Abstract
The V proteins of some paramyxoviruses have developed the ability to efficiently inactivate STAT protein function as a countermeasure for evading interferon (IFN) responses. Human parainfluenza virus type 4 (hPIV4) is one of the rubulaviruses, which are members of the family Paramyxoviridae, and has a V protein with a highly conserved cysteine-rich domain that is the hallmark of paramyxovirus V proteins. In order to study the function of the hPIV4 V protein, we established HeLa cells expressing the hPIV4A V protein (HeLa/FlagPIV4V). The hPIV4 V protein had no ability to reduce the level of STAT1 or STAT2, although it associated with STAT1, STAT2, DDB1, and Cul4A. It interfered with neither STAT1 and STAT2 tyrosine phosphorylation nor IFN-induced STAT nuclear accumulation. In addition, HeLa/FlagPIV4V cells are fully sensitive to both beta interferon (IFN-β) and IFN-γ, indicating that the hPIV4 V protein has no ability to block IFN-induced signaling. We further established HeLa cells expressing various chimeric proteins between the hPIV2 and hPIV4A V proteins. The lack of IFN-antagonistic activity of the hPIV4 V protein is caused by both the P/V common and V-specific domains. At least two regions (amino acids [aa] 32 to 45 and aa 143 to 164) of hPIV4 V in the P/V common domain and one region (aa 200 to 212) of the C terminus are involved in the inability to evade the IFN-induced signaling. Moreover, we established HeLa cells persistently infected with hPIV4 to make sure of the inability to escape IFN and confirmed that hPIV4 is the only paramyxovirus analyzed to date that can't evade the IFN-induced antiviral responses.
Human parainfluenza virus type 4 (hPIV4) is a member of the family Paramyxoviridae and is known to be an important agent of pediatric respiratory tract disease in infancy (26). hPIV4 is further divided into two antigenic subtypes, hPIV4A and hPIV4B, based mainly on in vitro hemadsorption inhibition characteristics and immunological reactivity (6). We previously identified the structural proteins of hPIV4A and 4B and sequenced all genes except the L gene (2, 18, 21, 22, 23). hPIV4 contains an mRNA-editing site, similar to other rubulaviruses. The unedited version of the “P” mRNA encodes the V protein, and addition of two G nucleotides at the editing site produces an mRNA that encodes the P protein. Therefore, the N-terminal 153 amino acids (aa) of the V and P proteins are common, and their C termini are unique (22). The C terminus of hPIV4 V protein contains seven invariant cysteine residues capable of binding two atoms of zinc, and C termini are approximately 50% identical among all paramyxovirus V proteins.
It has been demonstrated recently that the Paramyxovirinae, similar to other viruses, have evolved specific proteins that inhibit interferon (IFN)-induced innate antiviral responses through direct inhibition of cellular STAT proteins. The V proteins encoded by the rubulaviruses, simian virus 5 (SV5), SV41, mumps virus (MuV), and Newcastle disease virus (NDV, an avulavirus) block IFN signaling by targeting STAT1 for degradation (1, 7, 8, 15, 25, 32, 33, 36, 49, 50, 53, 54), whereas the V protein of human parainfluenza type 2 virus (hPIV2, a rubulavirus) targets STAT2 for degradation (32, 33, 35). Moreover, the V proteins of measles virus (morbillivirus), Nipah virus, and Hendra virus (henipaviruses) have been shown to inhibit IFN signaling by preventing STAT1 and STAT2 nuclear accumulation (34, 39, 40, 45). Sendai virus (SeV) and hPIV3 also block IFN signaling, and this anti-IFN ability has been shown to be a property of these respirovirus C proteins (10, 11, 12, 13, 14, 17, 20, 30, 46). The rubulavirus V protein-dependent degradation of STAT proteins involves degradation complexes that contain the V protein, STAT1, and STAT2 (and STAT3 in the case of mumps virus). A number of cellular proteins, including the UV-damaged DNA binding protein 1 (DDB1) and Cullin4A (Cul4A), that are subunits of an SCF-type ubiquitin ligase (49) are also required. The conserved seven cysteine residues play a critical role in specifically binding to DDB1 (1, 29). However, the binding between the V and STAT proteins occurs via a tryptophan-rich motif that lies just upstream of the cysteine cluster at the C terminus, and the cysteine residues are not required for this binding (31). The mechanism of STAT level reduction induced by rubulaviruses is not clearly understood yet.
In this study, we demonstrate that the hPIV4 V protein has no ability to escape IFN by using HeLa cells constitutively expressing the hPIV4A V protein. This article reports that the hPIV4 V protein forms a complex with STAT1, STAT2, DDB1, and Cul4A in vitro or in vivo. However, the hPIV4 V protein could not lower the intracellular levels of the STAT proteins like other rubulaviruses and NDV. We also established HeLa cells constitutively expressing various chimeric proteins between the hPIV2 and hPIV4A V proteins, and the levels of STAT proteins were analyzed. The failure of hPIV4 V protein to evade IFN responses is due to defects in both the P/V common domain and the V-specific domain. Moreover, we established HeLa cells persistently infected with hPIV4A or hPIV4B and analyzed whether these viruses have the ability to escape IFN. Our results further demonstrate that hPIV4 is the only paramyxovirus analyzed to date that can't evade the IFN-induced antiviral responses.
MATERIALS AND METHODS
Cells and viruses.
HeLa and human 2fTGH (gift from I. M. Kerr, Imperial Cancer Research Fund, London, United Kingdom) cells were grown in Eagle's minimal essential medium (MEM) supplemented with 10% fetal calf serum (FCS). BSR T7/5 (5) cells were cultured in Eagle's MEM supplemented with 10% FCS and 1 mg/ml G418 (Geneticin; GIBCO). hPIV4A (strain 68-340) and hPIV4B (strain 68-333) were used in this study.
Antibodies.
Anti-Flag monoclonal antibody (MAb) was purchased from SIGMA (St. Louis, MO). Anti-STAT1 p84/p91 (N terminus) MAb, anti-STAT3 MAb, and MAb to STAT1 phosphorylated at Tyr 701 [anti-STAT1pTyr(701) antibody] were obtained from BD Transduction Laboratories (Lexington, KY). Antibody to STAT2 (sc-476), antibody to Cul4A (sc-10782), MAb to actin (sc-8432), and MAb to hemagglutinin (HA) (sc-7392) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibodies against STAT2 phosphorylated at Tyr 689 [anti-STAT2pTyr(689) antibody] and against DDB1 were obtained from Upstate Biotechnology (Lake Placid, NY) and Zymed Laboratories (South San Francisco, CA), respectively. MAb against hPIV4A nucleoprotein (NP) (A130), which was cross-reactive with hPIV4B, was previously reported (19).
