Abstract
Infection with wild-type adeno-associated virus (AAV) is common in humans, but very little is known about the in vivo biology of AAV. On a molecular level, it has been shown in cultured cells that AAV integrates in a site-specific manner on human chromosome 19, but this has never been demonstrated directly in infected human tissues. To that end, we tested 175 tissue samples for the presence of AAV DNA, and when present, examined the specific form of the viral DNA. AAV was detected in 7 of 101 tonsil-adenoid samples and in 2 of 74 other tissue samples (spleen and lung). In these nine samples, we were unable to detect AAV integration in the AAVS1 locus using a sensitive PCR assay designed to amplify specific viral-cellular DNA junctions. Additionally, we used a second complementary assay, linear amplification-mediated-PCR (LAM-PCR) to widen our search for integration events. Analysis of individual LAM-PCR products revealed that the AAV genomes were arranged predominantly in a head-to-tail array, with deletions and extensive rearrangements in the inverted terminal repeat sequences. A single AAV-cellular junction was identified from a tonsil sample and it mapped to a highly repetitive satellite DNA element on chromosome 1. Given these data, we entertained the possibility that instead of integrated forms, AAV genomes were present as extrachromosomal forms. We used a novel amplification assay (linear rolling-circle amplification) to show that the majority of wild-type AAV DNA existed as circular double-stranded episomes in our tissues. Thus, following naturally acquired infection, AAV DNA can persist mainly as circular episomes in human tissues. These findings are consistent with the circular episomal forms of recombinant AAV vectors that have been isolated and characterized from in vivo transduced tissues.
Adeno-associated viruses (AAVs) are ubiquitous, noncytopathic, replication-incompetent members of the Parvoviridae family. AAV replication requires the presence of a helper virus, and this is usually one of the many serotypes of adenovirus. The epidemiology of AAV infection in humans was extensively studied after its initial description some 40 years ago (2, 3, 5, 19, 37). Two major conclusions were drawn from this work. First, many adults have antibodies reactive against one or more AAV serotypes (7, 15, 45), a finding which is entirely consistent with early and repeated exposures to AAV and adenoviruses throughout life. Second, even with this level of exposure, AAV does not cause any disease or other pathological condition in humans.
As noted above, AAV genomes are replicated and packaged into new infectious particles only in the presence of a helper virus. In the absence of helper virus, AAV is unique among viruses in its ability to direct site-specific integration of its genome into a specific locus (AAVS1) on human chromosome 19 (22, 23, 39). A similar locus has also been identified in nonhuman primates, and recently in rodents (14). Site specificity is mediated by virally encoded rep gene products via recognition and binding to similar viral and cellular sequences. Such sequence-specific interaction ultimately results in the insertion of head-to-tail proviral AAV DNA arrays that are characterized by rearrangement of viral inverted terminal repeat (ITR) and flanking cellular sequences (24, 33, 36, 50). The AAV DNA is harbored in this “latent” state until subsequent infection with a helper virus causes reactivation or “rescue” of the AAV genome, resulting in renewed viral replication and production of infectious particles.
To date, this unique property of site-specific integration has only been documented in transformed cultured cells and has never been demonstrated in tissues taken directly from humans. In fact, surprisingly little is known about the molecular events of AAV infection in vivo, either in humans or in permissive animal models. Only two studies have attempted to examine experimental wild-type AAV infection in nonhuman primates (1, 18). The first study established parameters for AAV infection and did not specifically address molecular characterization of the viral genome (1). In the second study, rhesus macaques were inoculated with wild-type AAV in the presence or absence of wild-type adenovirus (18). AAV DNA was found most readily in peripheral blood mononuclear cells in a subset of animals. Site-specific integration into the AAVS1 locus was apparently detected in a single animal using PCR amplification and dot blot hybridization. No attempt to confirm the site of integration or the molecular structure was reported.
Because of the interest in recombinant DNA delivery vectors based on wild-type AAV, a more thorough understanding the basic biology of wild-type AAV infection seems warranted. To that end, we have recently analyzed tissues from 175 children for the presence of AAV and adenoviral DNA (5a). In that study, we found nine samples that harbored AAV sequences. Seven of the nine were from tonsil-adenoid tissues removed at surgery, while the remaining two positives (lung and spleen) were normal samples retrieved from a tissue bank.
In the present study, we undertook evaluation of the molecular forms of the AAV DNA is these tissues using three distinct techniques. In contrast to the prevailing hypothesis, we were unable to identify wild-type AAV integration at the AAVS1 locus using a sensitive S1-specific PCR assay. When we broadened our search for viral-host sequence junctions using a modified linear amplification-mediated PCR assay (LAM-PCR), we found a single integration event on chromosome 1, but again were unable to find any AAVS1 insertions. Finally, using a novel linear rolling-circle amplification assay, we showed that the majority of the AAV genome sequences in these tissues were extrachromosomal circles.
MATERIALS AND METHODS
Cell propagation.
HeLa cells and Detroit 6 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin and streptomycin. 293 human embryonic kidney cells were grown in Eagle's minimal essential medium supplemented with 10% fetal bovine serum and penicillin and streptomycin. HeLa and 293 cells were purchased from the American Type Culture Collection (Rockville, MD) and Detroit 6 cells were obtained from R. Jude Samulski.
Human tissues and DNA isolation.
All tissue samples were acquired after approval from the Columbus Children's Hospital Institutional Review Board, and where required, informed consent was obtained. Fresh human tonsil and adenoid specimens (n = 101) were collected from children aged 2 to 13 years undergoing elective surgical excision at Columbus Children's Hospital. Additional human tissues (n = 74) representing normal liver, spleen, muscle, heart, and lung were obtained from the Cooperative Human Tissue Network at Columbus Children's Hospital. The ages of the individual subjects ranged from 0 to 30 years. Subjects under 6 months of age (n = 49) were analyzed but were considered unlikely to have been exposed to adenovirus and AAV infection. In addition, for 3 of the 74 samples, the age of the subjects could not be determined. Thus, there were 22 samples from individuals aged 6 months to 30 years available for analyses, and 6 of the 22 were from individuals older than 14 years. Samples were stored frozen at −80°C until DNA isolation.
Freshly thawed tissue (0.2 to 0.5 g) was digested for 15 h in 3 ml of digestion buffer (10 mM Tris, pH 8.0; 100 mM NaCl; 0.5% sodium dodecyl sulfate; 25 mM EDTA) supplemented with 2 mg/ml proteinase K at 55°C with constant agitation. DNA was subjected to three phenol-chloroform-isoamyl alcohol (25:24:1) extractions. A final chloroform extraction was performed, followed by DNA ethanol precipitation and resuspension in 10 mM Tris, pH 8.0.
Quantitative PCR and DNA hybridization.
AAV genome copy number in tissue samples was quantified using real-time TaqMan PCR analysis (ABI 7700, PE Applied BioSystems). The primers and probe set were selected following alignment of 255-bp cap DNA sequences ForCAPSS (bp 3004 to 3024): 5′-AACGACAACCACTACTTTGGC-3′ (50 nM); RevCAPSS (bp 3074 to 3054): 5′-AAGTGGCAGTGGAATCTGTTG-3′ (900 nM); probe (bp 3024 to 3050), [6-FAM]5′-CTACAGCACCCCCTGGGGGTATTTTGA-3′[6-carboxyfluorescein (FAM)-tetramethylrhodamine (TAMRA)-FAM] (270 nM). PCR conditions were: 50°C 2 min, 95°C 10 min, 40 cycles of 95°C 15 s, and 60°C 1 min using 250ng of human total cellular DNA in 1X TaqMan PCR master mix.
