Abstract
Host cell factors modulate retroviral infections. Among those, cyclophilin A (CypA) promotes virus infectivity by facilitating virus uncoating or capsid unfolding or by preventing retroviral capsid interaction with cellular restriction factors. In Aotus species, a retrotransposed copy of CypA inserted into the tripartite motif 5 (TRIM5) gene encodes a fusion protein which may block human immunodeficiency virus type 1 by targeting the incoming virus to ubiquitin-ligated degradation or by interfering with normal uncoating of the incoming particle, rendering those monkeys resistant to infection. In this study, we have extensively analyzed representative specimens from all New World primate genera and shown that the retrotransposed CypA copy is only present in Aotus. We have shown that this inserted copy diverged from its original counterpart and that this occurred prior to Aotus radiation, although no positive selection was observed. Finally, our data underscores the need for a precise taxonomic identification of primate species used as models for retroviral infections and novel antiviral approaches.
The zoonotic events involved in the transmission of human immunodeficiency virus type 1 (HIV-1) and HIV-2 from chimpanzees and sooty mangabeys to humans demonstrate the ability of lentiviruses in crossing species barriers (21). Although numerous examples of retroviral transmission across species have been reported to date (5, 13, 23), retroviral host ranges, in natura, are limited by restriction or enhancing factors in new hosts, which may block or promote the early steps of the life cycle of retroviruses inside cells (6). Some of these blocking factors include members of the tripartite motif 5 (TRIM5) protein family (25), contrary to factors that enhance infection, as is the case of cyclophilin A (CypA) (22). TRIM5 and CypA interact with the capsid (CA) portion of the Gag protein (3, 20). It has been suggested that CypA may facilitate disassembling of the virus capsid for reverse transcription (1). Conversely, one of the TRIM5 isoforms, TRIM5α, may target viral capsid to proteasome degradation before nuclear import or interfere with the uncoating of the incoming virus, while it has been postulated that CypA may promote infection by preventing the association of TRIM5α with the viral capsid (6).
An increasing amount of data on lentiviral infectivity has been provided by the lentiviral modulating factors of New World Primates (NWP), a group that, apparently, does not harbor lentiviruses in natura. NWP cells are only moderately infected by HIV-1 and SIVmac; owl monkey (Aotus trivirgatus) cells are not infected by HIV-1, while squirrel monkey cells are resistant to SIVmac (12). More recently, HIV-1 blockage in Aotus cells was explained by presence of a complete, in-frame insertion of CypA cDNA, between exons 7 and 8, in the TRIM5 locus originating a chimeric gene encoding a TRIM-CypA fusion protein (14, 18) capable of blocking HIV-1 replication in Aotus cells and in human cells transiently expressing its coding cDNA (14).
Although it has been postulated that this CypA insertion must have occurred by a transposition event that took place after the divergence of NWP from the Old World Primates (OWP) (18), the precise origin and timing of this event could not be traced along the NWP phylogeny because this chimeric gene has not been investigated in representative species of all NWP genera. Moreover, the extent of this retrotransposition in congeneric Aotus species and within species has not been analyzed. In fact, Aotus is a complex genus that was initially considered to be monotypic until Hershkovitz (9) recognized nine allopatric species divided into two groups: the gray-neck species group with A. brunbacki, A. trivirgatus, A. vociferans, and A. lemurinus (with two subspecies, A. lemurinus lemurinus and A. lemurinus griseimembra), and the red-neck group with A. nancymae, A. miconax, A. infulatus, A. azarae (with two subespecies, A. azarae azarae and A. azarae boliviensis), and A.nigriceps. Later, Groves (7) recognized a new species, A. hershkovitzi, belonging to the gray-neck species group.
