GKI HTI ITN GAY ACN GG-3′. The reverse transcriptase primer, EM17R, is 5′-TGGATCGCTGCAGGTAR NAD RTC RTC CAT RTA-3′. The primer regions encoding the amino acids shown in Fig. 1 are underlined, while the nonunderlined bases are the nonhomologous clamps.
FIG. 1.
Degenerate PCR primer design. The top half of the figure shows five retroelements and their amino acid sequences in the areas used for primer design. The Opal elements originated from our O. palustris small insert genomic DNA library. Each of the mysTR elements OpalH6 and OpalD3 covered only one area of the alignment. The bottom part of the figure shows the amino acids encoded by the primers at each of the six positions at each end of the region amplified. Dashes signify the internal region of approximately 892 bp. The underlined amino acids require additional degeneracies not covered in the Pro and EM primers of Herniou et al. (20).
PCR amplifications were similar to ones described previously (8) but with the following conditions. Amplifications of 50 μl contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 4 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate, 100 pmol of each primer, 200 ng of genomic DNA, and 1.25 U of AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA). Hot starts were performed in a GeneAmp PCR System 9600 (Applied Biosystems) by combining all components except the genomic DNA and the polymerase, heating to 70°C, and then adding these final two components before cycling. The cycling parameters were as follows: 70°C hotstart for 5 min; 94°C denaturation for 1.5 min; 4 cycles of 94°C for 0.5 min, 44°C for 0.5 min, ramp to 72°C at a rate of 0.2°C/min, and 72°C for 1 min; followed by 24 cycles of 94°C for 0.5 min, 52°C for 0.5 min with maximum ramp speed, and 72°C for 1 min; ending with a 7-min extension at 72°C.
Accession numbers of sequences used in the present study.
GenBank accession numbers of previously published sequences are: MPMV, NC_001550; MMTV, NC_001503; TvERV-D, AF224725; SMRV, M23385; MusD, AF246632; JSRV, ; RnERV-β1_NW_043429, NW_04329 (2); HERV-K10(HML2), M14123; RSV, NC_001407; and RV Rice rat, AY820125. GenBankaccession numbers of new elements appearing in the present study are DQ139724 to DQ139773.
RESULTS
Identification of an endogenous retrovirus family differentially amplified in O. palustris and S. hispidus.
The endogenous retrovirus family described below was initially identified during a phylogenetic screen for repetitive sequences amplified to higher copy numbers in O. palustris, a species with no LINE-1 activity, than in S. hispidus, the most closely related species known to retain LINE-1 activity. A total of 647 small insert clones covering approximately 750 kb of genomic DNA from an O. palustris library were screened, as was roughly the same amount of DNA in an S. hispidus library. During this process we identified three O. palustris clones that showed preferential hybridization to O. palustris genomic DNA on Southern blots and that showed greatest sequence similarity in Repbase to MYSERV. MYSERV is the consensus of an inactive family of endogenous retroviruses (ERVs) found in M. musculus and named after mys, the active but nonautonomous retroelement family we previously identified in Peromyscus species (51). Analysis of ORFs within these three ERV sequences, which we tentatively called mysTR elements, revealed domains for a retroviral aspartyl protease, a reverse transcriptase, and an integrase. Clones OpalB2 and OpalH6 contain single ORFs throughout their entire sequence of >1 kb and share an 819 bp overlapping region with 96.7% sequence identity, while clone OpalE11 shows sequence similarity to a more 3′ region in MYSERV.
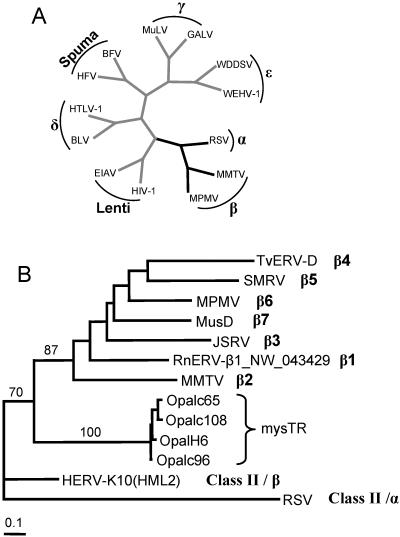
The relationship of these mysTR elements to the seven known genera of exogenous retroviruses (Fig. 2A) was explored by a combination of BLAST searches and phylogenetic analyses. BLAST searches with the elements showed relatively high similarities to HERV-K ERVs, which have been described in different classification schemes as class II elements (16) and endogenous betaretroviruses (24). Even higher homology was seen for an ERV fragment (AY820125) recently isolated from Oryzomys intermedius (15). The region encompassing the eight conserved domains of the reverse transcriptases of some of these mysTR elements was used in phylogenetic analyses with a group of retroelements encompassing the black branched region of the retrovirus tree in Fig. 2A. The maximum-likelihood tree in Fig. 2B includes OpalH6, three other mysTR elements described below, a HERV-K element, one element from each of 7 recently defined subgroups of betaretroviruses (2), and an alpharetrovirus, RSV. The bootstrap values shown on selected branches suggest that these mysTR elements are basal to the seven recently characterized subgroups of betaretroviruses, but the bootstrap value of 70 for inclusion of mysTR elements with those betaretroviruses to the exclusion of both HERV-K10(HML2) and RSV is relatively low. Amino acid-based trees (not shown) group the mysTR elements with RSV and place HERV-K10(HML2) in a clade with the other betaretroviruses. Throughout all analyses it seems clear that the mysTR elements fall within the class II retroelements, but the inclusion of mysTR elements as a new subgroup of the betaretroelements remains tentative.