Construction of expression plasmids.
To express the hPIV4A V protein in mammalian cells, cDNA expression vector (pCI-neo, Promega) that contained hPIV4A V cDNA fused to an N-terminal Flag epitope tag was constructed. Flag-tagged hPIV2 and various chimeric V protein expression vectors were generated in a manner similar to that described above. Human STAT1 and STAT2 genes fused to an N-terminal HA epitope tag were cloned into pTM1 (pTM-HA-hSTAT1 and -hSTAT2), which contains a T7 promoter and an encephalomyocarditis virus internal ribosome entry site (B. Moss, National Institutes of Health). All constructs were confirmed by nucleic acid sequencing.
Establishment of HeLa cells constitutively expressing Flag-tagged V proteins.
To obtain cell lines expressing each of the Flag-tagged V proteins, HeLa cells were transfected with each plasmid by using FuGENE 6 (Roche) according to the manufacturer's instructions. At 2 days after transfection, the culture media were changed to MEM containing 10% FCS, 1 mg/ml G418, and 0.2% agarose, and the cells were cultured for 3 weeks. Five independent clones that exhibited high expression levels were analyzed.
Cell extracts, immunoblotting, and immunoprecipitation.
For preparation of cell extracts, cells were lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.6% NP-40, and 4 mM phenylmethylsulfonyl fluoride). For immunoblotting, cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and analyzed by a Western blotting technique with appropriate antibodies as described previously (31). For immunoprecipitation of Flag epitope-tagged proteins, cell extracts were incubated with an anti-Flag M2 agarose affinity gel (Sigma, St. Louis, MO) for 6 h at room temperature. The agarose beads were washed three times with lysis buffer and then extracted with sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer for analysis by a Western blotting technique with appropriate antibodies.
Reporter gene assay with luciferase.
The IFN-α/β-responsive reporter plasmid p(9-27)4tk (−39) lucter, referred to here as pISRE(f)-luc, contains four tandem repeats of the IFN-inducible gene 9-27 IFN-stimulated response element fused to the firefly luciferase gene. pTK-r-luc, used as transfection standard, contains the herpes simplex virus thymidine kinase (TK) promoter region upstream of the Renilla luciferase gene (Promega). For the luciferase assays, 2fTGH cells were transfected with 1 μg of pCI-neo-V, 1 μg of pISRE(f)-luc, 0.3 μg of pTK-r-luc, and 7.5 μl of FuGENE 6. At 24 h posttransfection, the cells were treated with 1,000 U of recombinant IFN-α per ml or not treated. At 14 h after IFN treatment, the cells were harvested and assayed for firefly and Renilla luciferase activities (dual-luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the firefly luciferase values by those of the Renilla luciferase.
IFN susceptibility.
The monolayers of various cells were incubated with 10, 100, or 1,000 U of human IFN-α (hIFN-α), hIFN-β, or hIFN-γ for 24 h, and then the cells were infected with about 100 PFU of recombinant vesicular stomatitis virus-green fluorescent protein (rVSV-GFP) (gift from D. Kolakofsky, University of Geneva School of Medicine, Geneva, Switzerland), VSV, or Sindbis virus. At 12 h after infection with rVSV-GFP, GFP expression was analyzed using a fluorescence microscope. At 2 days after infection with VSV or Sindbis virus, plaque numbers were counted.
Immunofluorescence staining.
For STAT distribution experiments, HeLa or HeLa/FlagPIV4V cells were grown to 60% confluence and not stimulated or stimulated with 1,000 U of IFN-α/ml for 30 min to fixation. The cells were fixed with 3% paraformaldehyde for 30 min at room temperature and rinsed twice with phosphate-buffered saline (PBS). The cells were permeabilized with PBS-0.05% Tween 20 for 30 min and washed twice with PBS. The cells were then incubated for 60 min with antibody against STAT1 or STAT2 and washed three times with PBS. Next, the cells were incubated for 60 min with fluorescein isothiocyanate-labeled secondary antibodies and washed with PBS. Immunofluorescently stained cells were analyzed using a fluorescence microscope.
For detection of V or NP protein, the cells were grown to 60% confluence. Fixed and permeabilized cells were stained with anti-Flag or NP MAb as described above.
Establishment of persistent hPIV2, hPIV4A, or hPIV4B infection.
Monolayers of HeLa cells were infected with hPIV2, hPIV4A, or hPIV4B at a multiplicity of infection (MOI) of 0.01 to 1 and incubated with MEM supplemented with 5% FCS for 4 days. Subsequently, the cells were washed three times with MEM to remove the dead cells, and then cell cloning was carried out by limiting dilution method using 96-well plates. After about 3 weeks, the cloned cells were duplicatively subcultured, and 2 days after subculture, hemadsorption analysis using guinea pig erythrocytes for detecting the persistently virus-infected cells was carried out. HeLa cells persistently infected with SeV were established as described previously (16).
RESULTS
The hPIV4 V protein binds STATs, DDB1, and Cul4A without STAT degradation.
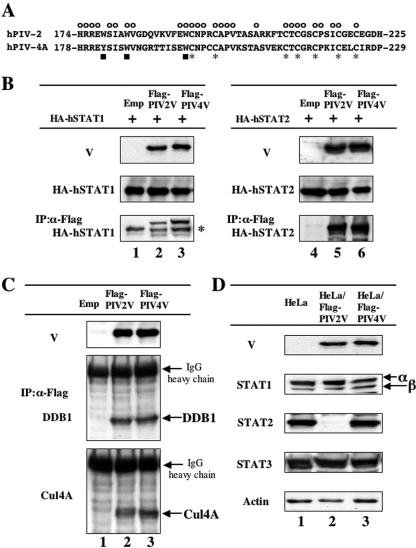
The V proteins encoded by the rubulaviruses SV5, SV41, hPIV2, and MuV block IFN-induced signaling by targeting STAT1 or 2 for degradation (1, 8, 25, 32, 33, 49, 50, 53, 54). hPIV4 is one of rubulaviruses and has a V protein possessing a highly conserved cysteine-rich domain and tryptophan-rich motif (Fig. 1A). To examine the potential of hPIV4 V for evasion of IFN-induced signaling, a cDNA encoding hPIV4A V protein was subcloned into a mammalian expression vector downstream of a Flag epitope tag. STAT protein targeting by SV5, MuV, and hPIV2 V proteins requires a multisubunit ubiquitin-ligase complex that includes cellular components STAT1, STAT2, DDB1, and Cul4A (49, 50). First, whether the hPIV4A V protein binds STAT proteins was studied. Flag-tagged V protein and HA-tagged human-origin STAT proteins were expressed in BSR T7/5 cells that contain cytoplasmic T7 RNA polymerase via transfected plasmids with T7 promoters. After immunoselection of cell lysates with an anti-Flag M2 agarose affinity gel, the precipitates were analyzed by Western blotting with anti-HA antibody. As shown in Fig. 1B, both hPIV2 and hPIV4A V proteins were capable of binding to both hSTAT1 and hSTAT2. To further determine if a similar STAT degradation complex was formed by the hPIV4A V protein, the binding capacity of hPIV4A V protein to DDB1 and Cul4A was examined. Flag-tagged V proteins were expressed in HeLa cells, and after immunoselection of cell lysates with an anti-Flag M2 agarose affinity gel, the precipitates were analyzed by Western blotting with anti-DDB1 or anti-Cul4A antibody. As shown in Fig. 1C, hPIV4A V proteins were capable of binding to both DDB1 and Cul4A, similar to the hPIV2 V protein, indicating that hPIV4 V protein associates with a similar STAT degradation complex.