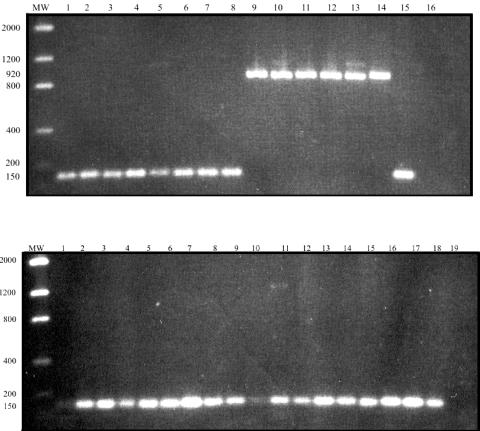
In this study, we analyzed representatives of all NWP genera, including representatives of several taxonomically characterized Aotus species of known geographic origin which were wild caught in different regions. For detecting the CypA insert, a single-round PCR was designed, resulting in fragments of different size from those observed when the insert was absent (Fig. 1). This was carried out with a forward primer (5′CAACGCTACTGGGGTAAGGAGA3′) annealing at the 3′ end of TRIM5 exon 7, and a reverse primer (5′CATGTTTTAAGATTTATATTTCTTCTTC3′) annealing at the 5′end of trim5 exon 8. When the CypA insert was present, a fragment of approximately 900 bp (corresponding to cyclophilin A cDNA) was amplified, while a smaller fragment of approximately 100bp was observed in agarose gels when the insert was not present. When studying representatives of all NWP genera (Table 1), the CypA insert was detected in all specimens of Aotus (A. infulatus, A. azarae, A. trivirgatus, and A. lemurinus) but was absent in the representatives of all other 15 genera of NWP. Identification of CypA inserts was confirmed by two-strand, direct DNA sequencing; reactions were carried out with the same primers used in the PCR assay and two internal primers (F1, 5′CAACGCTACTGGGGTAAGGAGA3′, and F2, 5′CATGTTTTAAGA TTTATATTTCTTCTTC3′). Samples were run in an ABI 377 automated DNA sequencer (Applied Biosystems, Foster City, CA); sequences were edited, translated into predicted amino acid sequences, and aligned with ClustalW (10).
FIG. 1.
PCR amplification of TRIM-CypA retrotransposition of representative New World monkey genera. Top panel, lanes: 1, Saimiri ustus; 2, Saimiri sciureus; 3, Cebus sp.; 4, Cebus sp.; 5, Cebus albifrons; 6, Cebus apella apella; 7, Cacajao melanocephalus; 8, Brachyteles arachnoides; 9, Aotus infulatus; 10, Aotus azarae boliviensis; 11, Aotus trivirgatus; 12, Aotus infulatus; 13, Aotus trivirgatus; 14, Aotus azarae; 15, Homo sapiens; 16, negative control. MW, molecular weight marker. Bottom panel, lanes: 1, Callithrix penicillata; 2, Callithrix geoffroyi; 3, Callithrix jacchus; 4, Callithrix kuhli; 5, Callithrix emiliae; 6, Callithrix argentata; 7, Callithrix humeralifer; 8, Callithrix (Cebuella) pygmaea; 9, Leontopithecus chrysopygus; 10, Leontopithecus rosalia; 11, Leontophitecus chrysomelas; 12, Saguinus fuscicollis; 13, Saguinus imperator; 14, Saguinus bicolor; 15, Alouatta seniculus; 16, Callicebus moloch; 17, Callicebus personatus; 18, Callicebus torquatus; 19, negative control.
TABLE 1.
New World monkey species analyzed for TRIMCyp retrotransposition
| Species | No. of specimens analyzed/species |
|---|---|
| 1. Callithrix penicillata | 3 |
| 2. Callithrix geoffroyi | 4 |
| 3. Callithrix jacchus | 2 |
| 4. Callithrix kuhli | 4 |
| 5. Callithrix emiliae | 3 |
| 6. Callithrix argentata | 1 |
| 7. Callithrix humeralifer | 4 |
| 8. Callithrix (Cebuella) pygmaea | 1 |
| 9. Leontopithecus chrysopygus | 3 |
| 10. Leontopithecus rosalia | 3 |
| 11. Leontophitecus chrysomelas | 4 |
| 12. Saguinus fuscicollis | 1 |
| 13. Saguinus imperator | 1 |
| 14. Saguinus bicolor | 1 |
| 15. Callimico goeldii | 1 |
| 16. Cebus sp. | 2 |
| 17. Cebus albifrons | 2 |
| 18. Cebus apella apella | 2 |
| 19. Saimiri ustus | 3 |
| 20. Saimiri sciureus | 2 |
| 21. Aotus infulatus | 2 |
| 22. Aotus azarae boliviensis | 3 |
| 23. Aotus trivirgatus | 2 |
| 24. Aotus lemurinus | 4 |
| 25. Callicebus personatus | 2 |
| 26. Callicebus torquatus | 2 |
| 27. Callicebus moloch | 2 |
| 28. Pithecia irrorata | 3 |
| 29. Chiropotes sp. | 1 |
| 30. Cacajao melanocephalus | 1 |
| 31. Alouatta seniculus | 1 |
| 32. Ateles paniscus | 1 |
| 33. Brachyteles arachnoides | 2 |
| 34. Lagothrix lagothrica | 1 |
| 35. Homo sapiensa |
Negative control.