FIG. 2.
Phylogenetic relationships of retroviruses. (A) Phylogeny adapted from Knipe et al. (24) of selected retroviruses with the names of all seven retroviral genera overlaid in boldface (16). The phylogeny is based on polymerase amino acid sequences, with all branches drawn of equal length. Branches leading to taxa present in part B are black. (B) Maximum likelihood tree of exogenous and endogenous retroviruses related to the mysTR family. The tree is based on nucleotide sequences encompassing reverse transcriptase domains 1 to 8 (23) and shows representative elements from each of the beta subgroups β1 to β7 (2), HERV-K10(HML2), and RSV, which is a member of the alpharetrovirus outgroup. The mysTR elements used here are the initial element isolated that contained all eight reverse transcriptase domains plus the three elements sequenced in their entirety. Bootstrap support values are listed above selected branches. The Opal prefix indicates isolation from O. palustris.
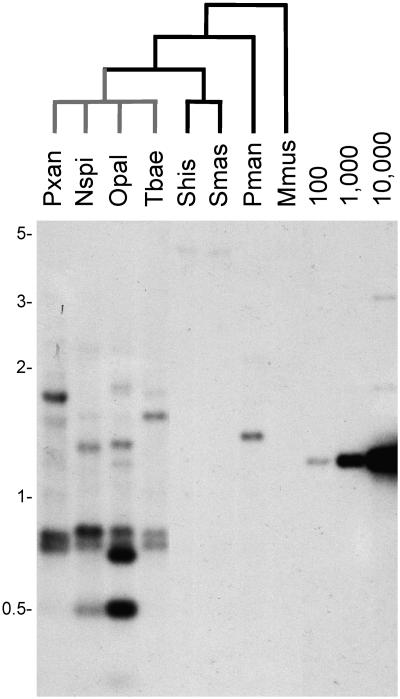
Both the identification of three mysTR clones after screening only 750 kb of genomic DNA, and such high sequence similarities, suggested a recent large-scale amplification of an ERV family. To confirm this assessment of a high copy number of mysTR endogenous retrovirus-like elements, the OpalH6 clone was used as a probe in the Southern hybridization to genomic DNA shown in Fig. 3. Comparison of the O. palustris (lane Opal) lane to the copy number control lanes, which contain various levels of the OpalH6 plasmid, shows that sequences similar to OpalH6 are at very high copy numbers. Moderate to high levels of hybridization to genomic DNA were also seen in lanes for the other three L1-inactive species, Phyllotis xanthopygus (lane Pxan), Neacomys spinosus (lane Nspi), and Thomosomys baeops (lane Tbae). Furthermore, these four species show differences in the intensity of hybridization and size of strongly hybridizing bands. The presence of each strongly hybridizing band indicates that hundreds or thousands of mysTR elements in the genome share two restriction sites. Strongly hybridizing bands that are the same in size and intensity across taxa suggest an amplification of that group of elements in a common ancestor of the species in question, while taxon-specific bands are a landmark of amplification after the divergence of the species (9, 27). These same four L1-inactive species show low levels of hybridization and no taxon-specific bands when probed with a LINE-1 probe (18). Therefore, the taxon-specific bands detected with the OpalH6 probe indicate that there was mysTR amplification after LINE-1 extinction and that this amplification continued in each lineage after these four species last shared a common ancestor.
FIG. 3.
Southern hybridization of rodent DNA with mysTR clone OpalH6. Genomic DNA from eight species of rodents was digested with DraI and probed with O. palustris clone OpalH6. Black branches of the phylogenetic tree lead to species showing LINE-1 activity, while gray branches lead to species with no LINE-1 activity. Lanes to the right contain controls for the indicated copy numbers per genome of the OpalH6 plasmid digested to release the mysTR sequence. The larger size bands in the control lanes represent a small percentage of incompletely digested control plasmids. The species names for each lane label are as follows: Pxan, Phyllotis xanthopygus; Nspi, Neacomys spinosus; Opal, Oryzomys palustris; Tbae, Thomosomys baeops; Shis, Sigmodon hispidus; Smas, Sigmodon mascotensis; Pman, Peromyscus maniculatus; and Mmus, Mus musculus.
Low levels of hybridization, or no detectable hybridization, are seen in the lanes containing genomic DNA from Sigmodon hispidus (Fig. 3, lane Shis), Sigmodon mascotensis (lane Smas), Peromyscus maniculatus (lane Pman), and Mus musculus (lane Mmus), all species that retain LINE-1 activity (“L1-active” species). This result could suggest that the unusual amplification of the mysTR ERV family occurred only in L1-inactive species and not in species that retained active LINE-1s. However, this difference could also be explained by the higher sequence similarity of the probe to sequences in the more closely related L1-inactive species compared to the more distantly related L1-active species.
Isolation and characterization of full-length mysTR endogenous retroviruses.
