Abstract
In a previous report we demonstrated that three injections of an rt-deleted noninfectious genome of the simian-human immunodeficiency virus SHIVKU2 induced protection against AIDS in macaques (D. K. Singh, Z. Liu, D. Sheffer, G. A. Mackay, M. Smith, S. Dhillon, R. Hegde, F. Jia, I. Adany, and O. Narayan, J. Virol 79:3419-3428, 2005). To make this DNA safer, we deleted two more genes, the integrase gene and vif, along with the 3′ long terminal repeat. We also replaced the gag, pro, and nef genes (SIVmac239 origin) with those of human immunodeficiency virus (HIV) type 1 strain SF2. The resultant construct, designated Δ4SHIVKU2 DNA, was used in this study to evaluate gene expression and immunogenicity in BALB/c mice. DNA-transfected human embryonic kidney epithelial cells (HEK 293) produced all of the major viral proteins and released p24 in the supernatant for 12 days. Inoculation of the vaccine DNA into the gastrocnemius muscles resulted in intense mononuclear cell infiltration at the inoculated sites and the production of viral p24 in myocytes, in infiltrating mononuclear cells, and in cells in the spleen and draining lymph nodes between 3 and 10 days postinoculation. Expression of p24 in the muscle cells peaked at day 7 and became undetectable after day 12. The same 12-day period of expression of p24 was observed in mice that were given a second injection 4 weeks after the first. Evaluation of immune responses in BALB/c mice revealed that the DNA induced enzyme-linked immunospot and antigen-specific proliferative cell-mediated immunity responses. The responses were stronger in mice that were coinjected with a second plasmid expressing granulocyte-macrophage colony-stimulating factor. Since new waves of viral antigen production could be induced with each boosting injection of the vaccine DNA, this DNA could be a safe and efficient agent to induce long-term protection against HIV.
An effective and safe vaccine against human immunodeficiency virus (HIV)/AIDS still remains elusive even after more than a decade of intense research. Although live vaccines are highly effective in preventing AIDS in macaques, they are not acceptable for use in humans because of concerns about reversion to pathogenicity by mutation or recombination. An alternative strategy using recombinant viral envelope glyco-protein was not effective in clinical trials. Further alternative strategies using viral DNA as a vaccine were therefore adopted. Initial clinical trials of certain of the DNA vaccines in humans have demonstrated only a limited degree of immunogenicity thus far (for a review, see reference 20), necessitating further improvement in DNA vaccine construction strategies. Attempts to improve the efficiency of DNA vaccines have been made either by optimization of HIV genes for codon usage in mammalian cells (4, 7, 18, 44, 45) or by modifying RNA structures through nucleotide changes facilitating high antigen expression in a Rev-independent manner (32). Other strategies tried to potentiate DNA vaccines by modulating and enhancing host immune response with the help of cytokine/chemokine adjuvants and T-cell costimulatory molecules (9, 10, 14, 32) or by different enhancers/promoters such as cytomegalovirus (37) or β-actin or muscle-specific desmin promoter (13) to obtain high expression of one or two fused genes of HIV (26). The use of these DNA moieties was followed by boosting injections with viral proteins expressed by various vector systems such as recombinant pox virus (31), modified vaccinia virus Ankara (2, 5, 16), and adenovirus (12, 24).
Two potential problems with current DNA approaches are that (i) it is still not known which of the viral proteins are essential for inducing optimum protection, since claims of efficacy using gag (8, 15), env (19, 25, 43), and tat (11, 21, 22) have been raised, and (ii) viral strategies that plan for the use of vectors expressing viral proteins will be cumbersome to apply in underdeveloped countries. To overcome the necessity for using a protein boost and yet achieve efficacy with a DNA vaccine, we adopted a new approach by using the DNA of the simian-human immunodeficiency virus SHIVKU2, which expresses all of the viral proteins with high efficiency but from which we deleted the rt gene to render the DNA noninfectious. The first prototype of this DNA, rtSHIVKU2, was used in an earlier study in which three injections of the DNA elicited protection of macaques against AIDS caused by heterologous SHIV 89.6P (35). The perceived advantage of this approach is to include all major genes of HIV in the vaccine in order to generate an optimal immune response that can suppress viral replication and provide efficient protection. We chose to use the simian immunodeficiency virus (SIV) promoter in the 5′ long terminal repeat (LTR) to drive viral genes because of its high efficiency of expression. Following up on this study, we deleted two more genes from rtSHIVKU2, the integrase gene (int) and vif, along with the 3′ LTR in order to minimize the integration potential of the DNA and to guarantee its lack of infectivity. We also replaced the gag, pro, and nef genes (SIVmac239 origin) of rtSHIVKU2 with those of HIV type 1 (HIV-1) (strain SF2) in order to make the DNA relevant as a vaccine against HIV. The resulting construct, designated Δ4SHIVKU2 DNA, was used in the pilot study described in this report to evaluate antigen expression and its immunogenicity in BALB/c mice. Transfection of 293 cell culture revealed high expression of all of the encoded viral proteins for approximately 12 days. Intramuscular injection of the DNA into mice resulted in transient high expression of p24 in skeletal muscle and in mononuclear cells infiltrating the injection site. This resulted in induction of cell-mediated immunity (CMI) responses that were further enhanced when mice were injected with a second plasmid expressing granulocyte-macrophage colony-stimulating factor (GM-CSF). This new DNA vaccine strategy could prove to be useful as a prophylactic as well as a therapeutic vaccine against AIDS.
MATERIALS AND METHODS
Construction of Δ4SHIVKU2 plasmid DNA.
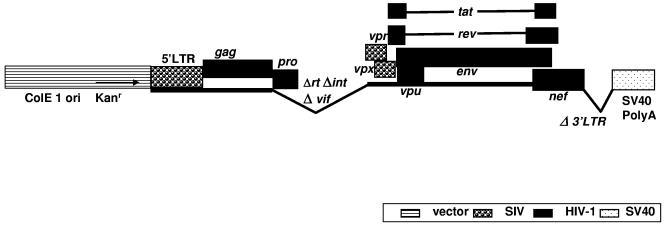
The procedures for construction of SHIVKU2 plasmid DNA have been described earlier (27). The insert sequences were derived from SHIVKU2 (GenBank accession no. AY751799) and HIV-1SF2. To replace the SIV gag, pol, and vif genes with those of HIV-1SF2, PCR site-directed mutagenesis was performed using forward 5′-GGTCACCATGTCAGATCCCAGGGAGAG-3′ and reverse 5′-GCATGAAGAGCGCTCGTTGGAGG-3′ primers to generate a BstEII site upstream of the ATG start codon of the SHIVKU2 vpx gene. Similarly, a BstEII site was created by PCR site-directed mutagenesis using forward 5′-AATCTCTAGCAGTGGCGCCCGAAC-3′ and reverse 5′-GCTGGTGACCCTTTCCATCCCTG-3′ primers at position 3007 downstream of protease-coding sequences. The 3.2-kb NarI-BstEII fragment (positions 831 to 4084 in SHIVKU2) was removed and replaced with the corresponding sequences in a 2.4-kb NarI-BstEII fragment from HIV-1SF2. (These positions are with reference to GenBank database accession no. K02007.) To replace SIV nef with that of HIV-1SF2, a series of PCR and standard molecular cloning steps were used. Briefly, an intermediate clone containing part of SHIVKU2 gp41 was made using the forward 5′-GTGAACGGATCCTTGGCAC-3′ (containing a BamHI site) and reverse 5′-GATATCTTATAGCAAAATCCTTTCC-3′ (containing a EcoRV site) primers. The HIV-1SF2 nef was amplified by PCR in a 1.4-kb fragment containing a SalI site at the 5′ end and a NotI site at the 3′ end of the HIV-1SF2 nef, using the forward 5′-GTCGACATGGGTGGCAAGTGGTC-3′ and reverse 5′-CGCCGGCGTCAGCAGTCTTTGTAGTAC-3′ primers. The resulting PCR fragment was cloned into pGEM-T (Promega). The HIV-1SF2 nef was then fused at the end of the SHIVKU2 gp41 gene. The 1-kb BamHI-SalI fragment of this clone was used to replace that of Δ1SHIVKU2 (Fig. 1). The resultant construct, named Δ4SHIVKU2, carries the vpx and vpr genes of SIVmac239 and the gag, pro, partial rt, vpu, tat, rev, env, and nef genes of HIV-1 under control of the SIVmac239 5′ LTR. The reverse transcriptase gene was truncated, and int, vif, and the 3′ LTR were deleted from SHIVKU2.