FIG. 1.
hPIV4 V protein complex formation with STAT1, STAT2, DDB1, and Cul4A without STAT degradation. (A) Amino acid sequences of the V-specific regions of hPIV2 and hPIV4A V proteins. The symbols above and below the sequences indicate the positions of conserved cysteine (asterisks), tryptophan (or tyrosine) (squares), and common (circles) residues. (B) Interaction of V proteins with STAT1 or STAT2. BSR T7/5 cells were transfected with either pTM-HA-hSTAT1 (lanes 1 to 3) or pTM-HA-hSTAT2 (lanes 4 to 6) plus pCI carrying the Flag epitope-tagged hPIV2 V gene (lanes 2 and 5), the hPIV4A V gene (lanes 3 and 6), or an empty pCI (lanes 1 and 4). Whole-cell extracts were prepared at 48 h posttransfection, and samples containing equal amounts of total protein were assayed by Western blotting for their levels of HA-hSTAT1 or HA-hSTAT2 and Flag-V proteins using α-HA and α-Flag antibodies, respectively. Other samples were first immunoprecipitated (IP) with Flag affinity gel (Sigma), and the selected material was then assayed by Western blotting for detection of hSTAT proteins. The asterisk on the right indicates the immunoglobulin heavy chain. (C) Interaction of V proteins with DDB1 or Cul4A. HeLa cells were transfected with pCI-Flag-PIV2V (lane 2), pCI-Flag-PIV4V (lane 3), or an empty pCI (lane 1). Whole-cell extracts were prepared at 48 h posttransfection, and samples containing equal amounts of total protein were assayed by Western blotting for detection of Flag-V proteins (α-Flag). Other samples were first immunoprecipitated (IP) with Flag affinity gel (Sigma), and the selected material was then assayed by Western blotting for detection of DDB1 and Cul4A proteins. (D) STAT levels in HeLa cells constitutively expressing V proteins. Samples of cytoplasmic extracts containing equal amounts of total protein of HeLa cells constitutively expressing Flag-PIV2 V (lane 2) or Flag-PIV4 V proteins (lane 3) or nonexpressing HeLa cells (lane 1) were assayed by Western blotting for detection of endogenous STAT1, STAT2, and STAT3 as well as V proteins using anti-Flag, anti-STAT1, anti-STAT2, and anti-STAT3 antibodies, as indicated at the left of each panel.
Subsequently, to investigate whether the mechanism of IFN antagonism used by SV5, MuV, and hPIV2 is shared by the hPIV4 V protein, the levels of STAT1 and 2 were analyzed in a newly established HeLa cell line constitutively expressing Flag-tagged hPIV4A V protein (HeLa/FlagPIV4V cells) as described in Materials and Methods. As a control, we also used a HeLa cell line constitutively expressing Flag-tagged hPIV2 V protein (HeLa/FlagPIV2V cells). The STAT2 levels in HeLa/FlagPIV2V cells were specifically reduced (Fig. 1D, lane 2), while HeLa/FlagPIV4V cells showed no reduction in their levels of either STAT1, STAT2, or STAT3 (Fig. 1D, lane 3). In addition, when hPIV4 V proteins without a Flag tag were expressed in HeLa cells, no reduction of the levels of STATs was observed (data not shown). These results indicate that the hPIV4 V protein has no ability to reduce the levels of STATs, although it associates with STAT1, STAT2, DDB1, and Cul4A.
hPIV4 V protein alters neither the levels of phosphorylated STAT nor STAT subcellular localization.
The SeV C protein blocks IFN signaling by inhibition of STAT2 phosphorylation (13). To examine the ability of the hPIV4 V protein to inhibit STAT activation, we examined the levels of phosphorylation of STAT1 and STAT2 in HeLa/FlagPIV4V cells. The cells were treated with 10, 100, or 1,000 U of IFN-α for the indicated time and then lysed. As shown in Fig. 2A, when the cells were treated with 1,000 U, the phosphorylation of STAT1 and STAT2 was not suppressed at all in HeLa/FlagPIV4V cells compared with that in the control HeLa cells. Intriguingly, no similar suppression in STAT1 (Fig. 2A) and STAT2 (data not shown) phosphorylation was observed at IFN doses of 10 or 100 U.
FIG. 2.
Levels of phosphorylated STATs or STAT subcellular localization in HeLa/FlagPIV4V cells treated with IFN. (A) HeLa or HeLa/FlagPIV4V cells were treated with 10, 100, or 1,000 U of IFN-α for the indicated times and assayed by Western blotting for detection of phosphorylated STAT1 or STAT2. The asterisk on the right indicates a cross-reacting host band. (B) After treatment with IFN-α (1,000 U) for 30 min or no treatment, the localization of STAT1 and STAT2 proteins was detected by indirect immunofluorescence staining with anti-STAT1 or anti-STAT2 MAb.
The V proteins of measles, Nipah, and Hendra viruses have been shown to inhibit IFN signaling by preventing STAT1 and STAT2 nuclear accumulation (34, 39, 40, 45). To examine the effects of hPIV4 V protein on STAT protein distribution, indirect immunofluorescence staining was used to visualize the subcellular localization of STAT1 and STAT2 in HeLa and HeLa/FlagPIV4V cells. The cells were treated with 1,000 U of IFN-α for 30 min or not treated. Treatment of cells with IFN-α resulted in a rapid nuclear translocation of both STAT1 and STAT2 in both cell types (Fig. 2B), indicating that the expression of hPIV4A V protein does not interfere with STAT nuclear accumulation in response to IFN-α. Therefore, these results demonstrate that the hPIV4 V protein has no ability to alter STAT phosphorylation and nuclear import.
hPIV4 V protein has no ability to evade the IFN response.