Nucleotide sequence accession numbers.
All nucleotide sequences were deposited in the GenBank database and were assigned the accession numbers DQ098851 to DQ098874.
Few amino acid substitutions were observed when comparing the transposed and nontransposed CypA sequences of NWP with the non-transposed sequences of OWP, indicating a very strong evolutionary conservation of this protein in the primate order. Interestingly, all Aotus CypA proteins showed an E84D substitution and lacked the last amino acid (E165) although we do not know whether these changes might represent signatures of Aotus CypA proteins or of all NWP genera. Other substitutions, like V29L, R69H and E81V, were also observed in several owl monkeys, with the exception of the previously reported A. trivirgatus sequences (AY646198, AY646199 and AY684997) and of A. lemurinus characterized herein. In fact, changes V2I and V20I were present only in specimens identified as A. trivirgatus sensu Hershkovitz (9), with a diploid chromosome number of 2n = 50, and captured in a locality within the geographic distribution of this species. Most interestingly, however, were changes V117A, I138V, S147C, and N149Y, which were present in all sequences derived from the retrotransposed CypA gene, but not in the sequence derived from the mRNA of the original (nontransposed) CypA gene of Aotus trivirgatus (Fig. 2). This finding confirmed a previous report showing sequence differences between the CypA gene and the retrotransposed insert of a same cell line (14), indicating that the retrotransposed gene evolved subsequently to the insertion event. Among the retrotransposed CypA substitutions, S147C created a new cysteine residue, which could alter potential intra- or intermolecular disulfide bonds. All amino acid residues previously associated in interactions with HIV-1 CA (H54, R55, N71, N102, and H126) and with cyclosporine (W121) (2, 4) were strictly conserved in all primate species, suggesting that all retrotransposed sequences are biologically functional.
FIG. 2.
Amino acid alignment of CypA proteins derived from sequences of original or retrotransposed CypA copies from New World monkeys (NWM) and representative species of OWP. Dots represent amino acid identities, whereas asterisks denote stop codons. Residues that are conserved among Aotus species (or NWM) are boxed in white, whereas residues which differ between the retrotransposed and the original CypA copy are boxed in gray. Residues which are known to interact with HIV-1 capsid or with cyclosporine are depicted by arrows.
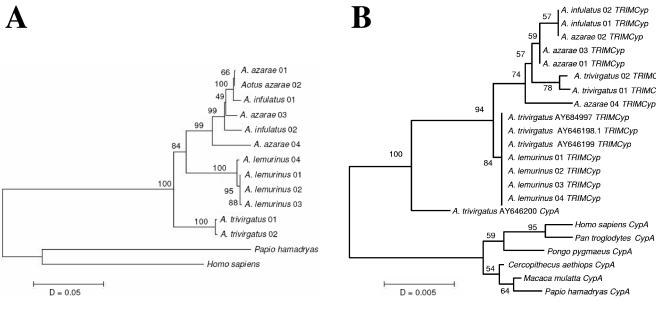
A phylogenetic CypA topology using six OWP sequences as outgroups, and including previous data on Aotus CypA, did not group specimens accordingly to their taxonomic status or geographic distribution. Retrotransposed CypA genes were tightly grouped in a well-supported clade divided in two branches. The divergence of the upper branch clearly indicated that A. infulatus specimens captured in Tucurui (3°45′S, 49°40′W) and A. azarae boliviensis specimens captured in the Samuel dam reservoir (8°45′S, 63°26′W) were clearly paraphyletic despite the fact that these animals were captured in regions that are very distant from one another. Moreover, two specimens of A. trivirgatus sensu Hershkovitz (9), captured north of the Rio Negro (Brazil), were also included in the upper branch. Conversely, the lower (retrotransposed) CypA branch showed seven collapsing, identical branches. These comprised (i) the previously reported cDNA insert of A. trivirgatus (AY646198); (ii) the previously reported A. trivirgatus insert (AY646199) isolated from genomic DNA of the same animal; (iii) the previously reported A.trivirgatus insert (AY684997) isolated from DNA of a kidney cell line (OMK); and (iv) the CypA sequences of four A. lemurinus from Colombia herein analyzed.