FIG. 1.
Schematic representation of the Δ4SHIVKU2 plasmid used in this study. This plasmid has tat, rev, env, and vpu genes from HIV-1 strain HXB2 and gag, pro, and nef from HIV-1 strain SF2. The 5′ LTR, vpr, and vpx are from SIVmac 239. Sequences from the rt, int, and vif genes and the 3′ LTR were deleted to make this plasmid noninfectious, and poly(A) sequences from SV40 were added.
The sequences for the vector were derived from plasmids pET-9a (Novagen) and pCMV-Tag2 (Stratagene) The 1.8-kb BamHI-XmmI fragment of pET-9a was replaced by the 547-bp BamHI-SspI fragment, and the recombinant plasmid was then cut with SalI-EcoRI, which resulted a 2.5-kb SalI-EcoRI fragment having simian virus 40 (SV40) polyadenylation signal sequences that was used as vector to make the final clone (Fig. 1).
Transfection of HEK 293 cells.
Transfection of Δ4SHIVKU2 DNA into HEK 293 cells was carried out using a cationic polymer polyethylenimine, ExGen 500, and the protocols provided by the manufacturer (catalog no. RO511; MBI Fermentas) for adherent cells. Approximately 1 ×105 to 3 ×105 cells/well in six-well plates were transfected with 4.75 μg of vaccine DNA and 15.5 μl of ExGen 500. Culture supernatant fluids were assayed on days 1, 2, 4, 6, 8, 10, 12, and 14 to assess the kinetics of synthesis and duration of production of Gag p24. Another set of transfected cultures was used for immunoprecipitation of different viral proteins.
Radioimmunoprecipitation assay.
We analyzed transfected cultures by a standard radioimmunoprecipitation method to assess and document expression of viral proteins encoded by Δ4SHIVKU2. Briefly, transfected cells were washed and starved in serum-, cystine-, and methionine-free medium for 2 h and labeled overnight by supplementing the medium with100 μCi/ml [35S]methionine (MP Biomedical Inc., Irvine, CA). Proteins from cell lysates and culture supernatants were precipitated using protein A-Sepharose (catalog no. 17-0780-01; Amersham Bioscience AB, Sweden), and sera from uninfected and HIV-infected persons or monoclonal antibodies (MAbs) against Nef or Vpu were used to precipitate various viral proteins. Immunoprecipitated samples were washed thrice in 1× radioimmunoprecipitation assay buffer, boiled in 2× sample reducing buffer, and separated on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels. Gels were fixed in 30:10 methanol-acetic acid fixative, treated with Amplify reagent (catalog no. NAMP100; Amersham Biosciences), mounted on a filter paper, and dried in a gel dryer. Dried gels were exposed to X-ray film, incubated at −80°C, and developed in an automated X-ray film developer. Autoradiographs were analyzed for the presence of different bands corresponding to Nef; Vpu; Env gp160, gp120, and gp41; and Gag p55 and p24.
Quantitation of p24 in the supernatant fraction of Δ4SHIVKU2 DNA-transfected cultures.
Supernatant fluids from transfected HEK 293 cells were analyzed with a capture enzyme-linked immunosorbent assay (ELISA) kit (Coulter Laboratories, Hialeah, FL) for quantity and duration of secretion of HIV p24 Gag protein. A standard curve was prepared for each assay as per the manufacturer's instructions. p24 concentrations were determined from the optical density at 450 nm plotted against a standard curve by linear regression analysis.
Localization of p24 in transfected cells.
HIV p24 was localized in transfected HEK 293 cells by indirect immunofluorescence using mouse anti-HIV p24 monoclonal antibody (catalog no. 13-112-100; Advanced Biotechnologies Inc., Columbia, MD) and goat anti-mouse immunoglobulin G (IgG) conjugate (AlexaFluor 488) (catalog no. A-11017; Molecular Probes, Oregon). Briefly, HEK cells were grown on coverslips, transfected with Δ4SHIVKU2, and then incubated at 37°C. Two days later, the cells were fixed with 1% paraformaldehyde for 15 min, washed and permeabilized in 80% methanol at −20°C for 1 h, and incubated with primary and secondary antibodies at 1:200 dilutions at room temperature for 1 h each. Stained coverslip preparations were mounted on clean glass slides, and images were collected using a Nikon TE300 fluorescence microscope and Spot imaging system.
Inoculation of mice.
BALB/c mice were injected intramuscularly with the plasmid DNA at concentrations of between 1 and 2 μg/μl. DNA solutions were prepared in phosphate-buffered saline (PBS) (0.1 M, pH 7.4). Each mouse was injected with a total of 100 μl of DNA solution, with 50 μl in each gastrocnemius muscle. Control mice for each protocol received saline instead of DNA in gastrocnemius muscles.
Histology and immunohistochemistry.
Injected mice were exsanguinated and perfused with saline, followed by perfusion with 4% paraformaldehyde. Gastrocnemius muscles, spleens, and draining lymph nodes were collected in 4% paraformaldehyde and processed for histological and immunohistological analyses by paraffin embedding. Approximately 5-μm-thick sections were stained with hematoxylin and eosin. Adjacent tissue sections were also stained with MAb against HIV Gag p24 to localize viral protein, using an immunoperoxidase technique. Other sections were reacted with antibodies against macrophages (F4/80) (anti-murine rat F4/80 pan macrophage monoclonal antibody; catalog no. RDI-T2006x, clone BM-8 RDI; Research Diagnostics Inc., NJ), dendritic cells (CD11c) (anti-mouse CD11c antibody; catalog no. RDI-MCD11C-HL; Research Diagnostics Inc., NJ), and T cells (CD3) (anti-mouse rat monoclonal antibody; catalog no. CBL 1302; Cymbus Biotechnologies Ltd., Chandlers Ford, Hants). Sections were viewed under a Nikon TE300 microscope, and images were collected using a Spot imaging system.
Processing of spleens for splenocytes.