To confirm the effect of hPIV4 V on IFN-induced signaling, we analyzed the biological activity of the hPIV4 V protein with respect to IFN-induced antiviral state. HeLa, HeLa/FlagPIV2V, and HeLa/FlagPIV4V cells were treated with hIFN for 24 h before being challenged with rVSV-GFP or Sindbis virus. The susceptibilities of these cells to IFN were monitored based on their ability to prevent rVSV-GFP replication, as measured by GFP expression. In HeLa/FlagPIV2V cells, viral replication of rVSV-GFP was not inhibited, though the cells were treated with hIFN-α (Fig. 3A). In contrast, when HeLa and HeLa/FlagPIV4V cells were treated with hIFN-α, rVSV-GFP replication was completely suppressed (Fig. 3A).
FIG. 3.
Interferon susceptibility in HeLa cells expressing Flag-tagged V proteins. (A) GFP expression in HeLa cells following VSV-GFP infection. The cells untreated or pretreated with 1,000 U of IFN-α for 24 h were infected with VSV-GFP. (B) Confluent monolayers of cells were inoculated with 10 or 100 U of IFN-β or IFN-γ, and after incubation for 24 h, these cells were infected with Sindbis virus. After 2 days of further incubation, plaque numbers were counted. All bars express percentages of values for plaque reduction rate.
Subsequently, IFN sensitivity was determined by a plaque reduction method using hIFN-β or hIFN-γ and Sindbis virus as a challenge virus. As shown in Fig. 3B, HeLa/FlagPIV2V cells are resistant to IFN-β, while they are sensitive to IFN-γ. These results are consistent with our previously reported findings that hPIV2 blocks IFN-α/β signaling but not IFN-γ signaling (33). On the other hand, HeLa/FlagPIV4V cells are fully sensitive to both IFN-β and IFN-γ (Fig. 3B).
Thus, these results indicate that the hPIV4 V protein has no ability to evade the IFN response and that any unknown anti-IFN mechanisms other than STAT degradation don't work in HeLa cells expressing the hPIV4 V protein.
Both the P/V common domain and the V-specific domain of hPIV4 V are involved in the inability to evade the IFN response.
The entire hPIV4 V protein is 24.6% identical in amino acid sequence to the hPIV2 V protein. However, these V proteins have 53.8% amino acid sequence identity in the 52 aa of the C terminus that have a conserved cysteine-rich domain and tryptophan-rich motif. In contrast, the P/V common domains are 15.6% identical (22). Here, we examined which domain of hPIV4 V protein accounts for the lack of the ability using a reporter gene assay with luciferase or a Western blot assay of HeLa cell extracts constitutively expressing V proteins. Two chimeric V proteins were created: (i) the N-terminal 154 amino acids (P/V common domain) of hPIV4 V protein were replaced with the corresponding N-terminal 164 amino acids of hPIV2 V protein (PIV2/4), and (ii) the C-terminal 75 amino acids (V-specific domain) of hPIV4 V protein were replaced with the corresponding C-terminal 61 amino acids of hPIV2 V protein (PIV4/2) (Fig. 4A).
FIG. 4.
Both the P/V common domain and the V-specific domain of hPIV4 V are involved in the inability to interdict IFN-induced signaling. (A) Schematic diagram of the chimera V proteins. Open boxes marked with T's, open boxes, and hatched boxes indicate the Flag epitope tag, amino acids of the hPIV4A V protein, and amino acids of the hPIV2 V protein, respectively. Inhibition of IFN-induced signaling and degradation of STAT2 are summarized in the right panel. (B) Effects of V proteins on IFN-α-stimulated gene activation. 2fTGH cells were transfected with a reporter plasmid [pISRE(f)-luc] and a V expression vector (pCI) or relevant control (see the text), along with pTK-r-luc as an internal reference for transfection efficiency. After incubation for 24 h, the cells were treated with IFN-α (1,000 U/ml) for 14 h or not treated and then lysed. The levels of both firefly (f) and Renilla (r) luciferase activities were determined. Data represent the mean values of the normalized luciferase activities from triplicate samples. GFP alone was used as negative control. (C) STAT1 and STAT2 levels in HeLa cells constitutively expressing V proteins. Samples of cytoplasmic extracts containing equal amounts of total protein of HeLa cells constitutively expressing V proteins (listed above each lane) or nonexpressing HeLa cells (lane 1) were assayed by Western blotting for detection of endogenous STAT1 and STAT2 as well as V proteins using anti-Flag, anti-STAT1, and anti-STAT2 antibodies, as indicated on the left of each panel. The asterisk on the right indicates a cross-reacting host band.
We examined the abilities of the chimeric V proteins to interdict IFN-α-induced signaling to an IFN-responsive reporter gene. 2fTGH cells were transfected with a luciferase reporter construct and V protein expression plasmids and subsequently treated with IFN-α under the protocol indicated in Fig. 4B. In cells transfected with a GFP expression plasmid as an irrelevant control, firefly (f) luciferase activity was increased ca. eightfold by treatment with IFN-α. Expression of hPIV2 V, as positive control, suppressed the activation of the IFN-α-responsive promoter. In contrast, hPIV4 V and both chimera V (PIV2/4 and PIV4/2) proteins tested had lost the ability to interdict IFN signaling (Fig. 4B).
Next, HeLa cell lines constitutively expressing chimera V proteins were established to examine the effect of the V proteins on endogenous STAT1 and STAT2 proteins (Fig. 4C). Expression of hPIV2 V protein led to a loss of cellular STAT2 (Fig. 4C, lane 2). Both chimera V-expressing HeLa cells (PIV2/4 and PIV4/2) showed no reduction in their levels of either STAT1 or STAT2, like hPIV4 V-expressing cells (Fig. 4C, lanes 3 and 5). As summarized in Fig. 4A, these results suggest that both the P/V common domain and the V-specific domain of hPIV4 V protein are involved in the inability to evade IFN-induced signaling.
The regions aa 32 to 45 and aa 143 to 164 of hPIV4 V are involved in the inability to evade IFN-induced signaling.
The V proteins of hPIV2, SV5, and SV41 exhibit characteristic nuclear localization patterns, as described previously (38, 52). Strikingly, the hPIV4 V protein was detected exclusively in the cytoplasm (Fig. 5B, panels 1 and 2), showing that the subcellular distribution of hPIV4 V protein differs significantly from that of the other rubulavirus V proteins, though the mumps virus V protein was also detected in the cytoplasm (50). The chimera PIV2/4 V protein was detected in the nuclei (Fig. 5B, panel 10), while the chimera PIV4/2 V protein was localized in the cytoplasm (data not shown), showing that the P/V common domain of hPIV4 V protein is important for its cytoplasmic localization. We therefore decided to determine whether there is a correlation between the distribution of V protein and the ability of V protein to interdict IFN signaling and which residues in the P/V common domain are important in interdicting IFN signaling.