In order to look more closely at this putative ERV family, an O. palustris genomic DNA cosmid library was constructed and probed with a mixture of the OpalH6 and OpalB2 clones. The mysTR elements in three cosmid clones hybridizing to these probes were sequenced and found to be of approximately 7.8 kb, each of which contained both left and right LTRs of 438 bp. Figure 4 presents maps of these three closely related elements, showing positions of stop codons within all three forward reading frames and revealing substantial ORFs. Blastp searches of translation products of these elements revealed retroviral coding regions within the ORFs, suggesting these elements are endogenous retroviruses which evolved from an exogenous form. Each element contains sequences encoding retroviral-like Gag, protease, and polymerase proteins, followed by about 2 kb containing no large ORFs before the right LTR. No regions were found with similarity to an envelope gene.
Examination of the Opalc96 and Opalc65 maps shows that removal of only a few stop mutations and/or frameshift mutations would return their gag, pro, and pol regions to a single ORF. Yet the distribution of stop codons and level of frameshifts in each element leave open the question of whether there have been debilitating mutations since autonomous transposition or the elements moved nonautonomously. These elements also require no frame shift between the protease and reverse transcriptase regions, a relatively unusual feature they share with the O. intermedius ERV fragment (15).
The putative primer binding site for the Opalc96 element gave significant matches to tRNAlys genes in BLAST searches, suggesting this tRNA as the most likely primer for reverse transcription. The same regions in the Opalc65 and Opalc108 elements did not yield significant BLAST results but their sequence similarity with Opalc96 in this region suggests that they use the same tRNA primer. Use of tRNAlys is consistent with classification of mysTR elements as endogenous betaretroviruses because the majority of betaretroviruses appear to utilize a tRNAlys as a primer (16, 24).
The time since insertion of an ERV into the genome can be estimated from the divergence between the left and right LTRs, assuming that there has been no gene conversion at the LTRs since retrotransposition. The LTRs are identical upon insertion into the genome and because both LTRs accumulate random mutations, the time since insertion is one half the sequence distance between those LTRs divided by the neutral mutation rate. The divergence between the left and right LTRs for Opalc96 is 1.14%, and for Opalc108 it is 0.68%. We have only 284 bp of the left LTR for the Opalc65 element because it was at the extreme edge of the insert DNA in the clone from which it was derived, but the divergence between the regions in common for the left and right LTRs of Opalc65 is 0.35%. If we assume a neutral mutation rate for rodents of approximately 0.01/Myr (i.e., 1% per million years [see reference 42 and references therein]), then all three elements likely inserted intotheir present locations within the last few hundred thousand years.
Distance analyses of these three elements show them to be quite closely related throughout their entire lengths, including their LTRs. The overall divergence between the most closely related pair (Opalc65 and Opalc108) is only 2.6%, while these elements differ from Opalc96 by 11.4 and 12.1%, respectively. Within the reverse transcriptase gene, divergences between these three elements range from only 3.1 to 5.3%, and Fig. 2 shows that these elements group phylogenetically into one closely related family.
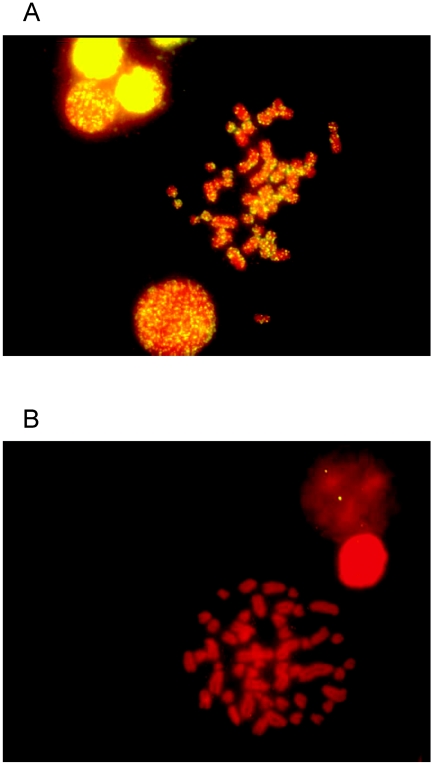
The identification of this endogenous retrovirus family and its apparent high copy number raise the question of how this family is dispersed in the genome of its host. Dispersed distribution in the genome is typical of retrotransposition, whereas accumulation in a block or in heterochromatin might indicate that the element was being amplified by an alternative mechanism such as unequal crossing over. The majority of the Opalc65 element indicated in Fig. 4 was used as a probe for in situ hybridization to O. palustris and S. hispidus chromosomes. Figure 5A is a photograph of a typical hybridization to O. palustris, indicating that the mysTR family has been highly amplified and is dispersed throughout all of the chromosomes, further suggesting that these elements have been amplified by retrotransposition. No hybridization was detected to S. hispidus (Fig. 5B), but as in the Southern hybridization, this could be due to either low copy number of mysTR elements in this species or divergence from the probe.
FIG. 5.
Fluorescent in situ hybridization of mysTR element Opalc65 to O. palustris (A) and S. hispidus (B) metaphase chromosomes. High levels of probe signal (yellow) indicate hybridization to large numbers of mysTR elements in the O. palustris chromosomes but not the S. hispidus chromosomes, while red signal (propidium iodide) is indicative of DNA staining.