Spleens of mice were aseptically collected and pressed between glass slides to harvest splenocytes. Cells were collected in Hanks-EDTA solution, erythrocytes were lysed using BD lysis solution, and mononuclear cells were counted in a hemocytometer. A portion of the splenocytes were used to isolate CD4+ or CD8+ T cells for use in enzyme immunospot (ELISPOT) assays, and another portion were used in antigen-specific proliferation assays. The fractions of splenocytes containing either CD4+ or CD8+ T cells were negatively isolated using MACS high-gradient magnetic separation columns of type LS (Miltenyi Biotech, Auburn, CA) and BD Image anti-human CD4+ and CD8+ MSC magnetic particles (BD Biosciences-Phar-Mingen) according to the manufacturer's instructions.
ASP of CD4+ T cells.
The antigen-specific proliferation (ASP) assay was described previously (23). Viral antigen consisted of a live SHIV vaccine virus stock that was UV irradiated for 30 min and heated at 56°C for 2 hours. The antigen was mixed with freshly collected triplicate samples of splenocytes, using105 cells/well in 96-well tissue culture plates in 200 μl of R-10 medium. The cells were cultured for 4 days at 37°C and then pulsed with 1 μCi of [3H]thymidine/well. Eighteen hours later, the cells were harvested and the [3H]thymidine incorporation was determined by liquid scintillation counting (Packard). Stimulation indices (SI) were calculated as mean counts per minute in stimulated wells divided by mean counts per minute in control wells. An SI value of >3 was considered significant.
ELISPOT assay.
We used a quantitative ELISPOT assay that measured gamma interferon (IFN-γ) production by splenocytes responding to groups of overlapping 15-mer peptides, with 11 amino acid overlaps, spanning the entire molecules of selected viral proteins. We used HIV Gag (HXB2) peptide (catalog no. 7872-7904), HIV Env (MN) peptide (catalog no. 6451), consensus HIV Tat (catalog no. 5138), consensus HIV Rev (catalog no.6445), and consensus HIV Nef (catalog no. 5189), all kindly provided by the AIDS Research and Reference Reagent Program. Pools of five peptides were used in the assays. Millipore multiscreen IP opaque hydrophobic high-protein-binding 96-well plates (0.45 μm) were coated overnight at 4°C with 50 μl/well of 10 μg/ml of anti-mouse IFN-γ antibodies (MAb AN18; MABTECH, Stockholm, Sweden). The unbound antibodies were removed on the following day by washing four times with PBS. The plates were blocked for 2 h at room temperature with R10. Fifty microliters of each peptide pool was then added to each of two wells, except for negative and positive control wells. R10 culture medium and 0.5 μg/well of concanavalin A were used for negative and positive controls, respectively. Fifty microliters of the splenocyte suspension (2 × 106/ml) was added to each well and the plates incubated for 18 h at 37°C. After unbound cells were discarded, the plates were washed three times with PBS, followed by three more washes with PBS-0.1% Tween 20. Fifty microliters of biotinylated anti-IFN-γ antibodies (2 μg/ml; catalog no. R46A2; MABTECH, Stockholm, Sweden) was then added to each well and the plates incubated for 3 h at room temperature. The plates were then processed as follows: six rinses with PBS-0.1% Tween 20, addition of 50 μl/well of Vectastain AB, incubation for 1 hour at room temperature, six more rinses, color development by treatment with 100 μl/well of Nova-Red for 4 min at room temperature, and a rinse in running tap water. Spots were counted with a stereomicroscope and reported as the number of spots per 106 splenocytes.
Antibody binding assay.
We analyzed sera from immunized mice for antibodies against different viral proteins in binding ELISA. Briefly, 96-well plates were coated with pools of HIV-1 HXB2 Gag, Env, Nef, Rev, and Tat peptides (obtained from the NIH AIDS Research and Reference Reagent Program) at a concentration of 5 μg/ml in PBS, pH 7.4, at 50 μl/well at 4°C overnight. The plates were blocked with PBS containing 10% fetal calf serum for 1 h at 37°C, washed with PBS containing 0.05% Tween 20. The serum samples, added in serial dilutions, were incubated at 37°C for 2 h. After being washed, plates were incubated with horseradish peroxidase-conjugated anti-mouse IgG (1:3,000 dilution; catalog no. AP308P; Chemicon International, Temecula, CA) for 1 h at 37°C. The plates were washed again and incubated with tetramethylbenzidine substrate reagent (Sigma, St. Louis, MO) for 30 min. The color reaction was stopped with 1 M H2SO4, and the plates were read at 450 nm using an ELISA plate reader (Dynatech MR4000).
Statistical analysis.
We employed the Wilcoxon rank sum test to assess statistical significance of the CMI responses of mice immunized with different doses of vaccine DNA or coinjected with GM-CSF DNA during priming and boosting, using the program Statistix 8 from Analytical Software Company, Tallahassee, Fla.
RESULTS
Immunoprecipitation studies.
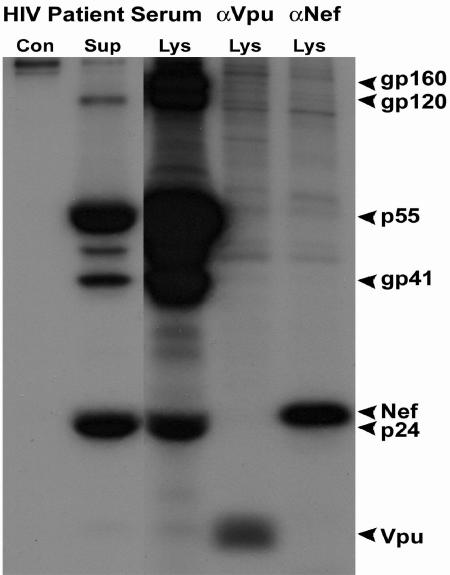
To evaluate expression and correct orientation of all of the viral proteins encoded in this DNA, we transfected HEK 293 cells with Δ4SHIVKU2 DNA and analyzed viral proteins by immunoprecipitation assays. The results indicated expression of envelope gp120, gp41, and their gp160 precursor; Gag p24 and its p55 precursor; and Nef and Vpu in the cell lysate fraction. The separation of these proteins on 10% SDS-polyacrylamide gels indicated their reported sizes, suggesting correct processing and posttranslational modifications. Envelope gp120 and gp41, capsid protein p24, and p55 were also detected in abundance in the supernatant fraction of the transfected culture. The SDS-polyacrylamide gels are shown in Fig. 2.
FIG. 2.
Immunoprecipitation of major viral proteins in HEK 293 cells transfected with Δ4SHIVKU2 DNA. Viral proteins precipitated by an HIV patient serum and by anti-Vpu and anti-Nef antibodies from transfected HEK 293 cells at 36 h posttransfection are shown. Both Env and Gag proteins with their precursors and Nef and Vpu were precipitated from the cell lysate fraction. Env gp120 and gp41 and Gag p55 and p24 were precipitated from the supernatant fluid (Sup). Con, cell lysate from nontransfected HEK 293 control cells.
Transfection of HEK 293 cells with Δ4SHIVKU2 DNA results in the transient expression of viral p24.