For hybridization analyses, DNA was fractionated on 0.8% agarose gels, dehydrated and subjected to in-gel hybridization or transferred to a nylon membrane using a vacuum manifold dot blot apparatus. DNA hybridization conditions were 65°C for 16 h in buffer containing 6× (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) SSC, 1× Denhardt's reagent, and 200 μg/ml sonicated herring sperm DNA. Rehydrated gels and nylon membranes were washed twice at 60°C in 2× SSC, 0.2% sodium dodecyl sulfate for 30 min, and then twice at 60°C in 0.2× SSC, 0.2% sodium dodecyl sulfate for 30 min.
AAVS1-PCR.
To detect AAV integration in the AAVS1 locus, nested PCR was performed using 1 μg of total cellular DNA as template with primers specific for the human AAVS1 locus and AAV rep or cap genes (20, 43). The reaction conditions for first-round PCR were as follows: 25 pmol of each primer, 200 mM deoxynucleotide triphosphates, 5 U Herculase Hotstart DNA polymerase (Stratagene) with Herculase reaction buffer. The AAV-specific primers used in first-round PCR were as follows: cap; CAPGSP1 (bp 4320 to 4349), 5′-GTCTGTTAATGTGGACTTTACTGTGGACAC-3′, or rep (bp 479 to 455); REPGSP1, 5′-CAGGGGTGCCTGCTCAATCAGATTC-3′. The AAVS1-specific primer was AAVS1-1R, 5′-ATGGCTCCAGGAAATGGGGGTGTG-3′ (43). The PCR cycling conditions were as follows: denaturation at 95°C for 90 s, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 6 min, with a final extension of 72°C for 10 min.
For nested PCR, 1 μl of the first-round PCR was used as template with the same reaction conditions as noted for the first-round PCR using the following primers: AAV-specific primers, cap (bp 4357 to 4379); CAPGSP2, 5′-GTGTATTCAGAGCCTCGCCCCAT-3′, rep (bp 404 to 427); REPGSP2, 5′-TCCCATTCCTTCTCGGCCACCCAG-3′, and AAVS1-specific primer AAVS1-2R; 5′-CACCAGATAAGGAATCTGCC-3′ (43). To verify the integrity of the AAVS1 locus and our ability to amplify AAVS1 sequences, we performed control PCRs using specific primers spanning 2.2 kb of the AAVS1 locus that included the Rep binding site. The control primers were: AAVS1-F4, 5′-GATTTCCACTGGGCGCCT-3′ and AAVS1-1R (above). PCR products were cloned into a TOPO-TA cloning vector (Invitrogen Corp.) and sequenced using an ABI 727 capillary electrophoresis automatic sequencer (PE Applied BioSystems) by the Columbus Children's Research Institute Sequencing Core Laboratory.
LAM-PCR.
Linear amplification-mediated (LAM)-PCR was used to isolate sequences contiguous with AAV genomes in samples of total cellular DNA representing a variety of human tissues. The AAV sequence was first amplified from 1 μg of genomic DNA using with a single, biotinylated primer corresponding to either the 5′ region of rep (bp 545 to 519); REPBIO, 5′[BioTEG]-CTCCGGGGCCTTACTCACRCGGCGCCA-3′, or the 3′ region of cap (bp 4257 to 4286); CAPBIO, 5′-[BioTEG]GCTGCAGAARGARAACAGCAAACGCTGGAA-3′. For the internal human erythropoietin (epo) control, the HepoBIO primer was used: 5′-[BioTEG]-TGGTTTCAGTTCTTGTCAATGAGGTTG-3′. The reaction conditions were as follows: 0.25 pmol of primer, 200mM deoxynucleotide triphosphates, 5 U Herculase Hotstart DNA polymerase (Stratagene Corp.) with the Herculase reaction buffer, and overlaid with mineral oil. The cycling conditions were as follows: denaturation at 95°C for 90 s, followed by 50 cycles of 95°C for 30 s, 65°C for 30 s, and 72°C for 6 min, with a final extension of 72°C for 10 min. This cycle was repeated with the addition of 5 U of fresh Herculase Hotstart DNA polymerase for a total of 100 cycles of linear amplification.
The biotinylated PCR products were captured using 200 μg of streptavidin-coated, magnetic beads (Dynabeads kilobase Binder kit, Dynal Inc.) in conjunction with a magnetic particle concentrator (Dynal) according to the manufacturer's protocol. After binding, the beads were washed twice with 100 μl of 10 mM Tris-HCl (pH 8.0) and incubated with 2 U of Klenow (Roche), 300 mM deoxynucleotide triphosphates (Invitrogen Corp.) and random hexanucleotide mix (Roche Applied Science) in a 20-μl reaction volume for 1 h at 37°C to create double-stranded DNA. Following incubation, the DNA-beads were washed twice as described above and incubated for 2 h at 37°C with 5 U of a blunt-cutting restriction endonuclease (EcoRV, PvuII, or StuI: New England Biolabs) and 5 U of a restriction enzyme that generates a 3′ overhang end and cuts once within the AAV genome (SalI). After restriction enzyme digestion, the beads were washed as above, and a blunt-end, double-stranded DNA adaptor (100 pmol of the GenomeWalker Adaptor, Universal GenomeWalker Kit, Clontech) was ligated to the restricted DNA in a 10-μl volume containing 20 U of T4 DNA ligase and the accompanying ligation buffer (New England Biolabs) at 16°C for 16 h.
The DNA-beads were washed as described above and used as the template in a nested PCR. The first round of PCR used 25 pmol of each primer, 200 mM deoxynucleotide triphosphates, 5 U of Herculase Hotstart DNA polymerase (Stratagene) with the Herculase reaction buffer using the same cycling conditions as above with the exception that only 30 cycles were performed. The primer specific to the adaptor (AP1, 5′-GTAATACGACTCACTATAGGGC-3′) was supplied in the Universal Genome Walker kit. The AAV-specific primers used in first-round PCR were as follows: cap, CAPGSP1, 5′-GTCTGTTAATGTGGACTTTACTGTGGACAC-3′; or rep, REPGSP1, 5′-CAGGGGTGCCTGCTCAATCAGATTC-3′. The epo control primer used was HepoGSP1, 5′-GACCCCATGAGAGCCAGAGGCCAG-3′. For nested PCR, 1 μl of first-round PCR was used as the template with the same reaction conditions as for first-round PCR with the following primers: adaptor primer-AP2, 5′-ACTATAGGGCACGCGTGGT-3′; cap, CAPGSP2, 5′-GTGTATTCAGAGCCTCGCCCCAT-3′; rep, REPGSP2, 5′-TCCCATTCCTTCTCGGCCACCCAG-3′; epo, HepoGSP2, 5′-CATCCTGTCTTCATGGTTCCCAC-3′. PCR products were cloned into the TOPO-TA cloning vector (Invitrogen Corp.) for sequence analysis.
Linear rolling-circle amplification.
To prepare genomic DNA samples for linear rolling-circle amplification, 1 μg of genomic DNA was first digested in a 15-μl volume with a restriction enzyme that does not cut within the AAV genome (SpeI for human tissue samples; EcoRV for Detroit 6 and plasmid spike samples). The resulting linearized DNA was digested by incubation with 10 U of Plasmid-Safe ATP-Dependent DNase (Epicenter Technologies) in a 25-μl final volume for 16 h at 37°C in 33 mM Tris (pH 7.8), 66 mM potassium acetate, 10mM magnesium acetate, 0.5 mM dithiothreitol, and 2 mM ATP. The nuclease was then heat inactivated for 30 min at 70°C. The resulting template DNA (2.5μl, equivalent to 100 ng) was mixed in a final volume of 15 μl in 10 mM Tris HCl, pH 8.0, with 300 pmol each of two AAV cap-specific primers: temp1 (bp 2882 to 2898), 5′-ATTGGCATTGCGATTCC-3′; and temp2 (bp 2926 to 2910), 5′-TGGTGATGACTCTGTCG-3′. Each primer contained a phosphothiorate linkage between the last 3′ base to increase primer stability. The reaction was heated to 95°C for 3 min, then cooled slowly to 4°C to allow primer annealing, and mixed with 15 μl of φ29 DNA phage polymerase (TempliPhi, Amersham Biosciences) reaction buffer containing φ29 polymerase, and incubated for 18 h at 30°C. Amplified products were heat inactivated at 65°C for 10 min and then digested with EcoRI for subsequent dot blot hybridization.