Comparison of endogenous mysTR-related elements from three mammalian species.
The Southern hybridization shown in Fig. 2 indicated an unusually high copy number for mysTR elements in O. palustris and suggested a large increase in this family′s copy number in the rodent species that have lost LINE-1 activity compared to the species that have retained LINE-1 activity. Yet the lower hybridization seen in the L1-active species might be due to lower sequence similarity of the O. palustris probe to elements in those other species rather than to copy number differences. In order to avoid potential bias and compare endogenous retroviruses related to the mysTR family in multiple mammalian species, we used PCR amplification to compare an internal region of endogenous retroviruses from O. palustris, S. hispidus, and P. maniculatus, an outgroup to the initial two species.
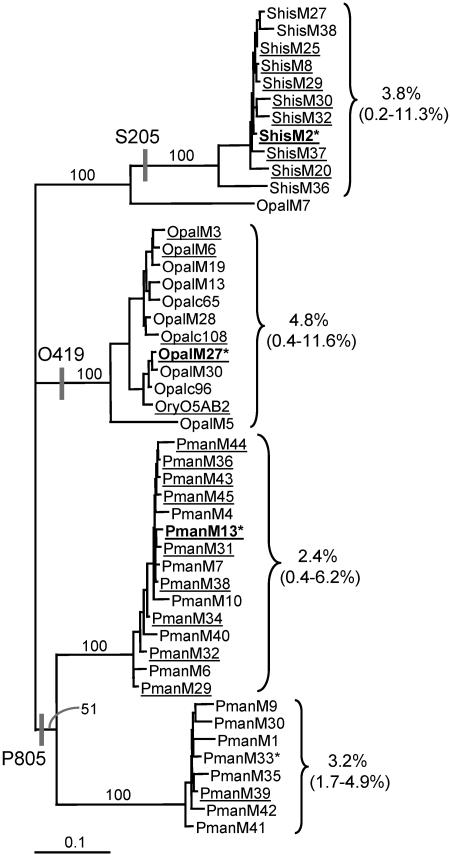
A conserved region in the 3′ portion of the protease gene and another conserved region in the 3′ portion of a conserved reverse transcriptase domain of betaretroviruses were used to design PCR primers for the same areas as those used by Herniou et al. (20) but containing modifications to allow amplification of a wider range of elements. O. palustris genomic DNA was amplified and initial phylogenetic analysis was carried out on 16 resultant clones. All of these clones contained ERV sequences showing a diverse range of endogenous retroviruses. The majority of the sequences (nine clones) grouped in the O. palustris clade bracketed in the likelihood tree in Fig. 6. The tree also shows that the initial B2 clone and the c65, c96, and c108 elements group within this clade. The relatively short branches connecting the majority of these elements show that they are closely related, suggesting recent activity. Clones which contain a single ORF and are therefore more likely to have been isolated from a recently inserted, autonomous ERV, are underlined. Within the bracketed O. palustris clade, uncorrected nucleotide sequence distances to nearest neighbors for the 471-bp reverse transcriptase region are less than 2.2% in nearly all cases, and the average sequence distance to all neighbors is 4.8%. Amplification and analysis of 16 elements from S. hispidus also showed grouping within one clade of relatively closely related ERVs. Among most of the S. hispidus elements, reverse transcriptase sequence distances to nearest neighbors are <2.2% and the average for the entire bracketed group is 3.8%.
FIG. 6.
Phylogenetic analysis of mysTR elements from three rodent species. The likelihood tree was constructed by using the pro/pol region averaging 892 bp between primers Pro17F and EM17R and rooted with the P. maniculatus sequences. Bootstrap support values are given for selected branches. Values to the right of the brackets are the reverse transcriptase region average uncorrected pairwise sequence distances between all elements within each group, while the values in parentheses are the minimum and maximum pairwise distances. ShisM, OpalM, and PmanM indicate S. hispidus, O. palustris, and P. maniculatus elements, respectively, isolated after degenerate PCR. Elements listed in boldface were used as probes for dot blot hybridizations, whereas elements marked with an asterisk were used as probes in library hybridizations. Underlined elements contain single ORFs throughout their entire length. The species specific oligonucleotides S205, O419, and P805 used as probes in dot blot hybridizations are mapped onto the tree with vertical bars.
Initial analysis of clones containing amplified P. maniculatus ERVs showed the majority of them to be divided between two distinct clades. A total of 25 elements were examined in order to get a more detailed view of each of these clades as seen in Fig. 6. The close relationships seen within each of the 2 clades are similar to those found among the majority of the O. palustris and S. hispidus elements, suggesting recent activity as with the other species.
The average reverse transcriptase nucleotide divergences between clades varied from 16.4 to 23%. Interestingly, the average divergence between elements in the bracketed O.palustris clade and the main P. maniculatus clade (16.4%) is less than the average divergence between the O. palustris clade and the S. hispidus clade (21.8%), even though O. palustris is more distantly related to P. maniculatus than it is to S. hispidus. The question remains open whether these differences may be due to lineage sorting, different rates of evolution of mysTR elements in different species, or horizontal transfer due to multiple exogenous infections of a retroviral form.
Copy numbers of mysTR-related endogenous retroviruses.