FIG. 5.
Subcellular distribution and the ability to interdict IFN signaling of chimera V proteins. (A) Schematic diagram of the chimera V proteins. Shown is an illustration of the chimera protein described in the legend to Fig. 4A. The chimera proteins are named using amino acid numbers of the N terminus of hPIV4A/hPIV2 (PIV4-aa/2) or the amino acid numbers of substitution [PIV2+4(substitution number) or PIV4+2(substitution number)]. Subcellular distribution, inhibition of IFN signaling, and degradation of STAT2 are summarized on the right panel. (B) Subcellular distribution of chimera V proteins as detected by indirect immunofluorescence staining with a MAb against the Flag epitope. (C) Effects of V proteins on IFN-α-stimulated gene activation. A reporter gene assay with luciferase was performed as described in the legend to Fig. 4B. Chimera 1, PIV2; chimera 2, PIV4; chimera 3, PIV4-17/2; chimera 4, PIV4-31/2; chimera 5, PIV4-46/2; chimera 6, PIV4-114/2; chimera 7, PIV2+4(66-72); chimera 8, PIV4+2(46-88)/2; and chimera 9, PIV2+4(143-164). (D) STAT1 and STAT2 levels in HeLa cells constitutively expressing V proteins. Western blotting was performed as described in the legend to Fig. 4C. The asterisk on the right indicates a cross-reacting host band. Numbers under the figure correspond to chimera numbers.
To study the functional domains directing cytoplasmic accumulation of hPIV4 V protein, the various chimera V proteins were constructed as illustrated in Fig. 5A. These chimera V proteins were expressed by transfection to HeLa cells, and the distribution of the expressed proteins was analyzed by indirect immunofluorescence staining with monoclonal antibody to the N-terminal Flag epitope tag. As demonstrated in Fig. 5B, PIV2+4(66-72) V (chimera number 7) and PIV4+2(46-88)/2 V (chimera number 8) proteins were detected in the cytoplasm and nuclei, respectively, indicating that the amino acids 46 to 88, especially 66 to 72, of hPIV4 V protein play an important role in the cytoplasmic distribution.
We next examined the abilities of the various chimera V proteins to interdict IFN-induced signaling to an IFN-responsive reporter gene. As demonstrated in Fig. 5C, the protein with the 31-amino-acid replacement of the N-terminal portion of hPIV2 V with amino acids of the hPIV4A V protein (chimera number 4) kept the ability to interdict IFN signaling, whereas a protein with a further 14-amino-acid replacement (chimera number 5) showed no inhibitory effect on IFN-induced signaling (Fig. 5C). Interestingly, PIV2+4(143-164) V protein (chimera number 9), that is, the protein with aa 143 to 164 of hPIV4A V protein substituted for the corresponding region of hPIV2 V, could not block IFN-induced signaling (Fig. 5C). These findings suggest that at least two regions, that is, aa 32 to 45 and aa 143 to 164, of hPIV4 V are involved in the inability to evade IFN-induced signaling.
The PIV4-114/2 V protein (chimera number 6) that had a replacement of hPIV2 V aa 1 to 113 with those of the hPIV4A V protein demonstrated cytoplasmic distribution, and it lost the ability to interdict IFN signaling. Intriguingly, the PIV2+4(66-72) V protein (chimera number 7) that had a replacement of hPIV2 V aa 66 to 72 with those of hPIV4A V protein demonstrated cytoplasmic distribution, but it kept the ability to interdict IFN signaling. These data show that the distribution of the V protein is unrelated to the inhibition of the IFN signaling.
HeLa cell lines constitutively expressing various chimera proteins were analyzed to examine their effect on endogenous STAT1 and STAT2 protein levels (Fig. 5D). Expression of PIV4-17/2 (chimera number 3), PIV4-31/2 (chimera number 4), and PIV2+4(66-72) (chimera number 7) V proteins led to a loss of cellular STAT2 (Fig. 5D, lanes 3, 4, and 7). Subcellular distribution, inhibition of IFN signaling, and degradation of STAT2 are summarized on the right panel of Fig. 5A, showing that the protein with a replacement of the N-terminal 31 aa of hPIV2 V protein with those of the hPIV4 V protein retains the ability to block IFN signaling and that inhibition of IFN signaling occurs irrespective of the V protein distribution.
Amino acids 200 to 212 of the C terminus are important for the inability to block IFN-induced signaling and to degrade STATs.
As described above, the C-terminal 52 amino acids are very similar between hPIV2 and hPIV4 V proteins (Fig. 6A). To identify the functional region in the C terminus of hPIV4 V protein responsible for the inability to inhibit IFN-induced signaling, the various chimera V proteins were constructed as illustrated in Fig. 6B. PIV2-209/4 (chimera number 5) and PIV2+4(178-195) (chimera number 7) V proteins block the signaling (Fig. 6C, lanes 5 and 7), whereas PIV2-177/4 (chimera number 3), PIV2-195/4 (chimera number 4), and PIV2+4(196-208) (chimera number 8) V proteins do not (Fig. 6C, lanes 3, 4, and 8). These results suggest that the region of aa 200 to 212 of the hPIV4 V protein (corresponding to hPIV2 V aa 196 to 208) is involved in the inability to interdict IFN signaling. In addition, the region of aa 178 to 195 (corresponding to hPIV2 V aa 174 to 191) and the region of aa 209 to 222 (corresponding to hPIV2 V aa 204 to 218) of hPIV4 V protein are found to be functionally intact. We described previously that Phe 207 (F207) of hPIV2 V protein is important for the ability to block IFN-induced signaling (32). Amino acid 211 of hPIV4 V protein, which corresponds to hPIV2 V protein F207, is Glu. Therefore, we constructed the PIV2+4(196-208)+E211F V protein (Fig. 6B, chimera number 7), but this point mutation did not recover the ability to block IFN-induced signaling (Fig. 6C, lane 9). Noteworthy is that an extra cysteine was found in the region of aa 200 to 212. Thus, we also constructed PIV2+4(196-208)+C200R, but this mutant V protein has no effect on the ability (data not shown). These data suggested that some amino acids other than F211 and C200 in the region of aa 200 to 212 are important for the inability to block IFN-induced signaling. We also described previously that S216 of hPIV2 V protein is essential for STAT2 degradation (24). As shown in Fig. 6C and D, lane 5, PIV2-209/4 V protein (chimera number 5) interdicts the signaling and lowers the intracellular levels of STAT1. PIV2-209/4+K220S (chimera number 6) (corresponding to hPIV2 V S216) V protein also interdicts the signaling but lowers the STAT2 level. As summarized in Fig. 6B, these data show that the C-terminal 17 aa of hPIV4 V protein are substitutable for those of hPIV2 V protein, and the chimera hPIV2 V protein lowers the intracellular levels of STAT1, like SV5, SV41, and MuV V proteins.