RESULTS
Identification of AAV sequences in human tissues.
As part of a study of the molecular epidemiology of AAV infection in children (5a), we tested 175 tissues for the presence of AAV DNA by PCR. From tonsil-adenoid tissues, we identified 7 AAV positive samples (designated as T17, T32, T40, T41, T70, T71, and T88). Samples T17 and T32 contained both AAV and adenovirus sequences. Two (lung and spleen, LG15 and S17, respectively) of 74 additional tissues were also AAV positive. From these nine AAV positive samples, entire AAV capsid gene sequences were generated and analyzed. Sequence analysis revealed that eight of the nine isolates were closely related to AAV2, while the other isolate shared significant homology to the recently described AAV2/3 hybrid clade (16).
Molecular characterization of AAV DNA in tissues.
Having identified AAV DNA sequences within these human tissues, we turned our attention to characterization of the molecular form of the DNA within the cell. Prevailing wisdom suggested that AAV DNA would be found integrated within the AAVS1 locus on human chromosome 19 (23, 39). As a first step, we determined (using quantitative PCR) the AAV genome copy number in tissues; values ranged from 80 to 33,000 copies/μg of total DNA. Given these values, it was not surprising that attempts to characterize AAV DNA within total cellular DNA by standard restriction enzyme digestion and Southern blot analysis were unsuccessful (data not shown). This led us to develop alternative assays to delineate the predominant viral form(s) present within these human tissues.
Analysis of integration into the AAVS1 locus.
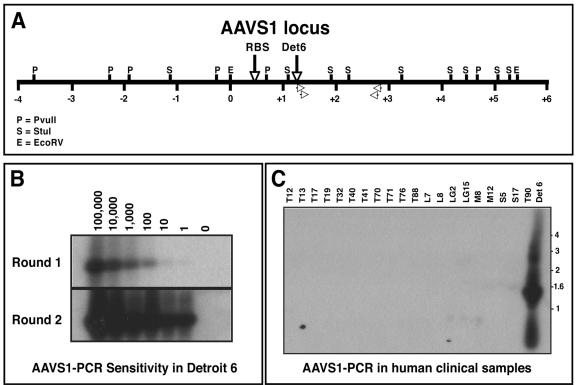
To directly examine AAV DNA insertion into the AAVS1 locus, we developed a set of nested PCR primers to amplify AAV/AAVS1 viral-cellular junctions. For a positive control, we used the well-characterized Detroit 6 cell line in which head-to-tail tandem integration of AAV2 into the AAVS1 locus had been described (Fig. 1A) (21). Quantitative PCR revealed that Detroit 6 cells contained five viral genome copies per cell, and this value was used to define the sensitivity of the assay. As seen in Fig. 1B, we readily detected a single cell equivalent (five AAV copies) as evidenced by the amplification of the expected 1.5-kb PCR product. Importantly, identical assays of the tissue DNA samples failed to yield a PCR product (Fig. 1C). DNA integrity was validated using a control AAVS1 PCR amplification and all samples amplified the expected 2.2-kb fragment (data not shown).
FIG. 1.
AAVS1 PCR on human samples. (A) Restriction map schematic of the human AAVS1 locus. The Rep binding site (RBS) and Detroit 6 (Det6) integration site are designated by vertical arrows. The location of the AAVS1-specific primers used in the nested PCRs are also shown (open arrowheads). AAVS1 locus numbering follows the convention of Kotin et al. (21), whereby the EcoRI site defining base pair 0 is denoted (positive integers to the right, negative integers to the left). (B) Southern blot hybridization analysis of AAVS1-PCR products using Detroit 6 cellular DNA. Decreasing copies of Detroit 6 genomes (100,000 to 1) were spiked into 1 μg of naïve human genomic DNA and subjected to nested AAVS1 PCR. The PCR products were fractionated on an agarose gel and Southern hybridization was performed using an AAVS1-specific probe. The expected 1.5-kb AAV-AAVS1 junction fragment is readily detected by nested PCR using a single Detroit 6 cell junction equivalent. (C) Southern blot AAVS1-PCR on human clinical samples. AAVS1-PCR was performed on 1 μg of genomic DNA isolated from various human clinical samples. Southern blot hybridization using an AAVS1-specific probe failed to detect any signal (except in Detroit 6 control DNA).
LAM-PCR analysis of AAV integration.
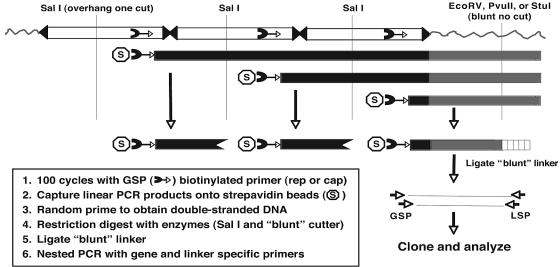
Our failure to detect AAVS1 integration by direct PCR led us to consider the possibility that AAV genomes were either (i) randomly integrated elsewhere into the host genome or (ii) not integrated. To address these possibilities, we adapted a linear amplification mediated PCR (LAM-PCR) method that was previously used to map retroviral vector integration sites (40). The assay (outlined in Fig. 2) used a single AAV-specific, biotinylated primer to amplify linear fragments (100 cycles) that extended into unknown flanking sequences. The linear DNA fragments were captured on streptavidin beads, converted to double-stranded DNA, and digested with restriction enzymes to create a blunt DNA end in the flanking sequence. A blunt-ended linker was then ligated to the free end that generated an anchoring point for nested PCR primers.
FIG. 2.
Schematic of linear amplification mediated PCR (LAM-PCR) to detect random AAV integrants. To isolate DNA sequences flanking integrated wild-type AAV genomes, linear PCR (100 cycles) was performed on total cellular DNA (1 μg) using an AAV-specific biotinylated primer homologous to a conserved region in the cap or rep gene (cap is shown). PCR products were captured on streptavidin beads and converted to double-stranded DNA by random hexanucleotide priming. The double-stranded DNA was then digested with a blunt-cutting restriction enzyme (EcoRV, PvuII, or StuI) to generate a substrate for ligation of a blunt linker (vertical box) to the end of the digested DNA. To reduce the level of competing internal AAV-AAV junctions, double stranded fragments were also digested with SalI, which created an incompatible end for ligation of the blunt adaptor. The resulting DNA fragments were subjected to two rounds of PCR using AAV and linker-specific primers. PCR products were analyzed by Southern hybridization and cloned into an appropriate PCR vector for subsequent analysis.