The phylogenetic analyses summarized above suggested an unusual amount of recent activity within this family of mysTR elements in all three species but did not allow estimation of the copy numbers of these elements in each species. Three methods were used to determine copy numbers for the mysTR subfamilies in each species.
The first method utilized species-specific oligonucleotide probes for quantitative dot blot hybridization. Oligonucleotides O419, S205, and P805 were designed for the mysTR elements bracketed in Fig. 6 based on shared, derived characters that could be mapped to the indicated branches. Each oligonucleotide was designed to a sequence region that was conserved within the target clade but gave at least 2 bp of mismatch with any of the elements outside of the target clade.
One of the most surprising results from these hybridizations was the unusually high copy numbers for these elements in all three of the species examined (Fig. 7). The minimal copy number estimated by this approach is of 1,800 mysTR elements in S. hispidus. It is quite interesting that O. palustris, the sister species that has lost LINE-1 activity shows a copy number of 10,500. This is six times higher than the copy number in S. hispidus, which has retained LINE-1 activity. However, the outgroup, P. maniculatus, shows an intermediate copy number of 4,300, raising the question of whether any correlation exists between ERV copy number and LINE-1 activity. One possible source for a portion of the differences in hybridization seen with these oligonucleotide probes could be due to the necessity to design each probe to a different area within the 892-bp region of analysis in order to find appropriate variation. Different levels of sequence conservation within each area could give rise to hybridization to slightly different numbers of elements.
FIG. 7.
MysTR copy numbers in three rodent species. Copy numbers for recently inserted mysTR elements were determined in O. palustris, S. hispidus, and P. maniculatus by three methods. Bars: □, numbers determined using oligonucleotide probes in dot blot hybridizations; ▨, numbers determined using 926-bp DNA probes in dot blot hybridizations; ▪, numbers determined using 926-bp DNA probes for library hybridizations. Clones identified by library hybridizations were sequenced and counted only if their divergence from probes was ≤10%.
In order to avoid this complication, dot blot hybridizations were also done with DNA probes derived from the entire 926-bp region amplified from a recently inserted mysTR element from each species (Fig. 6, elements in boldface). The same general trends were seen with these longer probes as were seen with the oligonucleotide probes (Fig. 7). O. palustris showed the highest copy number of 7,300, whereas S. hispidus showed a copy number of 700, roughly 10-fold lower. P. maniculatus again showed an intermediate copy number of 5,300, but this number was comparatively closer to the copy number seen in O. palustris than was seen when hybridizations with oligonucleotide probes were compared.
Each dot blot probed during these hybridizations contained genomic DNA from all three species, and in every case probes were either unable to detect elements in other species, or detected only a small fraction of the elements across species. The copy numbers determined here must thus be primarily due to elements inserted since divergence of the species rather than before species divergence. This leads to the necessary conclusion that each species has independently experienced a recent unusual amplification of the mysTR family.
Copy numbers were additionally determined from the incidence of mysTR clones in the libraries containing small DNA inserts constructed for each species. Young elements from each clade (marked with asterisks in Fig. 6) were used as probes in low-stringency colony hybridizations to each library. Potential mysTR clones were sequenced, and the incidence of those which diverged by ≤10% from their probe was used to calculate the copy numbers shown in Fig. 7. With this independent technique an even higher difference was seen between O. palustris and the other species, but the same trends were present: O. palustris showed a copy number of 12,000, while S. hispidus and P. maniculatus showed copy numbers of 360 and 4,020, respectively.
When the values obtained by each of these methods were averaged, we found that there has been an unprecedented amplification of approximately 10,000 recently inserted members of the mysTR endogenous retrovirus family in O. palustris. Independent recent insertions in the two L1-active species account for 1,000 elements in S. hispidus and 4,500 elements in P. maniculatus.
DISCUSSION
Mammalian genomes harbor many families of repetitive sequences which are unique in their genomic distributions, evolutionary histories, mechanisms of replication, and degree of activity. It is not clear why some elements, such as LINE-1s, have maintained activity in nearly all mammals, while others, such as SINEs and ERVs, tend to be limited in their phylogenetic distribution and subject to more frequent extinction events. Even with respect to ERVs, it is not clear why some genomic invasions result in only a limited number of copies and a limited phylogenetic distribution, whereas others, such as intracisternal A-particle (IAP) elements, are more widely distributed and prolific in some genomes. The large phylogenetic distances between well-characterized mammalian genomes make it more difficult to address these questions.
In the present study we have identified members of a group of endogenous retroviruses we call mysTR. This group includes an endogenous retrovirus fragment that was recently isolated by PCR from O. intermedius (15). Comparison of that fragment to the elements isolated in the present study shows it to be most closely related to the O. palustris elements here, showing an average divergence from these O. palustris elements of 7% in the shared part of the reverse transcriptase region. These mysTR elements are related to an inactive group of Mus endogenous retroviruses represented by the MYSERV consensus sequence found in Repbase, which has some sequence similarity to (and is named for) mys, an active nonautonomous retrotransposon we identified previously in Peromyscus species (51). MYSERV_RN is a similar family documented in the Rattus genome (40). The evolutionary relationship between the Murinae (Mus and Rattus) MYSERV elements on one hand and the Sigmodontinae (Oryzomys, Sigmodon and Peromyscus) mys and mysTR elements on the other hand is at this point unclear. MYSERV and mysTR have larger tracts of similarity to each other than either has to mys. Although it is apparent that MYSERV and mysTR have a common ancestor, it is not clear whether this ancestor was an endogenous element present in the lineage that gave rise to these two groups of rodents very roughly 18 million years ago (46) or whether the two rodent lineages were independently infected with related exogenous viruses.