FIG. 6.
The amino acids of the C-terminal region important for IFN-induced signaling and STAT degradation. (A) V-specific regions of hPIV2 and hPIV4A V proteins. The symbols below the sequence indicate the positions of conserved cysteine (asterisks) residues, tryptophan (or tyrosine) (squares) residues, and insertion of point mutations. (B) Schematic diagram of the chimera V proteins described in the legend to Fig. 4A. The chimera proteins are named for amino acid numbers of the N terminus of hPIV2/hPIV4A (PIV2-aa/4) or the amino acid number of the substitution [PIV2+4(substitution number)] or point mutation (+mutation). The asterisk indicates the position of the mutated residue. Inhibition of IFN signaling and degradation of STAT are summarized on the right panel. (C) Effects of V proteins on IFN-α-stimulated gene activation. A reporter gene assay with luciferase was performed as described in the legend to Fig. 4B. Chimera 1, PIV2; chimera 2, PIV4; chimera 3, PIV2-177/4; chimera 4, PIV2-195/4; chimera 5, PIV2-209/4; chimera 6, PIV2-209/4+K220S; chimera 7, PIV2+4(178-195); chimera 8, PIV2+4(196-208); and chimera 9, PIV2+4(196-208)+E211F. (D) STAT1 and STAT2 levels in HeLa cells constitutively expressing V proteins. Western blotting was performed as described in the legend to Fig. 4C. The asterisk on the right indicates a cross-reacting host band. Numbers under the figure correspond to chimera numbers.
hPIV4 cannot evade IFN-induced antiviral responses.
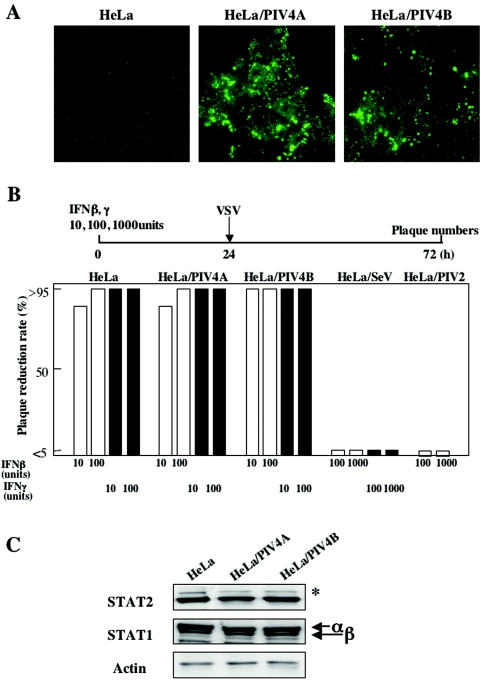
There is a possibility that hPIV4 has other functions to escape the IFN-induced antiviral responses. However, as hPIV4 virus growth is very limited, cells can't be infected with hPIV4 at high MOI. Thus, we established HeLa cells persistently infected with hPIV4A or hPIV4B (HeLa/PIV4A and HeLa/PIV4B cells). To confirm hPIV4 persistent infection, the expression of the hPIV4A or hPIV4B NP protein was detected using anti-hPIV4A NP MAb that was cross-reacted with hPIV4B NP protein (Fig. 7A).
FIG. 7.
Interferon susceptibility and STAT levels in HeLa cells persistently infected with hPIV4. (A) Detection of NP proteins in cloned cells persistently infected with hPIV4A or hPIV4B. Cells were immunostained with a MAb against the NP protein. (B) Interferon susceptibility was performed as described in the legend to Fig. 3B except that VSV was used as a challenge virus. HeLa/SeV or HeLa/PIV2 cells (HeLa cells persistently infected with Sendai virus or hPIV2 virus) were used as positive controls. (C) STAT1 and STAT2 levels. Western blotting was performed as described in the legend to Fig. 4C. The asterisk on the right indicates a cross-reacting host band.
Subsequently, we analyzed the susceptibility of the persistently infected cells to IFN. The cells were pretreated with 10, 100, or 1,000 U of IFN-β or IFN-γ for 24 h and were then infected with approximately 100 PFU of VSV. As shown in Fig. 7B, HeLa/SeV cells were resistant to 1,000 U of both IFNs, and HeLa/PIV2 cells were also resistant to 1,000 U of IFN-β. On the other hand, HeLa/PIV4A and HeLa/PIV4B cells were fully sensitive to both IFN-β and IFN-γ. In the HeLa/PIV4A and HeLa/PIV4B cells, a decrease of neither the STAT1 level nor the STAT2 level was observed (Fig. 7C). Furthermore, we established several other lines of HeLa cells persistently infected with hPIV4A or 4B. All of the cell lines are incapable of reducing the levels of STATs and can't evade 10 U IFNs (data not shown). These results show that hPIV4A and hPIV4B are not capable of inhibiting the IFN-induced antiviral responses.
DISCUSSION
The IFN-antagonistic functions of members of the subfamily Paramyxovirinae are associated with viral proteins encoded by the phosphoprotein (P) gene and in most cases are carried out by one of the accessory proteins, V or C. The C proteins of Sendai virus and hPIV3 (genus Respirovirus) counteract IFN signaling, whereas SV5, SV41, hPIV2, and MuV (genus Rubulavirus), NDV (genus Avulavirus), and measles virus (genus Morbillivirus) have been shown to counteract IFNs by using V protein. For Nipah virus (genus Henipavirus), all three proteins encoded by the P gene, V, W, and C proteins, have been shown to have IFN-antagonistic activity (37, 39, 40, 42). It has recently been shown that the SV5, hPIV2, and MuV V proteins bind DDB1 and Cullin4A, and this complex directly induces polyubiquitylation of STAT proteins (1, 49, 50). NDV, the nonmammalian virus, was previously classified as a Rubulavirus but has recently been reclassified in the new Avulavirus genus. It has also been shown that the expression of the NDV V protein degraded the STAT1 protein in 2fTGH and Vero cells (15). hPIV4 is a member of the Rubulavirus genus and expresses a V protein that is characterized by a highly conserved cysteine-rich C-terminal domain. Since the hPIV4 V protein binds STAT1, STAT2, DDB1, and Cullin4A, like other rubulavirus (SV5, hPIV2, and MuV) V proteins, we expected the hPIV4 V protein to have the ability to degrade STAT. Unexpectedly, the hPIV4 V protein has no ability to degrade STAT1 and STAT2. Furthermore, the hPIV4 V protein couldn't prevent STAT nuclear translocation or inhibit STAT phosphorylation. Consequently, we analyzed the biological activity of the hPIV4 V protein with respect to IFN-induced antiviral effect using Sindbis virus or rVSV-GFP. HeLa/FlagPIV4V cells were found to be fully sensitive to IFN-α, IFN-β, and IFN-γ. These results suggest that hPIV4 V protein has no ability to escape IFN via any mechanisms.