In order to assess the transfection efficiency of the vaccine DNA, we examined the cultures 2 days after transfection of cells by staining with anti-HIV p24 monoclonal antibody and anti-mouse IgG conjugate. Approximately 20% of the cells were positive (data not shown). We further tested the transfected cultures for duration of production of the viral protein by measuring p24 concentrations in supernatant fluids. These fluids were collected at 4, 8, 12, 16, 20, and 24 h and every 2 days thereafter for 14 days and were analyzed using p24 ELISA. p24 was first detected at 16 h posttransfection. The cells produced p24 for 12 days, with peak concentrations of >18 ng/ml between days 2 and 8 (data not shown).
Intramuscular injection of Δ4SHIVKU2 DNA elicits local inflammatory responses.
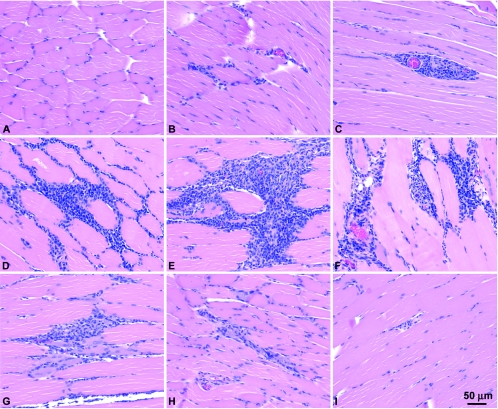
To evaluate histological changes at the site of injection of the DNA, we injected 24 mice with 200 μg of Δ4SHIVKU2 DNA into gastrocnemius muscles and killed three animals each on days 2, 3, 5, 7, 10, 12, 15, and 20 after inoculation. A control group of mice received injections of saline instead of DNA. Histological examination of the muscle sections from this group revealed that a cellular inflammatory response started at 2 days postinjection, with peak intensity between 5 to 12 days (Fig. 3D to G). However, mild cellular infiltration persisted at the injection sites in the muscles for up to 20 days. We did not see infiltration of mononuclear cells in saline-injected muscles (Fig. 3A). Coinjection of pGM-CSF resulted in more intense infiltration of mononuclear cells, which caused a larger infiltrate at the injection site (Fig. 4B).
FIG. 3.
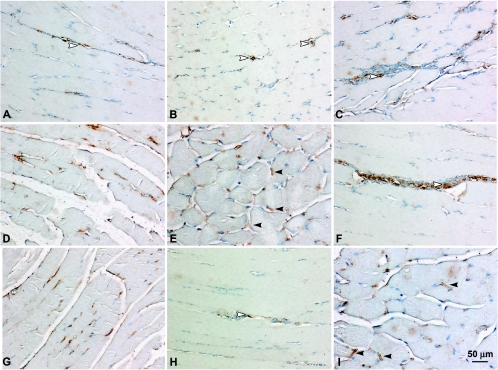
Histological examination of muscles injected with Δ4SHIVKU2 DNA. The DNA elicited an intense mononuclear infiltration at the injected sites. Cellular infiltration was concentrated mostly around the fascicles along with endomysium and in severe cases along with perimysium. Panel A represents muscle sections from control saline-injected mice. Panel B shows initiation of mononuclear cell infiltration into the muscles of injected mice at day 2 postinjection. Panel C shows mild mononuclear cell infiltration around blood vessels in muscles of injected mice at day 2 postinjection. Panels D to F show intense infiltration of mononuclear cells at day 5 (D), day 7 (E), and day 10 (F) in the muscle. Panels G to I show gradual resolution of mononuclear cell infiltration at days 12, 15, and 20, respectively.
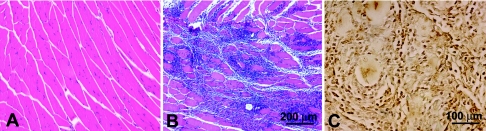
FIG. 4.
Histological examination of muscles coinjected with pGM-CSF and Δ4SHIVKU2 DNA. The coinjection of pGM-CSF DNA elicited massive mononuclear infiltration at the injected sites that resulted in a larger infiltrate than was elicited by vaccine DNA alone (Fig. 3). Panel A shows no infiltration in muscle sections from control mouse. Panel B shows a representative area of mononuclear infiltration in the muscle coinjected with pGM-CSF DNA at day 7 postinjection. Panel C shows that the majority of the cells in the infiltrates in the muscle coinjected with pGM-CSF and vaccine DNA were macrophages.
Cell phenotypes participating in the inflammatory response.
We selected serial histological sections of muscles from mice injected with vaccine DNA with or without GM-CSF and stained them with F4/80 to detect macrophages, anti-CD11c antibody to detect dendritic cells, and anti-CD3 antibody to detect lymphocytes. The results of immunostaining revealed that the majority of the cells in the infiltrates of both groups were macrophages (Fig. 4C), with a low percentage of dendritic cells (data not shown). We did not find a significant percent increase in dendritic cells when GM-CSF was coinjected with vaccine.
Expression of vaccine DNA in tissues.
Sections from muscles, spleens, and draining lymph nodes from the mice injected with vaccine DNA were used to evaluate expression of viral antigen at different time intervals postimmunization. We stained 5-μm-thick sections of these tissues with MAb against p24. Examination of these sections showed viral antigen in the sarcoplasmic reticulum of skeletal muscle cells and in the cytoplasm of a few of the mononuclear cells in cellular infiltrates. A small quantity of viral antigen appeared in a few cells as early as day 3 postinjection (Fig. 5A and B). By day 5, increased numbers of muscle cells were expressing antigen in the sarcoplasmic reticulum and cytoplasm of infiltrating cells (Fig. 5C and D). Increased amounts of antigen appeared to be accumulated in the sarcoplasmic reticulum, resulting in formation of sarcoplasmic reticular sacs, by day 7 (Fig. 5E and G). Antigen expression peaked around this time (Fig. 5E and G). By day 12, very few cells containing viral antigen were observed (Fig. 5H). By days 15 and 20, no antigen was detected in muscles.
FIG. 5.
Immunohistochemical localization of Gag p24 in muscles of mice injected with Δ4SHIVKU2 DNA. Gag p24 was localized in the sarcoplasmic reticulum of skeletal muscle cells and in the cytoplasm of infiltrating mononuclear cells in the muscle. Panel A shows Gag p24 in the cytoplasm of some of the infiltrating cells at day 3. Panel B shows Gag p24 in the sarcoplasmic reticulum of skeletal muscle cells (arrowheads). Panels C and D show Gag p24 in the sarcoplasmic reticulum of skeletal muscle cells and in the cytoplasm of infiltrating mononuclear cells (arrowhead) at day 5. Panels E and F show Gag p24 in the sarcoplasmic reticular sacs (closed arrowheads) and in the cytoplasm of mononuclear cells (open arrowhead) at day 7. Panels G and H show p24 in muscles at days 10 and 12 postinjection. Panel I shows Gag p24 expression peaking in the skeletal muscle cells at day 7 after the second injection of vaccine DNA.
To evaluate whether the viral antigen could be reexpressed in mice receiving serial injections of the DNA, we injected another group of 24 mice with 200 μg of vaccine DNA and administered a second injection of 100 μg of vaccine DNA 4 weeks after the first injection. Three mice each were killed at 2, 3, 5, 7, 10, 12, 15, and 20 days after the second DNA injection, and muscles and spleens were used to detect viral protein by immunoperoxidase staining using MAb against p24. We detected the same kinetics of antigen expression, with peak production between 3 and 10 days, as we had observed in other animals that received a single injection of the DNA. Again, p24 was not detected at the 15-day sampling time. Figure 5I shows the intensity of antigen production at day 7. This was similar to the result with the first injection (Fig. 5E to G).