Our analyses show that mysTR elements are class II retroelements, with an additional tentative classification as betaretroviruses. It would not be surprising if these elements are indeed beta elements, given that endogenous retroviruses from this group are among the most common ERVs deposited in the mouse and rat genomes since their divergence from humans (40). Activity of this group is also consistent with the work of Baillie et al. (2), who showed the existence of multiple groups of endogenous betaretroviruses in a number of mammalian species and suggested that the murid rodents have played a role in the global distribution of betaretroviruses. The recent study of class II ERVs, which included the isolation of the O. intermedius fragment (15), raised the possibility that these elements may be more closely related to the lentiviruses than to the classic betaretroviruses, but our analyses do not support this assignment (data not shown). This discrepancy could be due to the fact that the regions of analysis in the two studies are only partially overlapping, so ancient recombination events may have led to different histories for the different regions. Alternatively, the immense distances that separate these genera of the retroelements can lead to problems with phylogenetic reconstruction, such as long branch attraction, even when there appears to be statistical support for specific topologies (12, 48). Additional work will be needed to clarify the relationships among these retroelements, all of which appear to be class II elements.
The occurrence we have seen here of very high copy numbers ranging from roughly 1,000 to 10,000 with such low element divergences is unprecedented for an ERV family. Most ERV families with average nucleotide distances between individual elements of <20% have small group sizes, ranging from 5 to 50 copies with a few ranging into the upper hundreds (5, 7, 10, 16). A recent search of the mouse and rat genome sequences for endogenous betaretroviruses has resulted in the characterization of a number of previously unknown groups (2), but even with that grouping scheme which allowed elements with polymerase gene nucleotide identities as low as 53%, the largest copy number for any group was 60. With very few exceptions, previously described groups of ERVs numbering in the thousands show much higher sequence divergences than those found here and have been deposited over expanses of time ranging from tens of millions of years to the majority of the mammalian radiation (34, 35, 40, 43). A total of 90% or more of the elements in these large groups are single LTRs that have arisen by recombination between left and right LTRs and excision of the body of the original element. All of our estimations of mysTR copy numbers were based on internal sequences rather than LTRs. A notable exception to the above divergences for high-copy-number groups are the IAP elements, with around a thousand copies found in the mouse and hamster genomes (25, 26). Thus, even the mysTR copy number of 1,000 in S. hispidus would be considered exceptional, and the copy number of recently active elements in O. palustris is 10-fold higher than any previously documented ERV group.
The absence of an env gene in the three full-length mysTR elements analyzed here may shed some light on the exceptional copy numbers of mysTR. A similar situation is seen in two active ERV families with high copy numbers in Mus genomes, the IAP elements and the ETn/MusD group (5, 16, 26). The former are largely devoid of env genes, while the latter are completely devoid of env genes, and both families show relatively high copy numbers. The 3′ region of mysTR also shows homology with MYSERV, which is devoid of an env gene. This leads to two possibilities. Either that region in both families represents an ancient env region which has undergone mutational decay to the point of being unrecognizable (Fig. 4), or an ancient recombination in the ancestor of both families replaced the env gene with DNA of unknown origin. Either scenario suggests a long period of mysTR evolution within its host genomes. The gag, pro, and pol regions have been maintained by natural selection; the env region has not. Thus, unlike exogenous retroviral invasions that produce a small number of copies before they “burn out,” repeated mysTR amplifications may have come from an element coadapted to its host for millions of years.
It is not clear whether loss of the env gene has been a passive process or whether there has been positive selection for loss of gene function. Because the env gene is not needed for retrotransposition, it may have simply accumulated mutations due to lack of selection. Alternatively, loss of env could have been a selected event. Elements lacking an env gene may be less detrimental to their host because they would no longer be able to produce infectious viruses. Survival of those hosts could then allow continued retrotransposition to lead to higher copy numbers. Selection could also have occurred at the level of the elements rather than at the level of the host. Loss of env may be an important part of the process that turns some ERVs into well-adapted retroelements.
The present study was initiated to search for repetitive sequences whose amplification was correlated with the loss of LINE-1 activity in a mammalian species, O. palustris. In the course of this search we found the mysTR family, which is amplified to substantially higher levels in the L1-inactive species than in the L1-active species, S. hispidus. However, subsequent determination that mysTR elements are at an intermediate level in the L1-active outgroup, P. maniculatus, raises the question of whether there is any relationship between loss of LINE-1 activity and these unprecedented ERV expansions. These events may be merely coincidental. Alternatively, the initial activity of the mysTR family in the ancestor of all three of these species may have added an additional parasitic burden or taken over an unknown function that set the stage for subsequent loss of LINE-1 activity in O. palustris. One proposed function of LINE-1 elements has been their involvement as way stations for propagation of the X chromosome inactivation signal (31). The recent finding of a decreased density of LTRs in a region of the human X chromosome escaping inactivation versus the same region in the mouse X chromosome which undergoes X inactivation has led to the suggestion that LTRs may also be involved in the spreading of silencing (50). Since the great majority of the elements detected in each species were inserted after divergence from their common ancestor, each species has undergone independent mysTR expansions. Determination of LINE-1 and mysTR activity in additional species of related rodents will allow us to see if mysTR expansion is indeed correlated with a decline in LINE-1 activity.