Among negative-strand RNA viruses, several different IFN-subverting strategies that target a variety of components of the IFN system have been identified. For example, the influenza virus NS1 protein and the Ebola virus VP35 protein prevent production of IFN by inhibiting the activation of the transcription factor IFN regulatory factor 3 (3, 9, 47). Respiratory syncytial virus, which encodes neither a C nor a V protein (genus Pneumovirus, subfamily Pneumovirinae, family Paramyxoviridae), targets the IFN production pathway through two nonstructural proteins, NS1 and NS2 (4, 41, 43, 51). We next tested whether hPIV4 has an IFN-antagonistic function through another protein(s). As hPIV4 virus growth is very limited, and it is not possible to infect at a high MOI, we established HeLa cells persistently infected with hPIV4 and analyzed the biological activity with respect to IFN signaling inhibition using VSV. HeLa/PIV4A and HeLa/PIV4B cells were found to be sensitive to IFN-β and IFN-γ and kept the levels of endogenous STAT1 and STAT2. These results suggest that hPIV4 does not have any IFN-subverting strategies. The inability to evade IFN may be related to the limited replication of hPIV4.
In this study, we constructed a series of chimera V proteins to identify the region of hPIV4 V protein responsible for the inability to evade IFN. Both the P/V common domain and the V-specific domain of hPIV4 V were found to be involved in the inability to evade the IFN response. We further demonstrated that the hPIV4 and hPIV2 V proteins have at least four functionally interchangeable regions: two are located in the P/V common region of the hPIV4 V protein, aa 1 to 31 and 66 to 72, and the other two are located in the V-specific region, aa 178 to 199 and the C-terminal 17 amino acids. In addition, it is inferred that at least two regions (aa 32 to 45 and aa 143 to 164) of the hPIV4 V protein in the P/V common domain and one region (aa 200 to 212) of the C terminus are involved in the inability to evade IFN-induced signaling. Andrejeva et al. (1) reported that an SV5 V protein with a deletion of the first 85 amino acids at the N terminus and the V protein of CPI- (which is a strain of SV5 with mutations at the N terminus) failed to bind with DDB1. Both hPIV4 and hPIV2 V proteins can bind with DDB1, but replacement of only the N-terminal 31 amino acids of hPIV2 V proteins with those of hPIV4 allows the protein to retain the ability to degrade STAT. Therefore, a large part of the N terminus of V protein is required for STAT degradation, not only for binding with DDB1. The P/V common region of hPIV4A shows 15.6, 13.0, 16.2, and 15.6% homologies with those of hPIV2, MuV, SV5, and NDV, respectively. The P/V common region of hPIV2 shows 26.6, 31.8, and 16.2% homologies with those of MuV, SV5, and NDV, respectively (22). The divergence between hPIV4 and hPIV2 V proteins is similar to that between hPIV2 and NDV V proteins. Therefore, it is very strange that only hPIV4 V protein has no function for STAT degradation. We previously reported that not only seven conserved cysteine residues and a Trp-rich motif (W/Y-X3-W-X9-W, located just upstream of the Cys cluster) but also the Phe 207 residue within the V-specific region are important for STAT degradation (32). The hPIV4 V protein has seven conserved cysteine residues and a Try-rich motif (within aa 178 to 199) but not Phe 211 (corresponding to hPIV2 V Phe 207). Furthermore, the hPIV4 V protein has one more Cys 200 which is located in the region aa 200 to 212 involved in inability to interdict IFN signaling. As PIV2+4(196-208)+E211F and PIV2+4(196-208)+C200R V proteins failed to lower STAT protein levels, these results indicate that not only Phe 211 but also some residues within aa 200 to 212 are important for V function. The C-terminal 17 amino acids are replaceable between hPIV2 and hPIV4 V proteins, and the hPIV2 V protein and the chimeric hPIV2 with a replacement of 17 amino acids of hPIV4 V protein lower the levels of STAT2 and STAT1, respectively. K220 of hPIV4 V protein and S216 of hPIV2 V protein determine STAT tropism. This result is therefore in agreement with those recently published by Kozuka et al. (24), who came to a similar conclusion by using various chimeric proteins between the hPIV2 and SV41 V proteins.
The accessory protein of paramyxovirus is a multifunctional protein. For SeV, the C protein acts in a positive manner in promoting early steps in intracellular replication when it is present at low concentration (27). The SV5 V protein slows progression of the cell cycle (28). The changes in the cell cycle did not occur in the cells expressing the SV5 P protein or the V protein lacking its unique C terminus. The changes in the cell cycle can be partially restored by coexpression of DDB1 (28). Moreover, the SV5 and NDV V proteins play an important role in blocking apoptosis (36, 44). hPIV2 V protein is important in promoting virus growth but also in anti-IFN activity (32). The other function(s) of hPIV4 V protein thus remain to be investigated.
Acknowledgments
This study was supported in part by a Grant-in-aid for Scientific Research from the Ministry of Education, Science, Culture, Sports, and Technology of Japan.