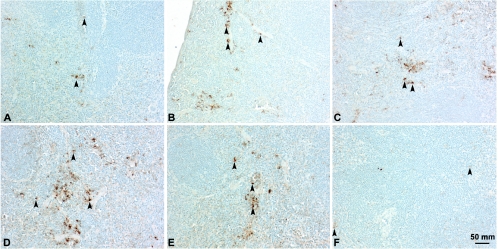
Evaluation of sections of spleen at different time points revealed expression of antigen with same kinetics as was observed in muscles. Gag p24 was detected in the cells in the red pulp between days 3 and 10. A few positive cells were present at day 3 (Fig. 6A). The number of positive cells increased during the following days, peaking at day 7 (Fig. 6C and D) and tapering off thereafter. Sections examined at day 15 were negative for the viral antigen. Antigen was also recognized in cells of sinuses of draining lymph nodes, with similar kinetics (data not shown).
FIG. 6.
Immunohistochemical localization of Gag p24 in vascular sinusoids of spleens of mice injected with Δ4SHIVKU2 DNA. Gag p24 was localized in the cytoplasm of mononuclear cells in the red pulp area at different days postinjection. Panel A shows Gag p24 in the cytoplasm of a few cells in the sinusoids at day 3 (arrowheads). Panel B shows Gag p24 in an increased number of cells (arrowheads) at day 5. Panels C and D show Gag p24 in an increased number of mononuclear cells (arrowheads) at day 7. Panels E and F show Gag p24 in the cells in vascular sinusoids at days 10 and 12, respectively.
Analysis of CMI responses in BALB/c mice immunized with Δ4SHIVKU2 DNA.
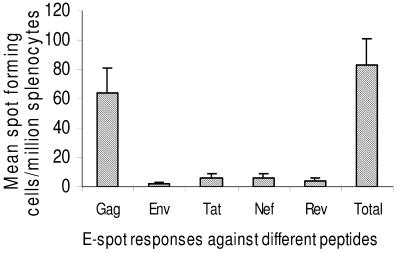
We injected six mice with 200 μg of vaccine DNA to evaluate CMI responses of mice against different viral genes encoded in this DNA vaccine. Splenocytes from vaccinated and nonvaccinated control groups were stimulated with Gag, Env, Rev, Tat, and Nef in ELISPOT assay to enumerate viral peptide-specific IFN-γ-producing cells. The result of the ELISPOT experiment with this group showed that the animals developed significant ELISPOT responses to Env, Gag, Tat, Rev, and Nef. The control peptide ELISPOT count was subtracted from the ELISPOT response elicited by Gag, Env, Tat, Rev, and Nef presented in Fig. 7. The ELISPOT counts for Gag peptides were highest (64 ± 17 out of a total of 83 ± 18 spots/106 splenocytes), indicating immunodominance of Gag over other viral proteins (Fig. 7).
FIG. 7.
ELISPOT (E-spot) responses in BALB/c mice immunized with single injection of Δ4SHIVKU2 DNA against different viral peptides. The numbers of IFN-γ-producing cells per million splenocytes reacting to different viral peptides (Gag, Env, Tat, Rev, and Nef) were counted in an ELISPOT assay. The control peptide ELISPOT count was subtracted from the ELISPOT responses elicited by Gag, Env, Tat, Rev, and Nef. The highest number of cells reacted to Gag peptides, indicating immunodominance of this protein. Error bars indicate standard deviations.
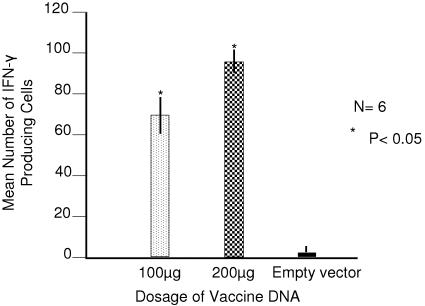
We then evaluated the immunogenicity of this DNA in BALB/c mice at different doses. We injected one group of six mice with 200 μg and another with 100 μg of Δ4SHIVKU2 DNA into gastrocnemius muscles. All animals were boosted twice with injections of 100 μg of DNA in the same muscles at 4 and 6 weeks after the priming injection. Control mice received injections of saline instead of DNA. All mice were killed 1 week following the last boost, and splenocytes were analyzed for IFN-γ-producing cells against Gag peptides by ELISPOT assay. Since ELISPOT responses were mostly against Gag, we performed ELISPOT analysis using only Gag peptides in a further immunogenicity study. The results showed that the animals receiving 200 μg of DNA developed significantly higher ELISPOT responses (84.7/106 splenocytes; P = 0.02) than those receiving 100 μg of DNA (58.3/106 splenocytes) (Fig. 8).
FIG. 8.
ELISPOT responses in BALB/c mice immunized with Δ4SHIVKU2 DNA. The number of IFN-γ-producing cells per million splenocytes from groups of mice injected with different doses of DNA was counted in an ELISPOT assay. The group receiving 200 μg DNA had significantly higher (P < 0.05) ELISPOT counts than the group receiving 100 μg DNA. Error bars indicate standard deviations.
Effect of GM-CSF on immunogenicity of the vaccine DNA.
We analyzed antibody responses of mice injected with vaccine DNA with or without GM-CSF DNA. We injected 36 mice with 200 μg vaccine and 100 μg of GM-CSF DNA in gastrocnemius muscles. The animals were then divided into two groups of 18. One group was boosted with injections of 100 μg vaccine DNA alone and the other with 100 μg of vaccine and 100 μg of GM-CSF DNA in the same muscles twice at 4 and 6 weeks, respectively, after the priming injection. A control group of mice received injections of saline instead of DNA. Six mice from each of the immunized groups were killed after 4, 8, and 12 weeks following the last boost. Sera and spleens were harvested for antibody and ELISPOT analysis. Binding antibody analysis revealed that the mice developed only a minimal amount of antibodies even after two boosts. Coinjection of pGM-CSF did not change this response
Splenocytes from these mice were analyzed for antigen-specific cell proliferation and viral peptide-specific IFN-γ-producing cells by ELISPOT assays. The results showed that the mice that were boosted with vaccine DNA alone developed ELISPOT responses that persisted at almost equivalent titers at all three sample periods. The mean ELISPOT titers for this group of animals were 58, 96, and 98 per 106 splenocytes at 4, 8, and 12 weeks, respectively. In contrast, the vaccinated animals that received GM-CSF DNA in the boosting injections showed ELISPOT titers that exceeded those caused by DNA alone, and the titers increased with time, reaching maximum values at the 12-week sampling point. The mean ELISPOT titers for this group of animals were 65, 117, and 142 per 106 splenocytes at 4, 8, and 12 weeks, respectively. The ELISPOT data are shown in Fig. 9. Examination of splenocytes for ASP T-cell responses showed that the two immunization protocols did not yield statistically different results. Both protocols elicited ASP responses (SI of >3 to 23.3) that persisted throughout the 12-week study. The stimulation indices for both groups of animals are shown in Table 1.