The identification of such a recent and probably ongoing expansion with widely varying levels of amplification in the three species examined here presents a unique opportunity to look into recent bursts of ERV activity in a group of related rodents that have undergone an extremely large species expansion (11, 44). By applying additional ERV and LINE-1 screens both on these species and on a wider range of species within this group of rodents, we should be able to dissect alternative hypotheses in the ebb and flow of unusual retroelement expansions as they are played out in related host species.
Acknowledgments
We thank Kent Edmonds for generously providing live specimens and tissue of O. palustris used in this study and The Museum of Texas Tech University for providing S. hispidus, O. palustris, and P. maniculatus tissues. We thank Armando Martinez and Kiana Bush for technical assistance.
This study was supported by a grant from the National Institutes of Health (GM38737 to H.A.W.). Analytical resources were provided by INBRE (RR016454) and COBRE (RR016448) grants from the National Institutes of Health.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. Green Publishing/Wiley-Interscience, New York, N.Y.
- 2.Baillie, G. J., L. N. van de Lagemaat, C. Baust, and D. L. Mager. 2004. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J. Virol. 78:5784-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, R. J., M. Hamilton, and D. A. Parish. 2003. Preparations of mammalian karyotypes under field conditions. Occasional Papers Museum Texas Tech Univ. 228:1-8. [Google Scholar]
- 4.Baker, R. J., and H. A. Wichman. 1990. Retrotransposon mys is concentrated on the sex chromosomes: implications for copy number containment. Evolution 44:2083-2088. [DOI] [PubMed] [Google Scholar]
- 5.Baust, C., L. Gagnier, G. J. Baillie, M. J. Harris, D. M. Juriloff, and D. L. Mager. 2003. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse. J. Virol. 77:11448-11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belshaw, R., V. Pereira, A. Katzourakis, G. Talbot, J. Paces, A. Burt, and M. Tristem. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 101:4894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benit, L., J.-B. Lallemand, J.-F. Casella, H. Philippe, and T. Heidmann. 1999. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J. Virol. 73:3301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantrell, M. A., R. A. Grahn, L. Scott, and H. A. Wichman. 2000. Isolation of markers from recently transposed LINE-1 retrotransposons. BioTechniques 29:1310-1316. [DOI] [PubMed] [Google Scholar]
- 9.Casavant, N. C., L. Scott, M. A. Cantrell, L. E. Wiggins, R. J. Baker, and H. A. Wichman. 2000. The end of the LINE? lack of recent L1 activity in a group of South American rodents. Genetics 154:1809-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costas, J. 2003. Molecular characterization of the recent intragenomic spread of the murine endogenous retrovirus MuERV-L. J. Mol. Evol. 56:181-186. [DOI] [PubMed] [Google Scholar]
- 11.Engel, S. R., K. M. Hogan, J. F. Taylor, and S. K. Davis. 1998. Molecular systematics and paleobiogeography of the South American sigmodontine rodents. Mol. Biol. Evol. 15:35-49. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27:401-410. [Google Scholar]
- 13.Furano, A. V. 2000. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog. Nucleic Acids Res. Mol. Biol. 64:255-294. [DOI] [PubMed] [Google Scholar]
- 14.Furano, A. V., D. D. Duvernell, and S. Boissinot. 2004. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 20:9-14. [DOI] [PubMed] [Google Scholar]
- 15.Gifford, R., P. Kabat, J. Martin, C. Lynch, and M. Tristem. 2005. Evolution and distribution of class II-related endogenous retroviruses. J. Virol. 79:6478-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gifford, R., and M. Tristem. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291-315. [DOI] [PubMed] [Google Scholar]
- 17.Goodier, J. L., E. M. Ostertag, and H. H. Kazazian, Jr. 2000. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 9:653-657. [DOI] [PubMed] [Google Scholar]
- 18.Grahn, R. A., T. A. Rinehart, M. A. Cantrell, and H. A. Wichman. 2005. Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet. Genome Res. 110:407-415. [DOI] [PubMed] [Google Scholar]
- 19.Han, J. S., S. T. Szak, and J. D. Boeke. 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429:268-274. [DOI] [PubMed] [Google Scholar]
- 20.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchison, C. A., III, S. C. Hardies, D. D. Loeb, W. R. Shehee, and M. H. Edgell. 1989. LINEs and related retroposons: long interspersed repeated sequences in the eucaryotic genome, p. 593-617. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 22.IHGSC. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 23.Jacobo-Molina, A., and E. Arnold. 1991. HIV reverse transcriptase structure-function relationships. Biochemistry 30:6351-6356. [DOI] [PubMed] [Google Scholar]