Nested PCR was performed using primers specific for AAV and linker sequences, and products were cloned and sequenced. To validate the LAM-PCR assay, we mapped the known insertion site of AAV2 in Detroit 6 cells (Fig. 1A) using a variety of blunt-cutting restriction enzymes (StuI, PvuII, and EcoRV). Regardless of the blunt cutting restriction enzyme employed, clones possessing identical viral/cellular junctions were obtained, which matched the published insertion site (21). LAM-PCR sensitivity was determined by preparing 10-fold serial dilutions of Detroit 6 DNA (ranging from 1.6 to 160,000 copies of AAV-cellular junctions) in a background of 1 μg of genomic DNA isolated from a naïve human tonsil. We were able to amplify as few as 160 AAV-cellular junctions using either a cap (Fig. 3) or rep biotinylated primer (data not shown). As an internal control for DNA integrity and purity, a human erythropoietin (epo) gene-specific LAM-PCR primer was designed and yielded the expected 0.5-kb epo LAM-PCR product (when used with the linker specific primer) in all DNA samples assayed (Fig. 3, bottom panel).
FIG. 3.
LAM-PCR validation and sensitivity using Detroit 6 cells. Southern blot hybridization was performed on LAM-PCR products to determine assay sensitivity. Naïve human total cellular DNA (1 μg), containing various spiked copies of Detroit 6 cell DNA, was used as the template for LAM-PCR using an AAV cap-specific primer and a ligated blunt linker. Positive Southern hybridization (using an AAVS1 probe) detected amplification of as few as 160 Detroit 6 viral-cellular junctions. Shown in the bottom panel is an ethidium bromide stained gel of a human erythropoietin (cap) LAM-PCR genomic fragment that was generated using the same DNA templates used for the AAV LAM-PCR analysis. All samples amplified the expected 500-bp cap genomic fragment following LAM-PCR with an cap gene-specific primer (see Materials and Methods), confirming the DNA integrity of the sample and the reproducibility of the LAM-PCR assay.
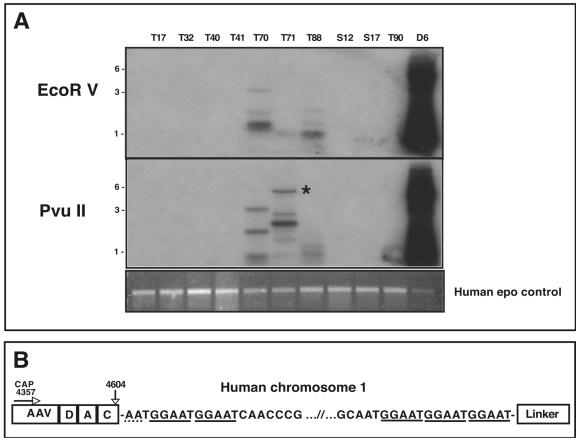
LAM-PCR was performed on the nine AAV positive human samples using both cap and rep primer sets, with an array of blunt cutting restriction enzymes (StuI, PvuII, and EcoRV) to maximize the chances of obtaining a blunt-cutting restriction site in close proximity to an integration site. Representative Southern blots of the resulting AAV specific LAM-PCR products (cap primer) are shown in Fig. 4A. Multiple hybridizing PCR products of various sizes (0.5 to 5.5 kb) were present in tissues T70, T71, and T88. As expected, the Detroit 6 template DNA also yielded fragments of the predicted size based on the specific blunt-cutting restriction enzyme employed (see Fig. 1A for restriction map). Moreover, the internal human erythropoietin control LAM-PCR primer amplified the expected DNA fragment, verifying DNA template quality (Fig. 4A).
FIG. 4.
LAM-PCR of positive human clinical samples. (A) Southern blot of LAM-PCR using AAV-cap primers. LAM-PCR was performed using 1 μg of total cellular DNA with either AAV rep or cap primer (cap is shown). The top panel shows LAM-PCR using EcoRV as the blunt-cutting restriction enzyme, while the middle panel shows LAM-PCR using PvuII. The DNA was hybridized with an AAV-specific probe. Detroit 6 DNA (D6) was used as a positive control and gave the predicted size based on the restriction analysis of the AAVS1 locus (see Fig. 1A). Positive hybridization of various sizes can clearly be seen in samples T70, T71, and T88. Analysis of the LAM-PCR products revealed that an approximate 5.5-kb band in sample T71 contained an AAV-cellular junction (starred). The bottom panel shows an ethidium bromide stained gel of the LAM-PCR products using a human epo gene-specific primer as an internal control. All sample amplified the expected 500-bp genomic fragment, confirming the DNA integrity of the samples and the reproducibility of the LAM-PCR assay. (B) Schematic of AAV-cellular junction from tissue T71. The 3′ AAV sequence is represented on the left ranging from bp 4357 (the position of the cap LAM primer) to the junction at bp 4604 in the AAV ITR. The AAV sequence in the LAM-PCR product contained the 3′ cap sequence as well as a complete D, A, and C ITR region. There appears to be only 2 bp of homology between the breakpoint of the AAV ITR and the chromosomal sequence (dotted underline). The LAM-PCR product contained approximately 5 kb of human chromosomal DNA that mapped to position 1q31.1, composed primarily of a (GGAAT)n repeat sequence (solid underline).
We successfully isolated 44 AAV-specific LAM-PCR products from six of the nine positive human samples (T17, T32, T40, T70, T71, and T88), and subsequently cloned and sequenced the fragments. From these clones, a single AAV integration event was documented within a 5.5-kb PvuII clone from tonsil sample T71 (Fig. 4). This clone contained 250 bp of AAV DNA followed by approximately 5 kb of chromosome 1 sequence that ended with the expected LAM-PCR blunt linker. The cellular sequence mapped to position 1q31.1 and is composed primarily of a (GGAAT)n pentameric repeat sequence (Fig. 4B). The viral junction breakpoint consisted of a single 3′ AAV ITR that contained a complete D, A, and C region, with the remainder of the ITR deleted (Fig. 4B). There were two base pairs (AA) of microhomology between the breakpoint of the AAV ITR and the cellular sequence.
The remaining LAM-PCR products (n = 43) consisted of either partial AAV sequence or AAV-AAV junctions separated by various amounts of intervening viral ITR sequence (summarized in Table 1). The fine structure of several AAV-AAV junction clones (isolated from T70) are shown in Fig. 5, the majority of which contain large ITR deletions. However, several clones (T70-11, T70-12, and T70-C1) appeared to contain a more fully intact ITR sequence, characterized by the presence of a double-D sequence structure (49). We were unable to obtain complete DNA sequence beyond the D regions, which was likely due to the extreme secondary structure present within the ITR cruciform.
TABLE 1.
Summary of LAM-PCR-positive clones isolated from clinical samples
| Tissuea | AAV-cellular junction | AAV fragmentb | Head-to-tail junction | Head-to-head junction | Tail-to-tail junction | Total isolates |
|---|---|---|---|---|---|---|
| T17 | 0 | 3 | 0 | 0 | 0 | 3 |
| T32 | 0 | 2 | 1 | 0 | 0 | 3 |
| T40 | 0 | 5 | 0 | 0 | 0 | 5 |
| T70 | 0 | 5 | 12 | 1 | 1 | 19 |
| T71 | 1 | 4 | 0 | 0 | 0 | 5 |
| T88 | 0 | 4 | 2 | 1 | 2 | 9 |
| Total | 1 | 23 | 15 | 2 | 3 | 44 |
Three of nine positive samples (T41, S17, and LG15) did not give rise to any detectable LAM-PCR product.
LAM-PCR products contained AAV sequence corresponding to a portion of a single AAV genome.
FIG. 5.
Schematic of AAV-AAV junctions isolated following LAM-PCR. A complete head-to-tail AAV ITR junction is shown at the top of the figure, along with the positions of the cap and rep primers used. Shown below are various AAV-AAV junctions isolated from tissue T70. The majority of junctions analyzed were in a head-to-tail orientation; however, tail-to-tail (T70-14) and head-to-head (T70-E9) orientations were observed. The breakpoints for each junction are given, with deleted sequence designated using a solid line. For several clones (T70-C1, T70-11, and T70-12), we were unable to obtain readable sequence after the D region from either side (designated as dotted lines), which was presumably due to strong secondary structure associated with the ITR structure.