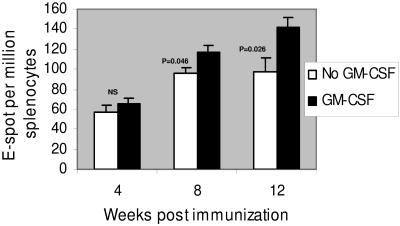
FIG. 9.
Effect of coinjections of GM-CSF DNA with Δ4SHIVKU2 DNA on immune responses in mice. Numbers of IFN-γ-producing splenocytes per million, at 4, 8, and 12 weeks, from groups of mice that were coinjected with either vaccine DNA or vaccine and GM-CSF DNAs during the boost are shown. The group that received the GM-CSF DNA boost had significantly higher (P < 0.05) ELISPOT counts than the group receiving only the vaccine DNA. NS, not significant. Error bars indicate standard deviations.
TABLE 1.
Antigen-specific T-cell proliferation
| Boosted with vaccine DNA | Mouse no. | Stimulation index at indicated weeka
|
||
|---|---|---|---|---|
| 4 | 8 | 12 | ||
| Without GM-CSF | 1 | 3.23 | 4.35 | 2.07 |
| 2 | 6.67 | 9.40 | 3.00 | |
| 3 | 6.64 | 5.14 | 1.63 | |
| 4 | 2.96 | 9.20 | 3.14 | |
| 5 | 9.35 | 3.10 | 3.00 | |
| 6 | 1.65 | 1.93 | 1.92 | |
| Mean ± SE | 5.08 ± 1.195 | 5.52 ± 1.27 | 2.46 ± 0.2695 | |
| With GM-CSF | 1 | 6.49 | 4.31 | 3.00 |
| 2 | 15.05 | 6.00 | 7.48 | |
| 3 | 3.02 | 4.43 | 13.87 | |
| 4 | 8.41 | 4.72 | 3.00 | |
| 5 | 6.34 | 5.42 | 1.18 | |
| 6 | 23.31 | 3.05 | 3.00 | |
| Mean ± SE | 10.37 ± 3.046 | 4.65 ± 0.4141 | 5.255 ± 1.92 | |
Stimulation indices of >3 were considered significant.
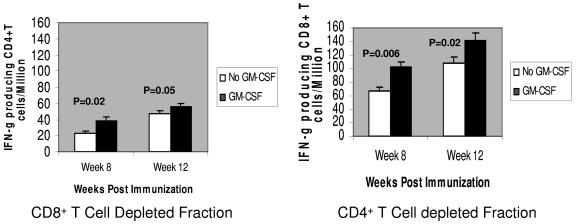
Evaluation of splenocytes of immunized mice depleted of CD4+ and CD8+ T cells at 8 and 12 weeks, respectively, showed that the ELISPOT response was induced mostly by CD8+ T cells and that the ASP was induced mostly by CD4+ T cells. The ELISPOT results for both cell subsets are shown in Fig. 10.
FIG. 10.
Cell phenotypes responsible for producing IFN-γ in mice injected with vaccine or with vaccine and GM-CSF DNA. Numbers of IFN-γ-producing CD4+ or CD8+ T cells per million splenocytes, at 8 and 12 weeks, from groups of mice that were coinjected with either vaccine DNA or vaccine and GM-CSF DNAs during the boost are shown. The CD4+ T-cell-depleted fraction had significantly higher numbers of IFN-γ-producing cells than the CD8+ T cell fraction. The group that received the GM-CSF DNA boost had significantly higher (P < 0.05) ELISPOT counts in both cell subsets than the group that received only vaccine DNA. Error bars indicate standard deviations.
DISCUSSION
In the present study, we used Δ4SHIVKU2 DNA in BALB/c mice to assess details of viral gene expression and host responses at tissue levels that led to development of CMI responses. This DNA is devoid of int, rt, vif, and the 3′ LTR and encodes Env, Gag, Vpu, Tat, Rev, and Nef of the virus. At least three of these proteins, Gag, Env, and Tat, either alone or in combination, have been claimed to be important for inducing protection against AIDS (1, 3, 17, 27). All three of these genes were expressed in our construct. In addition, we included nef and vpu in the vaccine as an additional potential advantage because of their ability to down-regulate expression of major histocompatibility complex class I (MHC-I) (6, 30, 33, 41). This could thus facilitate a longer duration of expression of viral proteins in cells in the body that became transfected by the DNA. Elicitation of high expression of viral antigen and recruitment of antigen-presenting cells at injection sites have been shown to be of great importance for induction of cellular immune responses (28, 36). The uptake of vaccine DNA and viral protein expression by skeletal muscle cells and mononuclear cells may facilitate MHC-I and MHC-II processing pathways that can support the development of both humoral and cellular immune responses (14, 37, 38, 42). Transfected muscle cells have been shown to independently activate CD8+ T cells to elicit strong cytotoxic T-lymphocyte responses via the MHC-I pathway (37-39). Involvement of skeletal muscle cells, therefore, is of utmost importance in initiation and maintenance of CMI responses, as transfected skeletal muscle cells not only may activate antiviral cytotoxic T lymphocytes directly but also can regulate the magnitude and duration of antigen-specific responses by serving as antigen reservoirs when boosted at regular intervals.
Our data confirmed that the Δ4SHIVKU2 DNA fulfilled these requirements in the BALB/c mice. Proof of expression of all of the viral genes was clearly illustrated in cell culture experiments. In mice, it was clear that the vaccine DNA was transferred directly into skeletal muscle cells without the help of any in vivo transfectant, use of a gene gun, or electroporation (29, 34, 40). The localization of viral p24 in infiltrating mononuclear cells in the injected gastrocnemius muscles, in the spleens, and in the draining lymph nodes of the animals suggested strongly that the viral protein produced in muscles was scavenged by macrophages and conveyed to the lymph node and spleen, where induction of immune responses was initiated. This in effect represented cross presentation of the viral antigen. Other studies on the site of injection of vaccine DNA have shown that viral RNA, detectable by reverse transcription-PCR, was present (45). Our study is the first report demonstrating an abundance of viral antigen in skeletal muscle, in macrophages that infiltrated the injected muscles, and in lymphoid tissues of the animals.
We undertook this study to document in vivo events after the inoculation of a DNA vaccine for the generation of immune responses. The kinetics of antigen production in skeletal muscle cells were similar to those observed in in vitro studies. An interesting finding of this study was that the 12-day duration of antigen production in skeletal muscle cells was repeated in mice that were given a second injection of the DNA. This suggested strongly that the DNA itself, rather than viral proteins, could be used as a boost for maintaining antiviral immune responses. Whether the relatively long duration of antigen production after the second injection was mediated by down-regulation of expression of MHC-I in the transfected myocytes is not known. Future studies will examine this question. This direct injection of Δ4SHIVKU2 DNA into muscles for viral antigen production along with cross presentation possibilities will presumably provide a further impetus for the generation and enhancement of the magnitude of antiviral cellular responses in inoculated animals. The results from ELISPOT and antigen-specific proliferation assays, showing a strong immune response that kept increasing in magnitude with time, are testimony to this effect. Apart from these inherent advantages of this DNA construct in viral protein expression and inducement of antiviral CMI, its potency was further enhanced by use of a molecular adjuvant, GM-CSF DNA, with priming and booster injections of vaccine DNA. This resulted in increasing ELISPOT titers with time. Taken together, our data suggest that this DNA construct could be an excellent vaccine candidate for use in humans.