REFERENCES
- 1.Andrejeva, J., E. Poole, D. F. Young, S. Goodbourn, and R. E. Randall. 2002. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 76:11379-11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bando, H., K. Kondo, M. Kawano, H. Komada, M. Tsurudome, M. Nishio, and Y. Ito. 1990. Molecular cloning and sequence analysis of human parainfluenza type 4A virus HN gene: its irregularities on structure and activities. Virology 175:307-312. [DOI] [PubMed] [Google Scholar]
- 3.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossert, B., and K. K. Conzelmann. 2002. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: a chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol. 76:4287-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canchola, J., A. J. Vargosco, H. W. Kim, R. H. Parrott, E. Christmas, B. Jeffries, and R. M. Chanock. 1964. Antigenic variation among newly isolated strains of parainfluenza type 4 virus. J. Hyg. 79:357-364. [DOI] [PubMed] [Google Scholar]
- 7.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for protease-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 10.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcin, D., J. B. Marq, L. Strahle, P. Le Mercier, and D. Kolakofsky. 2002. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295:256-265. [DOI] [PubMed] [Google Scholar]
- 13.Gotoh, B., K. Takeuchi, T. Komatsu, and J. Yokoo. 2003. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J. Virol. 77:3360-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gotoh, B., K. Takeuchi, T. Komatsu, J. Yokoo, Y. Kimura, A. Kurotani, A. Kato, and Y. Nagai. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459:205-210. [DOI] [PubMed] [Google Scholar]
- 15.Huang, Z., S. Krishnamurthy, A. Panda, and S. K. Samal. 2003. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J. Virol. 77:8676-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, M., T. Takeuchi, M. Nishio, M. Kawano, H. Komada, M. Tsurudome, and Y. Ito. 2004. Early stage of establishment of persistent Sendai virus infection: unstable dynamic phase and then selection of viruses which are tightly cell associated, temperature sensitive, and capable of establishing persistent infection. J. Virol. 78:11939-11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting virus RNA synthesis. J. Virol. 75:3802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komada, H., H. Bando, M. Ito, H. Ohta, M. Kawano, M. Nishio, M. Tsurudome, N. Watanabe, N. Ikemura, S. Kusagawa, X. Mao, M. O'Brien, and Y. Ito. 1995. Sequence analyses of human parainfluenza virus type 4A and type 4B fusion proteins. J. Gen. Virol. 76:3205-3210. [DOI] [PubMed] [Google Scholar]
- 19.Komada, H., M. Tsurudome, M. Ueda, M. Nishio, H. Bando, and Y. Ito. 1989. Isolation and characterization of monoclonal antibodies to human parainfluenza virus type 4 and their use in revealing antigenic relation between subtypes 4A and 4B. Virology 171:28-37. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu, T., K. Takeuchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 74:2477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo, K., H. Bando, M. Kawano, M. Tsurudome, H. Komada, M. Nishio, and Y. Ito. 1990. Sequence analyses and comparison of human parainfluenza type 4A and 4B NP protein genes. Virology 174:1-8. [DOI] [PubMed] [Google Scholar]
- 22.Kondo, K., H. Bando, M. Tsurudome, M. Kawano, M. Nishio, and Y. Ito. 1990. Sequence analysis of the phosphoprotein (P) genes of human parainfluenza type 4A and 4B viruses and RNA editing at transcript of the P genes: the number of G residues added is imprecise. Virology 178:321-326. [DOI] [PubMed] [Google Scholar]
- 23.Kondo, K., M. Fujii, T. Nakamura, H. Bando, M. Kawano, M. Tsurudome, H. Komada, S. Kusakawa, M. Nishio, and Y. Ito. 1991. Sequence characterization of the matrix protein genes of parainfluenza virus types 4A and 4B. J. Gen. Virol. 72:2283-2287. [DOI] [PubMed] [Google Scholar]
- 24.Kozuka, Y., Y. Yamashita, M. Kawano, M. Tsurudome, M. Ito, M. Nishio, H. Komada, and Y. Ito. 2003. Identification of amino acids essential for the human parainfluenza type 2 virus V protein to lower the intracellular levels of the STAT2. Virology 317:208-219. [DOI] [PubMed] [Google Scholar]
- 25.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 26.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 27.Latorre, P., T. Cadd, M. Ito, J. Curran, and D. Kolakofsky. 1998. The various Sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection. J. Virol. 72:5984-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 30.Malur, A. G., S. Chattopadhyay, R. K. Maitra, and A. K. Banerjee. 2005. Inhibition of STAT1 phosphorylation by human parainfluenza virus type 3 C protein. J. Virol. 79:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishio, M., D. Garcin, V. Simonet, and D. Kolakofsky. 2002. The carboxyl segment of the mumps virus V protein associates with Stat proteins in vitro via a tryptophan-rich motif. Virology 300:92-99. [DOI] [PubMed] [Google Scholar]
- 32.Nishio, M., M. Tsurudome, M. Ito, D. Garcin, D. Kolakofsky, and Y. Ito. 2005. Identification of paramyxovirus V protein residues essential for STAT protein degradation and promotion of virus replication. J. Virol. 79:8591-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palosaari, H., J.-P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 77:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 36.Park, M. S., A. Garcia-Sastre, J. F. Cros, C. F. Basler, and P. Palese. 2003. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 77:9522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez, J. J., J.-P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez, J. J., L. F. Wang, and C. M. Horvath. 2003. Hendra virus V protein inhibits interferon signaling by preventing STAT1 and STAT2 nuclear accumulation. J. Virol. 77:11842-11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw, M. L., A. Garcia-Sastre, P. Palese, and C. F. Basler. 2004. Nipah virus V and W proteins have a common STAT-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78:5633-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spann, K. M., K. C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 78:4363-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun, M., T. A. Rothermel, L. Shuman, J. A. Aligo, S. Xu, Y. Lin, R. A. Lamb, and B. He. 2004. Conserved cysteine-rich domain of paramyxovirus simian virus 5 V protein plays an important role in blocking apoptosis. J. Virol. 78:5068-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi, K., S. I. Kadota, M. Takeda, N. Miyajima, and K. Nagata. 2003. Measles virus V protein blocks interferon (IFN)-alpha/beta but not IFN-gamma signaling by inhibiting STAT1 and STAT2 phosphorylation. FEBS Lett. 545:177-182. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi, K., T. Komatsu, J. Yokoo, A. Kato, T. Shioda, Y. Nagai, and B. Gotoh. 2001. Sendai virus C protein physically associates with Stat1. Genes Cells 6:545-557. [DOI] [PubMed] [Google Scholar]
- 47.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reference deleted.
- 49.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 50.Ulane, C. M., J. J. Rodriguez, J.-P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 77:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valarcher, J. F., J. Furze, S. Wyld, R. Cook, K. K. Conzelmann, and G. Taylor. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 77:8426-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe, N., M. Kawano, M. Tsurudome, S. Kusagawa, M. Nishio, H. Komada, T. Shima, and Y. Ito. 1996. Identification of the sequences responsible for nuclear targeting of the V protein of human parainfluenza type 2 virus. J. Gen. Virol. 77:327-338. [DOI] [PubMed] [Google Scholar]
- 53.Yokosawa, N., S. Yokota, T. Kubota, and N. Fujii. 2002. C-terminal region of STAT-1α is not necessary for its ubiquitination and degradation caused by mumps virus V protein. J. Virol. 76:12683-12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]