Linear rolling-circle amplification of episomal DNA.
Given that two complementary methods failed to show integration of AAV DNA into the S1 locus, and that integration elsewhere seemed rare, we were left with the conclusion that the majority of the AAV DNA in the human tissues must be extrachromosomal. To directly address this possibility, we developed a novel assay using linear rolling-circle amplification to specifically amplify double-stranded, circular AAV genomes from total cellular DNA. A schematic of the approach is shown in Fig. 6. Total cellular DNA was initially digested with a restriction enzyme that was not predicted to cut within the AAV genome. This generated linear DNA fragments that were substrates for degradation by digestion with a novel exonuclease (Plasmid-Safe DNase) that selectively removes double-stranded and single-stranded linear DNA, as well as single-stranded circular DNA. Importantly, Plasmid-Safe DNase does not degrade double-stranded, circular DNA molecules, thus effectively enriching the DNA sample for free circular AAV genomes while degrading integrated AAV forms. We previously demonstrated that this exonuclease is able to efficiently degrade single-strand AAV genomes and all detectable replicative forms (10, 41). After digestion, remaining intact circular AAV episomes were amplified by isothermic rolling-circle amplification using φ29 phage DNA polymerase. AAV-specific primers (cap gene) were used to yield AAV-specific DNA consistent with high-molecular-weight head-to-tail amplicons.
FIG. 6.
Linear rolling-circle amplification for the detection of AAV episomes. Total cellular DNA was digested with a restriction enzyme that does not cut within the AAV genome. The DNA was then treated with Plasmid-Safe DNase, which degrades linear fragments but leaves circular, double-stranded DNA intact. The digestion reaction served as a template for linear rolling-circle amplification using AAV-specific primers and φ29 phage DNA polymerase. Large, linear concatameric arrays (U, uncut linear rolling-circle amplification DNA from tissue T88) were produced following amplification of circular AAV episomes. The linear arrays were subsequently digested into unit-length monomers by restriction enzyme digestion with an enzyme that cleaves the AAV genome once (labeled 1 in the figure). The unit-length fragment was then cloned into an appropriate vector for further sequence analysis.
T88 DNA (100 ng) was subjected to linear rolling-circle amplification, which resulted in the synthesis of high-molecular-weight concatameric AAV forms that upon digestion with a one-cut restriction enzyme generated the unit-length form (Fig. 6). Control linear rolling-circle amplification reactions using either linear or circular plasmid DNA spiked into naïve cellular DNA confirmed the assay specificity (Fig. 7). These results indicated that episomal wild-type AAV forms were present within sample T88. A minor hybridizing band with faster mobility was also observed (Fig. 6), which did not correspond in size to the predicted head-to-head or tail-to-tail orientation. This smaller DNA form was probably derived from deleted episomal forms, consistent with the AAV-AAV junctions isolated by LAM-PCR (Fig. 5). Moreover, five additional samples (T32, T40, T70, T71, and S17) also contained various amounts of linear rolling-circle amplification-amplifiable AAV DNA, which indicated the presence of similar episomal forms in these tissues (data not shown). The three samples that did not amplify (T17, T41, and LG15) tended to have a lower copy number of AAV DNA (Fig. 7) or lower quality of the total genomic DNA preparation.
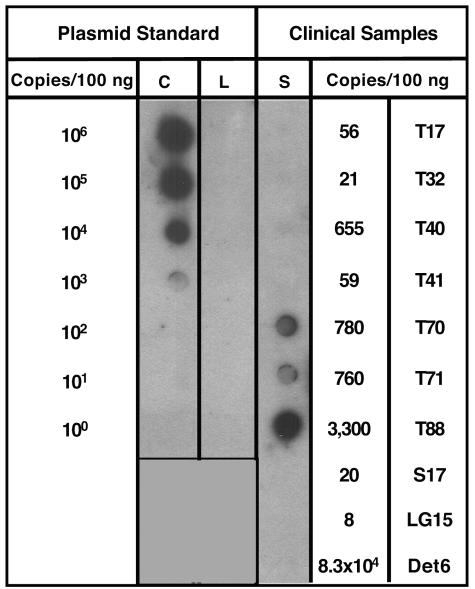
FIG. 7.
Wild-type AAV genomes are present predominantly as episomes. To determine the sensitivity of the linear rolling-circle amplification assay, 10-fold dilutions of circular (C) or linear (L) DNA plasmids containing the AAV2 rep and cap genes were spiked into 100ng of genomic DNA from a naïve human tonsil sample. Linear rolling-circle amplification was performed using cap-specific primers and the products were analyzed by dot blot Southern hybridization using an AAV-specific probe. Positive hybridization could be detected at 1,000 circular plasmid copy spike (representing episomal DNA), whereas linear plasmid DNA (representing integrated AAV) could not be amplified to any detectable level. Total cellular DNA (100 ng) from AAV-positive clinical samples (S) was then analyzed by linear rolling-circle amplification, and tissues T70, T71, and T88 showed positive amplification. The AAV genome copy numbers in these particular tissues were at or above the level of sensitivity of linear rolling-circle amplification as determined from the plasmid spike experiments. Detroit 6 (Det6) cells, which have 83,000 integrated AAV genomes in 100ng, failed to amplify the integrated AAV, further confirming linear rolling-circle amplification assay specificity.
AAV genomes persist predominantly as episomes in human tissues.
The ability to amplify circular AAV DNA led us to consider whether the linear rolling-circle amplification assay could be used to estimate the amount of AAV DNA that existed as extrachromosomal circles. As noted above, the linear rolling-circle amplification assay was validated by subjecting plasmid DNA (in supercoiled or linear form) to the assay using 10-fold serial dilutions of a 15-kb plasmid spiked into 100 ng of human genomic DNA from a naïve tonsil (Fig. 7, Plasmid Standard). Amplified DNA was observed by Southern dot blot hybridization using as few as 1,000 circular input copies, while linear plasmid DNA failed to yield amplified DNA at any input (up to 106 copies), thereby confirming linear rolling-circle amplification specificity. Furthermore, for Detroit 6 cells, which have 83,300 integrated AAV genomes in 100 ng, linear rolling-circle amplification failed to amplify the integrated AAV genome, providing further confirmation of assay specificity.
We analyzed the AAV-positive human samples for the presence of episomal AAV genomes using linear rolling-circle amplification in combination with Southern dot blot hybridization to visualize the amplified product (Fig. 7, Clinical Samples). Samples T70 and T71 demonstrated amplification at an intensity on the Southern blot similar to the 1,000 circular copy plasmid standard (Fig. 7). Based on the initial AAV genome copy number in these samples (T70 had 780 AAV copies/100 ngand T71 had 760 AAV copies/100 ng), these data suggested that the majority of the T70 and T71 AAV DNA existed in a circular conformation. Likewise, tissue sample T88 (3,300 AAV copies/100 ng) showed DNA amplification at a similar level as that detected for the 10,000 plasmid copy standard, which is also consistent with the majority of T88 AAV DNA also being in a circular conformation. The lower amplification levels observed in the remaining samples (T17, T32, and S17) did not permit an estimation of episomal prevalence using the Southern dot blot methodology.
DISCUSSION
The goal of this study was to define the molecular form of AAV genomes that we found in human tissue samples. Based on extensive in vitro cell culture data demonstrating preferential site-specific integration of wild-type AAV into AAVS1 (26, 33), our belief was that targeted integration would also occur in vivo. However, we were unable to show site-specific integration of AAV DNA using two complementary PCR-based assays. In addition, we went on to show that the major form of AAV DNA in these tissues was extrachromosomal and circular.