- 24.Knipe, D. M., P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.). 2001. Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 25.Kuff, E. L., J. E. Fewell, K. K. Lueders, J. A. DiPaolo, S. C. Amsbaugh, and N. C. Popescu. 1986. Chromosome distribution of intracisternal A-particle sequences in the Syrian hamster and mouse. Chromosoma 93:213-219. [DOI] [PubMed] [Google Scholar]
- 26.Kuff, E. L., and K. K. Lueders. 1988. The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res. 51:183-276. [DOI] [PubMed] [Google Scholar]
- 27.Lee, R. N., J. C. Jaskula, R. A. van den Bussche, R. J. Baker, and H. A. Wichman. 1996. Retrotransposon Mys was active during evolution of the Peromyscus leucopus-maniculatus complex. J. Mol. Evol. 42:44-51. [DOI] [PubMed] [Google Scholar]
- 28.Longmire, J. L., and N. C. Brown. 2003. pFOS-LA: a modified vector for production of random shear fosmid libraries. BioTechniques 35:50-54. [DOI] [PubMed] [Google Scholar]
- 29.Longmire, J. L., A. K. Lewis, N. C. Brown, J. M. Buckingham, L. M. Clark, M. D. Jones, L. J. Meincke, J. Meyne, R. L. Ratliff, F. A. Ray, R. P. Wagner, and R. K. Moyzis. 1988. Isolation and molecular characterization of a highly polymorphic centromeric tandem repeat in the family Falconidae. Genomics 2:14-24. [DOI] [PubMed] [Google Scholar]
- 30.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyon, M. F. 1998. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell. Genet. 80:133-137. [DOI] [PubMed] [Google Scholar]
- 32.Mager, D., and P. Medstrand. 2003. Retroviral repeat sequences, p. 57-63. In Nature encyclopedia of the human genome. Macmillan Publishers, Ltd./Nature Publishing Group. New York, N.Y.
- 33.Martin, S. L., and H. A. Wichman. 1993. Molecular approaches to mammalian retrotransposon isolation. Methods Enzymol. 224:309-322. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy, E. M., and J. F. McDonald. 2004. Long terminal repeat retrotransposons of Mus musculus. Genome Biol. 5:R14.1-R14.8 [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MGSC. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 36.Minin, V., Z. Abdo, P. Joyce, and J. M. Sullivan. 2003. Performance-based selection of likelihood models for phylogeny estimation. Syst. Biol. 52:674-683. [DOI] [PubMed] [Google Scholar]
- 37.Morrish, T. A., N. Gilbert, J. S. Myers, B. J. Vincent, T. D. Stamato, G. E. Taccioli, M. A. Batzer, and J. V. Moran. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31:159-165. [DOI] [PubMed] [Google Scholar]
- 38.Parish, D. A., P. Vise, H. A. Wichman, J. J. Bull, and R. J. Baker. 2002. Distribution of LINEs and other repetitive elements in the karyotype of thebat Carollia: implications for X-chromosome inactivation. Cytogenet. Genome Res. 96:191-197. [DOI] [PubMed] [Google Scholar]
- 39.Pickeral, O. K., W. Makalowski, M. S. Boguski, and J. D. Boeke. 2000. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.RGSPC. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428:493-521. [DOI] [PubMed] [Google Scholar]
- 41.Rinehart, T. A., R. A. Grahn, and H. A. Wichman. 2005. SINE extinction preceded LINE extinction in sigmodontine rodents: implications for retrotranspositional dynamics and mechanisms. Cytogenet. Genome Res. 110:416-425. [DOI] [PubMed] [Google Scholar]
- 42.She, J. X., F. Bonhomme, P. Boursot, L. Thaler, and F. Catzeflis. 1990. Molecular phylogenies in the genus Mus: comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP data. Biol. J. Linnean Soc. 41:83-103. [Google Scholar]
- 43.Smit, A. F. A. 1993. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 21:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, M. F., and J. L. Patton. 1999. Phylogenetic relationships and the radiation of Sigmodontine rodents in South America: evidence from cytochrome b. J. Mammalian Evol. 6:89-128. [Google Scholar]
- 45.Sprinzl, M., C. Horn, M. Brown, A. Ioudovitch, and S. Steinberg. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steppan, S., R. Adkins, and J. Anderson. 2004. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst. Biol. 53:533-553. [DOI] [PubMed] [Google Scholar]
- 47.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 48.Swofford, D. L., G. J. Olsen, P. J. Waddell, and D. M. Hillis. 1996. Phylogenetic inference, p. 407-514. In D. M. Hillis, C. Moritz, and B. K. Mable (ed.), Molecular systematics, 2nd ed. Sinauer Associates, Inc., Sunderland, Mass.
- 49.Teng, S. C., B. Kim, and A. Gabriel. 1996. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature 383:641-644. [DOI] [PubMed] [Google Scholar]
- 50.Tsuchiya, K. D., J. M. Greally, Y. Yi, K. P. Noel, J. P. Truong, and C. M. Disteche. 2004. Comparative sequence and x-inactivation analyses of a domain of escape in human xp11.2 and the conserved segment in mouse. Genome Res. 14:1275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wichman, H. A., S. S. Potter, and D. S. Pine. 1985. Mys, a family of mammalian transposable elements isolated by phylogenetic screening. Nature 317:77-81. [DOI] [PubMed] [Google Scholar]