Acknowledgments
The reagents HIV Gag (HXB2) peptide (catalog no. 7872-7904), HIV Env (MN) peptide (catalog no. 6451), consensus HIV Tat (catalog no. 5138), consensus HIV Rev (catalog no. 6445), and consensus HIV Nef (catalog no. 5189) were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This work was supported by Public Health Service grants NS040238, RR006753, AI051220, P20RR016443, GM060792, and P51RR000165.
REFERENCES
- 1.Amara, R. R., J. M. Smith, S. I. Staprans, D. C. Montefiori, F. Villinger, J. D. Altman, S. P. O'Neil, N. L. Kozyr, Y. Xu, L. S. Wyatt, P. L. Earl, J. G. Herndon, J. M. McNicholl, H. M. McClure, B. Moss, and H. L. Robinson. 2002. Critical role for Env as well as Gag-Pol in control of a simian-human immunodeficiency virus 89.6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J. Virol. 76:6138-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2002. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Vaccine 20:1949-1955. [DOI] [PubMed] [Google Scholar]
- 3.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barouch, D. H., T. M. Fu, D. C. Montefiori, M. G. Lewis, J. W. Shiver, and N. L. Letvin. 2001. Vaccine-elicited immune responses prevent clinical AIDS in SHIV(89.6P)-infected rhesus monkeys. Immunol. Lett. 79:57-61. [DOI] [PubMed] [Google Scholar]
- 6.Blagoveshchenskaya, A. D., L. Thomas, S. F. Feliciangeli, C. H. Hung, and G. Thomas. 2002. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 111:853-866. [DOI] [PubMed] [Google Scholar]
- 7.Bojak, A., J. Wild, L. Deml, and R. Wagner. 2002. Impact of codon usage modification on T cell immunogenicity and longevity of HIV-1 gag-specific DNA vaccines. Intervirology 45:275-286. [DOI] [PubMed] [Google Scholar]
- 8.Bojak, A., J. Wild, H. Wolf, and R. Wagner. 2002. Efficiency of a myogenic DNA vaccine is strictly dependent upon cellular localization of HIV-1 Pr55(gag). Vaccine 20:1980-1984. [DOI] [PubMed] [Google Scholar]
- 9.Calarota, S. A., M. Otero, K. Hermanstayne, M. Lewis, M. Rosati, B. K. Felber, G. N. Pavlakis, J. D. Boyer, and D. B. Weiner. 2003. Use of interleukin 15 to enhance interferon-gamma production by antigen-specific stimulated lymphocytes from rhesus macaques. J. Immunol. Methods 279:55-67. [DOI] [PubMed] [Google Scholar]
- 10.Calarota, S. A., and D. B. Weiner. 2004. Enhancement of human immunodeficiency virus type 1-DNA vaccine potency through incorporation of T-helper 1 molecular adjuvants. Immunol. Rev. 199:84-99. [DOI] [PubMed] [Google Scholar]
- 11.Caputo, A., R. Gavioli, and B. Ensoli. 2004. Recent advances in the development of HIV-1 Tat-based vaccines. Curr. HIV Res. 2:357-376. [DOI] [PubMed] [Google Scholar]
- 12.Casimiro, D. R., L. Chen, T. M. Fu, R. K. Evans, M. J. Caulfield, M. E. Davies, A. Tang, M. Chen, L. Huang, V. Harris, D. C. Freed, K. A. Wilson, S. Dubey, D. M. Zhu, D. Nawrocki, H. Mach, R. Troutman, L. Isopi, D. Williams, W. Hurni, Z. Xu, J. G. Smith, S. Wang, X. Liu, L. Guan, R. Long, W. Trigona, G. J. Heidecker, H. C. Perry, N. Persaud, T. J. Toner, Q. Su, X. Liang, R. Youil, M. Chastain, A. J. Bett, D. B. Volkin, E. A. Emini, and J. W. Shiver. 2003. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J. Virol. 77:6305-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cazeaux, N., Y. Bennasser, P. L. Vidal, Z. Li, D. Paulin, and E. Bahraoui. 2002. Comparative study of immune responses induced after immunization with plasmids encoding the HIV-1 Nef protein under the control of the CMV-IE or the muscle-specific desmin promoter. Vaccine 20:3322-3331. [DOI] [PubMed] [Google Scholar]
- 14.Chattergoon, M. A., V. Saulino, J. P. Shames, J. Stein, L. J. Montaner, and D. B. Weiner. 2004. Co-immunization with plasmid IL-12 generates a strong T-cell memory response in mice. Vaccine 22:1744-1750. [DOI] [PubMed] [Google Scholar]
- 15.Deml, L., A. Bojak, S. Steck, M. Graf, J. Wild, R. Schirmbeck, H. Wolf, and R. Wagner. 2001. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J. Virol. 75:10991-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Earl, P. L., L. S. Wyatt, D. C. Montefiori, M. Bilska, R. Woodward, P. D. Markham, J. D. Malley, T. U. Vogel, T. M. Allen, D. I. Watkins, N. Miller, and B. Moss. 2002. Comparison of vaccine strategies using recombinant env-gag-pol MVA with or without an oligomeric Env protein boost in the SHIV rhesus macaque model. Virology 294:270-281. [DOI] [PubMed] [Google Scholar]
- 17.Ensoli, B., and A. Cafaro. 2000. Control of viral replication and disease onset in cynomolgus monkeys by HIV-1 TAT vaccine. J. Biol. Regul. Homeost. Agents 14:22-26. [PubMed] [Google Scholar]
- 18.Gao, F., Y. Li, J. M. Decker, F. W. Peyerl, F. Bibollet-Ruche, C. M. Rodenburg, Y. Chen, D. R. Shaw, S. Allen, R. Musonda, G. M. Shaw, A. J. Zajac, N. Letvin, and B. H. Hahn. 2003. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res. Hum. Retroviruses 19:817-823. [DOI] [PubMed] [Google Scholar]
- 19.Gao, F., E. A. Weaver, Z. Lu, Y. Li, H. X. Liao, B. Ma, S. M. Alam, R. M. Scearce, L. L. Sutherland, J. S. Yu, J. M. Decker, G. M. Shaw, D. C. Montefiori, B. T. Korber, B. H. Hahn, and B. F. Haynes. 2005. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J. Virol. 79:1154-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giri, M., K. E. Ugen, and D. B. Weiner. 2004. DNA vaccines against human immunodeficiency virus type 1 in the past decade. Clin. Microbiol. Rev. 17:370-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hel, Z., J. M. Johnson, E. Tryniszewska, W. P. Tsai, R. Harrod, J. Fullen, J. Tartaglia, and G. Franchini. 2002. A novel chimeric Rev, Tat, and Nef (Retanef) antigen as a component of an SIV/HIV vaccine. Vaccine 20:3171-3186. [DOI] [PubMed] [Google Scholar]
- 22.Hel, Z., E. Tryniszewska, W. P. Tsai, J. M. Johnson, R. Harrod, J. Fullen, V. S. Kalyanaraman, J. D. Altman, J. McNally, T. Karpova, B. K. Felber, J. Tartaglia, and G. Franchini. 2002. Design and in vivo immunogenicity of a polyvalent vaccine based on SIVmac regulatory genes. DNA Cell Biol. 21:619-626. [DOI] [PubMed] [Google Scholar]