The possibility of positive selection at differing sites was tested using the CODEML program of the PAML package (24). Three alternative topologies were constructed using the OWP topology of Page and Goodman (15) and the parsimony topologies obtained with selected Aotus cyclophilin A sequence data (excluding specimens with identical sequences): 1- (((((Homo sapiens, Pan troglodytes), Pongo pygmaeus), (Papio hamadryas, Macaca mulatta)), Cercopithecus aethiops), (A. trivirgatus-AY646200 CypA mRNA, (A. trivirgatus AY684997-TRIMCyp, (A. azarae 04-TRIMCyp, ((A. infulatus 02-TRIMCyp, A. infulatus 01-TRIMCyp), (A. trivirgatus 01-TRIMCyp, A. trivirgatus 02-TRIMCyp)))))); 2- (((((Homo sapiens, Pan troglodytes), Pongo pygmaeus), (Papio hamadryas, Macaca mulatta)), Cercopithecus aethiops), (A. trivirgatus -AY646200 CypA mRNA, (A. trivirgatus AY684997-TRIMCyp, (A. azarae 04-TRIMCyp, A. infulatus 02-TRIMCyp, A. infulatus 01-TRIMCyp, (A. trivirgatus 01-TRIMCyp, A. trivirgatus 02-TRIMCyp))))); 3- (((((Homo sapiens, Pan troglodytes), Pongo pygmaeus), (Papio hamadryas, Macaca mulatta)), Cercopithecus aethiops), (A. trivirgatus -AY646200 CypA mRNA, (A. trivirgatus AY684997-TRIMCyp, ((A. azarae 04-TRIMCyp, A. infulatus 02-TRIMCyp, A. infulatus 01-TRIMCyp), (A. trivirgatus 01-TRIMCyp, A. trivirgatus 02-TRIMCyp))))). These topologies were tested assuming the same nonsynonymous/synonymous (dN/dS) substitution ratio for all branches (one-ratio model) and different dN/dS for branches (free-ratio model). No significant differences were observed between likelihood values obtained under these models, indicating absence of positive selection. Additionally, no evidence of positive selection for individual codon sites was found with CODEML using different codon substitution models (M0, M1, M2a, M3, M7, and M8).
Phylogenetic topologies relating the four Aotus species herein analyzed were clearly different when analyzing cytochrome b (1,140 bp) because a distance tree showed that the A. trivirgatus sensu Hershkovitz (9) were included in the most basal offshoot, with a sister branch splitting in a dichotomy, comprising A. lemurinus in one lineage and A. infulatus/ A. azarae boliviensis in another (Fig. 3). These two species, although strongly grouped, showed a paraphyletic arrangement, indicating the presence of a trans-specific, ancestral cytochrome b polymorphism. It is noteworthy that A. infulatus and A. azarae boliviensis were found to be karyotypically identical, and that their taxonomic status, as separate species, has been questioned (16, 17). This topology clearly indicated, however, that A. trivirgatus, A. lemurinus and A. infulatus/A. azarae boliviensis comprise three separate evolutionary lineages distributed in different geographic regions of South America. Distance estimates, considering synonymous substitutions, allowed for calculating the time when these Aotus lineages diverged from one another in approximately 4.5 million years before present (MYBP), in respect with human-chimpanzee synonymous cytochrome b substitutions and a divergence time of 6 MYBP.
FIG. 3.
Neighbor-joining nucleotide trees of New World monkey cytochrome b (A) and CypA/TRIM-CypA (B). Bootstrap values are based on 1,000 replicates. Representative species of OWP were used as outgroups.
The discordant CypA and cytochrome b topologies clearly rule out that these sequences have co-evolved in the Aotus genome. In fact, the subfamilies Cebinae and Aotinae (sensu Schneider [19]) split some 22 MYBP, which represents the upper time limit when the CypA retrotransposition might have occurred contrary to a lower limit of 4.5 MYBP. Once inserted, it is apparent that nucleotide substitutions at the CypA insert must have taken place before Aotus radiation, representing an ancestral polymorphism unrelated to the divergence of the three evolutionary lineages shown by the cytochrome b topology. This could also be due to the fact that mitochondrial DNA is more prone to be evolved by genetic drift than to the fact that it is maternally transmitted (11).