Although the precise mechanism of targeted AAV genome integration remains unknown, detailed knowledge regarding cis and trans requirements for integration are well documented. The sole viral cis elements necessary for AAVS1 integration are the 145-bp ITR, although enhanced integration has been observed with the inclusion of a portion of the p5 promoter (35). Integration is dependent on the large Rep proteins and involves dual recognition in cis of Rep binding sites within the viral ITR and the AAVS1 locus. Mutational analysis of the Rep78/68 proteins indicated that DNA binding, helicase, and site-specific endonuclease activities were all necessary for AAVS1 integration (25, 44). Both the AAVS1 Rep-binding site and trs sequence elements have also been shown to be essential for viral targeting to this locus via Rep78/68-dependent replication from the AAVS1 origin of replication (52).
Mechanistically, Rep protein multimerization is thought to be essential to bring both physical elements into proximity for recombination via limited DNA replication at the AAVS1 origin. Following integration, wild-type AAV proviral structures are predominantly found in head-to-tail concatameric arrays, and possess microhomology at the viral/cellular junctions (6, 27, 38, 50). As noted earlier, this process has been well documented in transformed cultured cells, and may occur in vivo. In a study by Hernandez et al. (18), nine rhesus macaques were inoculated with AAV in the presence or absence of wild-type adenovirus. Site-specific integration was apparently detected in 1 animal by PCR amplification of wild-type AAV-cellular DNA junctions via dot blot hybridization.
Recently, site-specific integration has been observed in an AAVS1 transgenic mouse model (36) using a recombinant adenovirus vector harboring an AAV ITR-containing vector. Under conditions of inducible AAV Rep protein expression, AAV vector genome integration into AAVS1 was clearly documented with this hybrid vector. This study reaffirms the requirement for Rep protein expression and AAV ITR elements for targeted integration, but whether this occurs at similar frequencies in vivo following wild-type AAV infection remains to be determined in this experimental model.
At the outset, we chose to use the well-characterized latently infected Detroit 6 cell line to validate and optimize an AAVS1 integration assay. In these cells, we detected single AAV-AAVS1 integration events in Detroit 6 genomic DNA spiked into a competing background of naïve total cellular DNA. When applied to our AAV-positive human tissues, we were unable to detect AAV-AAVS1 cellular junctions in any of the nine samples. Failure to observe AAVS1 targeted integration was unexpected, but similar results had been observed in vivo using an experimental model of AAV infection in nonhuman primates (18).
The absence of targeted integration did not preclude the possibility of random integration into the host genome. Several cellular pathways exist that could facilitate random integration of exogenous DNA into the host cell, including either homologous or illegitimate recombination via host cellular DNA repair enzymes (28, 48). In fact, for many common modalities of in vivo exogenous DNA transfer (i.e., naked plasmid DNA and adenovirus vectors) random integration of introduced DNA has been shown to occur (17, 34, 46, 47). Moreover, recent data suggested that the frequency of random integration is related to the amount of intranuclear exogenous input DNA (46). Increased levels of random integration of plasmid DNA into the host genome were observed following highly efficient in vivo electroporation of muscle tissue, which increased intracellular levels of plasmid DNA by 6- to 34-fold compared to standard DNA injection (46). Interestingly, random integration of recombinant AAV vectors in cultured cells has been shown to preferentially occur at sites of double-strand chromosomal DNA breaks, possibly via interaction between viral ITR and cellular nonhomologous end-joining repair enzymes at regions possessing microhomology (30, 42). Whether wild-type virus in vivo also integrates at such sites is unknown, but it is notable that the nonhomologous end-joining cellular enzyme Ku86 appears capable of binding to the AAV ITR in a Rep-independent manner in cultured cells (53).
To directly address the question of random integration, we developed a modified LAM-PCR integration assay to identify and amplify random insertion sites. The LAM-PCR technique was previously exploited to map retroviral vector insertion sites (40). We modified the assay to minimize detection of AAV-AAV head-to-tail junctions, which are characteristically formed following wild-type infection. By simultaneously using two different restriction enzymes that yield either overhanging or blunt ends, we were able to achieve a detection sensitivity of 1 in 160 viral-cellular integration events, again using control Detroit 6 cellular DNA. The specificity of the assay was confirmed by our ability to map the identical AAV2 insertion site in Detroit 6 cells as that previously published (4, 21). Using this method, we were able to isolate a single AAV integration event in sample T71 that mapped to the q arm of chromosome 1 (1q31.1). The sequence consisted of a highly repetitive pentameric sequence (GGAAT) characteristic of satellite II/III repetitive DNA. Similar purine tract sequences have been associated with illegitimate integration sites observed in vitro (48). Also, this integrant possessed 2 base pairs of microhomology with the cellular breakpoint sequence, which was similar to the microhomologies observed in cell culture at sites of recombinant AAV vector integration (29).
The fact that we detected a single, cell-specific (i.e., nonclonal) integration event encompassing 5 kb of flanking cellular DNA and possessing the expected blunt restriction site and 3′ linker sequence supported the notion that LAM-PCR is both robust and sensitive. In addition, our ability to amplify a control gene (epo) from all the samples suggested that the assay is reproducible and has sufficient sensitivity to detect a significant proportion of potential in vivo integration events.
Although integration events were rare, we readily identified LAM-PCR products that contained either AAV DNA fragments or consisted of AAV-AAV junction elements oriented predominantly in a head-to-tail manner (Table 1). These forms likely arose by one of several mechanisms. First, several of the AAV-AAV junction clones were isolated because the blunt cutting enzyme that was predicted to not cut within the AAV genome actually was present and therefore yielded the expected internal AAV-AAV junction product. Second, not all of the LAM-PCR products contained the expected blunt restriction site located just 5′ to the ligated linker. Hexamer primer annealing at the end of the single-stranded amplicon during conversion to a double-stranded template might have created such random blunt ends.
An alternative possibility is that double-stranded DNA breaks could have occurred prior to cleavage with the blunt-cutting restriction enzyme and would therefore be substrates for linker ligation. This underscored the importance of using multiple blunt-cutting enzymes in independent LAM-PCRs to increase the probability of obtaining viral-cellular LAM-PCR junction products. Regardless of the underlying cause, we identified multiple AAV-AAV junction sequences that were predominantly in a head-to-tail orientation. Sequence analysis of the products revealed structures with deleted sequence within the viral ITR and flanking rep and cap sequences, which are very similar to ITR rearrangements observed in vitro following wild-type AAV infection (20, 50).
These data caused us to consider an alternative possibility, that wild-type AAV DNA persisted in vivo as extrachromosomal elements. To explore this possibility, we adopted a technique known as linear rolling-circle amplification to specifically amplify circular in vivo AAV structures. In previous work, we had characterized the activity of a novel DNA exonuclease (Plasmid Safe DNase) that readily digests double- and single-stranded linear molecules, but not double-stranded circular episomes (10, 41). With this activity in mind, we used this exonuclease to enrich the AAV-positive samples for episomal AAV forms, while also removing greater than 99% of the total cellular DNA (linear) by digestion with a “no-cut” (does not cleave the AAV genome) restriction enzyme. We then employed an isothermal phage polymerase-dependent amplification process (originally commercialized to amplify plasmid DNA for high-throughput sequencing projects) to amplify circular templates via rolling-circle replication. The assay was validated using linear or circular plasmid DNA spiked into total cellular DNA. Amplification (105-fold) of low-copy circular plasmid inputs (1,000 copies) was easily detected by dot blot hybridization, while linear DNA (106 copies) failed to amplify.