- 23.Joag, S. V., Z. Q. Liu, E. B. Stephens, M. S. Smith, A. Kumar, Z. Li, C. Wang, D. Sheffer, F. Jia, L. Foresman, I. Adany, J. Lifson, H. M. McClure, and O. Narayan. 1998. Oral immunization of macaques with attenuated vaccine virus induces protection against vaginally transmitted AIDS. J. Virol. 72:9069-9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostense, S., W. Koudstaal, M. Sprangers, G. J. Weverling, G. Penders, N. Helmus, R. Vogels, M. Bakker, B. Berkhout, M. Havenga, and J. Goudsmit. 2004. Adenovirus types 5 and 35 seroprevalence in AIDS risk groups supports type 35 as a vaccine vector. AIDS 18:1213-1216. [DOI] [PubMed] [Google Scholar]
- 25.Letvin, N. L., Y. Huang, B. K. Chakrabarti, L. Xu, M. S. Seaman, K. Beaudry, B. Korioth-Schmitz, F. Yu, D. Rohne, K. L. Martin, A. Miura, W. P. Kong, Z. Y. Yang, R. S. Gelman, O. G. Golubeva, D. C. Montefiori, J. R. Mascola, and G. J. Nabel. 2004. Heterologous envelope immunogens contribute to AIDS vaccine protection in rhesus monkeys. J. Virol. 78:7490-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letvin, N. L., D. C. Montefiori, Y. Yasutomi, H. C. Perry, M. E. Davies, C. Lekutis, M. Alroy, D. C. Freed, C. I. Lord, L. K. Handt, M. A. Liu, and J. W. Shiver. 1997. Potent, protective anti-HIV immune responses generated by bimodal HIV envelope DNA plus protein vaccination. Proc. Natl. Acad. Sci. USA 94:9378-9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacGregor, R. R., R. Ginsberg, K. E. Ugen, Y. Baine, C. U. Kang, X. M. Tu, T. Higgins, D. B. Weiner, and J. D. Boyer. 2002. T-cell responses induced in normal volunteers immunized with a DNA-based vaccine containing HIV-1 env and rev. AIDS 16:2137-2143. [DOI] [PubMed] [Google Scholar]
- 28.McKay, P. F., D. H. Barouch, S. Santra, S. M. Sumida, S. S. Jackson, D. A. Gorgone, M. A. Lifton, and N. L. Letvin. 2004. Recruitment of different subsets of antigen-presenting cells selectively modulates DNA vaccine-elicited CD4+ and CD8+ T lymphocyte responses. Eur. J. Immunol. 34:1011-1020. [DOI] [PubMed] [Google Scholar]
- 29.Otten, G., M. Schaefer, C. Greer, M. Calderon-Cacia, D. Coit, J. Kazzaz, A. Medina-Selby, M. Selby, M. Singh, M. Ugozzoli, M. J. zur, S. W. Barnett, D. O'Hagan, J. Donnelly, and J. Ulmer. 2003. Induction of broad and potent anti-human immunodeficiency virus immune responses in rhesus macaques by priming with a DNA vaccine and boosting with protein-adsorbed polylactide coglycolide microparticles. J. Virol. 77:6087-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quaranta, M. G., B. Mattioli, L. Giordani, and M. Viora. 2004. HIV-1 Nef equips dendritic cells to reduce survival and function of CD8+ T cells: a mechanism of immune evasion. FASEB J. 18:1459-1461. [DOI] [PubMed] [Google Scholar]
- 31.Robinson, H. L., D. C. Montefiori, R. P. Johnson, M. L. Kalish, S. L. Lydy, and H. M. McClure. 2000. DNA priming and recombinant pox virus boosters for an AIDS vaccine. Dev. Biol. (Basel) 104:93-100. [PubMed] [Google Scholar]
- 32.Schneider, R., M. Campbell, G. Nasioulas, B. K. Felber, and G. N. Pavlakis. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71:4892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz, O., V. Marechal, S. Le Gall, F. Lemonnier, and J. M. Heard. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338-342. [DOI] [PubMed] [Google Scholar]
- 34.Selby, M., C. Goldbeck, T. Pertile, R. Walsh, and J. Ulmer. 2000. Enhancement of DNA vaccine potency by electroporation in vivo. J. Biotechnol. 83:147-152. [DOI] [PubMed] [Google Scholar]
- 35.Singh, D. K., Z. Liu, D. Sheffer, G. A. Mackay, M. Smith, S. Dhillon, R. Hegde, F. Jia, I. Adany, and O. Narayan. 2005. A noninfectious simian/human immunodeficiency virus DNA vaccine that protects macaques against AIDS. J. Virol. 79:3419-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sumida, S. M., P. F. McKay, D. M. Truitt, M. G. Kishko, J. C. Arthur, M. S. Seaman, S. S. Jackson, D. A. Gorgone, M. A. Lifton, N. L. Letvin, and D. H. Barouch. 2004. Recruitment and expansion of dendritic cells in vivo potentiate the immunogenicity of plasmid DNA vaccines. J. Clin. Investig. 114:1334-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, D. C., M. DeVit, and S. A. Johnston. 1992. Genetic immunization is a simple method for eliciting an immune response. Nature 356:152-154. [DOI] [PubMed] [Google Scholar]
- 38.Ulmer, J. B., R. R. Deck, C. M. Dewitt, J. I. Donnhly, and M. A. Liu. 1996. Generation of MHC class I-restricted cytotoxic T lymphocytes by expression of a viral protein in muscle cells: antigen presentation by non-muscle cells. Immunology 89:59-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulmer, J. B., and G. R. Otten. 2000. Priming of CTL responses by DNA vaccines: direct transfection of antigen presenting cells versus cross-priming. Dev. Biol. (Basel) 104:9-14. [PubMed] [Google Scholar]
- 40.Widera, G., M. Austin, D. Rabussay, C. Goldbeck, S. W. Barnett, M. Chen, L. Leung, G. R. Otten, K. Thudium, M. J. Selby, and J. B. Ulmer. 2000. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J. Immunol. 164:4635-4640. [DOI] [PubMed] [Google Scholar]
- 41.Williams, M., J. F. Roeth, M. R. Kasper, R. I. Fleis, C. G. Przybycin, and K. L. Collins. 2002. Direct binding of human immunodeficiency virus type 1 Nef to the major histocompatibility complex class I (MHC-I) cytoplasmic tail disrupts MHC-I trafficking. J. Virol. 76:12173-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolff, J. A., R. W. Malone, P. Williams, W. Chong, G. Acsadi, A. Jani, and P. L. Felgner. 1990. Direct gene transfer into mouse muscle in vivo. Science 247:1465-1468. [DOI] [PubMed] [Google Scholar]
- 43.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 44.zur Megede, J., M. C. Chen, B. Doe, M. Schaefer, C. E. Greer, M. Selby, G. R. Otten, and S. W. Barnett. 2000. Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J. Virol. 74:2628-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.zur Megede, J., G. R. Otten, B. Doe, H. Liu, L. Leung, J. B. Ulmer, J. J. Donnelly, and S. W. Barnett. 2003. Expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 subtype B Pol and GagPol DNA vaccines. J. Virol. 77:6197-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]