Considered together, these data suggest that AAV genomes rarely integrate into the host genome, but instead, persist as free circular forms. While we cannot rule out the possibility that the circular forms detected are integration intermediates, we believe this is unlikely for 2 reasons. First, most of our AAV-positive samples (seven of nine) did not contain detectable adenovirus sequences, which is consistent with a “latent” AAV infection. Although it is formally possible that other viruses (e.g., herpesviruses) could have been present as helpers, AAV has never been (to our knowledge) isolated along with herpesviruses from clinical samples. Second, it seems unlikely that for all the samples assayed, failure to detect targeted integration was due to the preferential isolation of preintegration “intermediates” at the complete exclusion of integrated forms.
Persistence of wild-type AAV genomes as circular episomes is entirely plausible. Complete double-D ITR structures (similar to those detected by our LAM-PCR assay; Fig. 5) contain sufficient cis-sequence information for the complete AAV life cycle (49). The double-D ITR structure, as part of a circular plasmid, is able to undergo resolution (in a Rep-independent process) in vitro to yield a linear no-end substrate. In the presence of helper virus coinfection, this no-end linear substrate undergoes a productive linear replication cycle after Rep-dependent nicking at the trs site. Thus, a double-D circularized AAV form could efficiently undergo productive lytic replication if provided with complete helper functions. Interestingly, an alternative AAV replication pathway involving circular duplex monomer genomes was recently identified (31, 32). These circular species appear to constitute up to 10% of monomer duplex intermediates of wild-type AAV. The circularization point of circular duplex monomer AAV genomes also appears to yield the double-D ITR element. This circular duplex monomer genome form was shown to replicate either along the standard linear strand-displacement pathway following resolution of the double-D ITR domain or by a mechanism that was able to maintain the integrity of the circular conformation.
These data, combined with the propensity of recombinant derivatives to assume circular monomeric or multimeric structures (9, 11, 13, 51), suggest that circularization in vivo may be the preferred result of interaction between cellular double-strand DNA repair enzymes and the incoming AAV genome. Whether in vivo circularization occurs via a single-stranded or double-stranded intermediate is currently unknown, but recent in vitro data suggests circularization likely occurs via a double-stranded intermediate (8).
In conclusion, we have shown that following naturally acquired infection, AAV DNA can persist mainly as circular episomes in human tissues. These findings are consistent with the circular episomal forms of recombinant AAV vectors that have been isolated and characterized from in vivo transduced tissues (12, 13). Since the wild-type and vector genomes share only the AAV ITR, it is tempting to conclude that the ITR is the predominant determinant of genome persistence in vivo, and that recombination events drive circle formation in both cases. While we were unable to demonstrate integration into the AAVS1 locus, more human samples should be studied to confirm these findings before we presume that site-specific integration directed by AAV is restricted to cells in culture. Moreover, it is now time to go beyond the straightforward identification and characterization of AAV DNA sequences in tissues and move on to understanding which cell types harbor these persistent AAV genomes.
Acknowledgments
We thank Mary Connell and Robin Bolek for expert technical assistance. We also thank Greg Wiet, Sue Hammond, and Janet Berry for assistance with tonsil tissue procurement, Steve Qualman and the Cooperative Human Tissue Network, and the CCRI Core Sequencing Laboratory for help with DNA sequencing.
This work was funded by National Institutes of Health (NIAID/DAIDS) grant 2-P01-AI56454.
REFERENCES
- 1.Afione, S. A., C. K. Conrad, W. G. Kearns, S. Chunduru, R. Adams, T. C. Reynolds, W. B. Guggino, G. R. Cutting, B. J. Carter, and T. R. Flotte. 1996. In vivo model of adeno-associated virus vector persistence and rescue. J. Virol. 70:3235-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atchison, R. W., B. C. Casto, and W. M. Hammon. 1965. Adenovirus-associated defective virus particles. Science 149:754-756. [DOI] [PubMed] [Google Scholar]
- 3.Atchison, R. W., B. C. Casto, and W. M. Hammon. 1966. Electron microscopy of adenovirus-associated virus (AAV) in cell cultures. Virology 29:353-357. [DOI] [PubMed] [Google Scholar]
- 4.Berns, K. I., T. C. Pinkerton, G. F. Thomas, and M. D. Hoggan. 1975. Detection of adeno-associated virus (AAV)-specific nucleotide sequences in DNA isolated from latently infected Detroit 6 cells. Virology 68:556-560. [DOI] [PubMed] [Google Scholar]
- 5.Blacklow, N. R., M. D. Hoggan, and W. P. Rowe. 1967. Isolation of adenovirus-associated viruses from man. Proc. Natl. Acad. Sci. USA 58:1410-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Chen, C. L., R. L. Jensen, B. C. Schnepp, M. J. Connell, R. Shell, T. J. Sferra, J. S. Bartlett, K. R. Clark, and P. R. Johnson. 2005. Molecular characterization of adeno-associated viruses infecting children. J. Virol. 79:14781-14192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. K., M. D. Hoggan, W. W. Hauswirth, and K. I. Berns. 1980. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J. Virol. 33:739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chirmule, N., K. Propert, S. Magosin, Y. Qian, R. Qian, and J. Wilson. 1999. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 6:1574-1583. [DOI] [PubMed] [Google Scholar]
- 8.Choi, V. W., R. J. Samulski, and D. M. McCarty. 2005. Effects of adeno-associated virus DNA hairpin structure on recombination. J. Virol. 79:6801-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, K. R., T. J. Sferra, and P. R. Johnson. 1997. Recombinant adeno-associated viral vectors mediate long-term transgene expression in muscle. Hum. Gene Ther. 8:659-669. [DOI] [PubMed] [Google Scholar]
- 10.Clark, K. R., T. J. Sferra, W. Lo, G. Qu, R. Chen, and P. R. Johnson. 1999. Gene transfer into the CNS using recombinant adeno-associated virus: analysis of vector DNA forms resulting in sustained expression. J. Drug Target. 7:269-283. [DOI] [PubMed] [Google Scholar]
- 11.Duan, D., P. Sharma, J. Yang, Y. Yue, L. Dudus, Y. Zhang, K. J. Fisher, and J. F. Engelhardt. 1998. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 72:8568-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan, D., P. Sharma, J. Yang, Y. Yue, L. Dudus, Y. Zhang, K. J. Fisher, and J. F. Engelhardt. 1998. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 72:8568-8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan, D., Z. Yan, Y. Yue, and J. F. Engelhardt. 1999. Structural analysis of adeno-associated virus transduction circular intermediates. Virology 261:8-14. [DOI] [PubMed] [Google Scholar]
- 14.Dutheil, N., M. Yoon-Robarts, P. Ward, E. Henckaerts, L. Skrabanek, K. I. Berns, F. Campagne, and R. M. Linden. 2004. Characterization of the mouse adeno-associated virus AAVS1 ortholog. J. Virol. 78:8917-8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erles, K., P. Sebokova, and J. R. Schlehofer. 1999. Update on the prevalence of serum antibodies (IgG and IgM) to adeno-associated virus (AAV). J. Med. Virol. 59:406-411. [DOI] [PubMed] [Google Scholar]
- 16.Gao, G., L. H. Vandenberghe, M. R. Alvira, Y. Lu, R. Calcedo, X. Zhou, and J. M. Wilson. 2004. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harui, A., S. Suzuki, S. Kochanek, and K. Mitani. 1999. Frequency and stability of chromosomal integration of adenovirus vectors. J. Virol. 73:6141-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez, Y. J., J. Wang, W. G. Kearns, S. Loiler, A. Poirier, and T. R. Flotte. 1999. Latent adeno-associated virus infection elicits humoral but not cell-mediated immune responses in a nonhuman primate model. J. Virol. 73:8549-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoggan, M. D., N. R. Blacklow, and W. P. Rowe. 1966. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proc. Natl. Acad. Sci. USA 55:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huser, D., S. Weger, and R. Heilbronn. 2002. Kinetics and frequency of adeno-associated virus site-specific integration into human chromosome 19 monitored by quantitative real-time PCR. J. Virol. 76:7554-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotin, R. M., and K. I. Berns. 1989. Organization of adeno-associated virus DNA in latently infected Detroit 6 cells. Virology 170:460-467. [DOI] [PubMed] [Google Scholar]
- 22.Kotin, R. M., R. M. Linden, and K. I. Berns. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11:5071-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotin, R. M., M. Siniscalco, R. J. Samulski, X. D. Zhu, L. Hunter, C. A. Laughlin, S. McLaughlin, N. Muzyczka, M. Rocchi, and K. I. Berns. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 87:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linden, R. M., P. Ward, C. Giraud, E. Winocour, and K. I. Berns. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 93:11288-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linden, R. M., E. Winocour, and K. I. Berns. 1996. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. USA 93:7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarty, D. M., S. M. Young, Jr., and R. J. Samulski. 2004. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu. Rev. Genet. 38:819-845. [DOI] [PubMed] [Google Scholar]
- 27.McLaughlin, S. K., P. Collis, P. L. Hermonat, and N. Muzyczka. 1988. Adeno-associated virus general transduction vectors: analysis of proviral structures. J. Virol. 62:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merrihew, R. V., K. Marburger, S. L. Pennington, D. B. Roth, and J. H. Wilson. 1996. High-frequency illegitimate integration of transfected DNA at preintegrated target sites in a mammalian genome. Mol. Cell. Biol. 16:10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, D. G., L. M. Petek, and D. W. Russell. 2004. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 36:767-773. [DOI] [PubMed] [Google Scholar]
- 30.Miller, D. G., L. M. Petek, and D. W. Russell. 2003. Hum. gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol. Cell. Biol. 23:3550-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musatov, S., J. Roberts, D. Pfaff, and M. Kaplitt. 2002. A cis-acting element that directs circular adeno-associated virus replication and packaging. J. Virol. 76:12792-12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musatov, S. A., T. A. Scully, L. Dudus, and K. J. Fisher. 2000. Induction of circular episomes during rescue and replication of adeno-associated virus in experimental models of virus latency. Virology 275:411-432. [DOI] [PubMed] [Google Scholar]
- 33.Muzyczka, N. 1992. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 158:97-129. [DOI] [PubMed] [Google Scholar]
- 34.Overturf, K., M. al-Dhalimy, C. N. Ou, M. Finegold, R. Tanguay, A. Lieber, M. Kay, and M. Grompe. 1997. Adenovirus-mediated gene therapy in a mouse model of hereditary tyrosinemia type I. Hum. Gene Ther. 8:513-521. [DOI] [PubMed] [Google Scholar]
- 35.Philpott, N. J., C. Giraud-Wali, C. Dupuis, J. Gomos, H. Hamilton, K. I. Berns, and E. Falck-Pedersen. 2002. Efficient integration of recombinant adeno-associated virus DNA vectors requires a p5-rep sequence in cis. J. Virol. 76:5411-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Recchia, A., L. Perani, D. Sartori, C. Olgiati, and F. Mavilio. 2004. Site-specific integration of functional transgenes into the human genome by adeno/AAV hybrid vectors. Mol. Ther. 10:660-670. [DOI] [PubMed] [Google Scholar]
- 37.Rose, J. A., M. D. Hoggan, and A. J. Shatkin. 1966. Nucleic acid from an adeno-associated virus: chemical and physical studies. Proc. Natl. Acad. Sci. USA 56:86-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutledge, E. A., and D. W. Russell. 1997. Adeno-associated virus vector integration junctions. J. Virol. 71:8429-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samulski, R. J., X. Zhu, X. Xiao, J. D. Brook, D. E. Housman, N. Epstein, and L. A. Hunter. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt, M., G. Hoffmann, M. Wissler, N. Lemke, A. Mussig, H. Glimm, D. A. Williams, S. Ragg, C. U. Hesemann, and C. von Kalle. 2001. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum. Gene Ther. 12:743-749. [DOI] [PubMed] [Google Scholar]
- 41.Schnepp, B. C., K. R. Clark, D. L. Klemanski, C. A. Pacak, and P. R. Johnson. 2003. Genetic fate of recombinant adeno-associated virus vector genomes in muscle. J. Virol. 77:3495-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song, S., Y. Lu, Y. K. Choi, Y. Han, Q. Tang, G. Zhao, K. I. Berns, and T. R. Flotte. 2004. DNA-dependent PK inhibits adeno-associated virus DNA integration. Proc. Natl. Acad. Sci. USA 101:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsunoda, H., T. Hayakawa, N. Sakuragawa, and H. Koyama. 2000. Site-specific integration of adeno-associated virus-based plasmid vectors in lipofected HeLa cells. Virology 268:391-401. [DOI] [PubMed] [Google Scholar]
- 44.Urabe, M., Y. Hasumi, A. Kume, R. T. Surosky, G. J. Kurtzman, K. Tobita, and K. Ozawa. 1999. Charged-to-alanine scanning mutagenesis of the N-terminal half of adeno-associated virus type 2 Rep78 protein. J. Virol. 73:2682-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walz, C. M., M. Nakamura, T. Fukunaga, Y. Jasiewicz, L. Edler, J. R. Schlehofer, and Y. Tanaka. 2001. Reduced prevalence of serum antibodies against adeno-associated virus type 2 in patients with adult T-cell leukaemia lymphoma. J. Med. Virol. 65:185-189. [PubMed] [Google Scholar]
- 46.Wang, Z., P. J. Troilo, X. Wang, T. G. Griffiths, S. J. Pacchione, A. B. Barnum, L. B. Harper, C. J. Pauley, Z. Niu, L. Denisova, T. T. Follmer, G. Rizzuto, G. Ciliberto, E. Fattori, N. L. Monica, S. Manam, and B. J. Ledwith. 2004. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 11:711-721. [DOI] [PubMed] [Google Scholar]
- 47.Wurm, F. M., and C. J. Petropoulos. 1994. Plasmid integration, amplification and cytogenetics in CHO cells: questions and comments. Biologicals 22:95-102. [DOI] [PubMed] [Google Scholar]
- 48.Wurtele, H., K. C. Little, and P. Chartrand. 2003. Illegitimate DNA integration in mammalian cells. Gene Ther. 10:1791-1799. [DOI] [PubMed] [Google Scholar]
- 49.Xiao, X., W. Xiao, J. Li, and R. J. Samulski. 1997. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J. Virol. 71:941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, C. C., X. Xiao, X. Zhu, D. C. Ansardi, N. D. Epstein, M. R. Frey, A. G. Matera, and R. J. Samulski. 1997. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 71:9231-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, J., W. Zhou, Y. Zhang, T. Zidon, T. Ritchie, and J. F. Engelhardt. 1999. Concatamerization of adeno-associated virus circular genomes occurs through intermolecular recombination. J. Virol. 73:9468-9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young, S. M., Jr., and R. J. Samulski. 2001. Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat. Proc. Natl. Acad. Sci. USA 98:13525-13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zentilin, L., A. Marcello, and M. Giacca. 2001. Involvement of cellular double-stranded DNA break binding proteins in processing of the recombinant adeno-associated virus genome. J. Virol. 75:12279-12287. [DOI] [PMC free article] [PubMed] [Google Scholar]