In summary, we have demonstrated that Aotus is the only genus of NWP harboring a CypA retrotransposition in TRIM5. Our results also indicate that the CypA retrotransposed copy present in Aotus has evolved subsequently to this insertion but prior to Aotus radiation. Although no positive selection was detected in the CypA gene as a whole or at specific codons, it is possible that ancient retroelements other than lentiviruses (such as endogenous retroviruses or retrotransposons) might have selected for amino acid substitutions seen today when comparing the original and the retrotansposed CypA copies. In addition, our data highlight the importance of an accurate classification of Aotus species in the study of restriction factors. Although it has been stated that Aotus trivirgatus is particularly resistant to HIV-1 infection (12), the specimens analyzed inthis report grouped with A. lemurinus rather than with A.trivirgatus sensu Hershkovitz (9). It is noteworthy that the striking variability among Aotus species accounts for different susceptibilities to the development of malaria upon infection by different Plasmodium variants (8). Therefore, a precise characterization of primate species with distinctive susceptibility patterns to lentiviruses will enable a faster and better understanding of the processes controlling retroviral restriction and their application in antiviral strategies.
Acknowledgments
This work was supported by INCA, Fundação Ary Frauzino, UFRJ, and CNPq grants 476155/2003-7 and 470290/2004-8 (Brazil).
REFERENCES
- 1.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braaten, D., H. Ansari, and J. Luban. 1997. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 71:2107-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colgan, J., H. E. Yuan, E. K. Franke, and J. Luban. 1996. Binding of the human immunodeficiency virus type 1 Gag polyprotein to cyclophilin A is mediated by the central region of capsid and requires Gag dimerization. J. Virol. 70:4299-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorfman, T., A. Weimann, A. Borsetti, C. T. Walsh, and H. G. Gottlinger. 1997. Active site residues of cyclophilin A are crucial for its incorporation into human immunodeficiency virus type 1 virions. J. Virol. 71:7110-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 6.Goff, S. P. 2004. Genetic control of retrovirus susceptibility in mammalian cells. Annu. Rev. Genet. 38:61-85. [DOI] [PubMed] [Google Scholar]
- 7.Groves, C. P. 1993. Order Primates, p. 243-277. In D. E. Wilson and D. M. Reeder (ed.), Mammal species of the world: a taxonomic and geographic reference. Smithsonian Institution Press, Washington, D.C.
- 8.Herrera, S., B. L. Perlaza, A. Bonelo, and M. Arévalo-Herrera. 2002. Aotus monkeys: their great value for anti-malaria vaccines and drug testing. Int. J. Parasitol. 32:1625-1635. [DOI] [PubMed] [Google Scholar]
- 9.Hershkovitz, P. 1983. Two new species of night monkeys, genus Aotus (Cebidae, Platyrrhini): a preliminary report on Aotus taxonomy. Am. J. Primatol. 4:209-243. [DOI] [PubMed] [Google Scholar]
- 10.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoelzer, G. A. 1997. Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene revisited. Evolution 51:622-626. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin, M. J., J. Rogers, J. E. Phillips-Conroy, J. S. Allan, R. C. Desrosiers, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J. Virol. 68:8454-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page, S. L., and M. Goodman. 2001. Catarrhine phylogeny: noncoding DNA evidence for a diphyletic origin of the mangabeys and for a human-chimpanzee clade. Mol. Phylogenet. Evol. 18:14-25. [DOI] [PubMed] [Google Scholar]
- 16.Pierczarka, J. C., and C. Y. Nagamachi. 1988. Cytogenetic studies of Aotus from Eastern Amazonia: Y/autosome rearrangement. Am. J. Primatol. 14:255-263. [DOI] [PubMed] [Google Scholar]
- 17.Pieczarka, J. C., R. M. de S. Barros, F. M. Faria, and C. Y. Nagamachi. 1993. Aotus from the Southwestern Amazon region is geographically and chromosomally intermediate between A. azarae boliviensis and A. infulatus. Primates 34:197-204. [Google Scholar]
- 18.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 19.Schneider, H., F. C. Canavez, I. Sampaio, M. A. M. Moreira, C. H. Tagliaro, and H. N. Seuánez. 2001. Can. molecular data place each neotropical monkey in its own branch? Chromosoma 109:515-523. [DOI] [PubMed] [Google Scholar]
- 20.Sebastian, S., and J. Luban. 2005. Trim5a selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp, P. M., E. Bailes, R. R. Chaudhuri, C. M. Rodenburg, M. O. Santiago, and B. H. Hahn. 2001. The origins of acquired immune deficiency syndromeviruses: where and when? Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:867-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sokolskaja, E., D. M. Sayah, and J. Luban. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 78:12800-12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Rensburg, E. J., S. Engelbrecht, J. Mwenda, J. D. Laten, B. A. Robson, T. Stander, and G. K. Chege. 1998. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: detection of a SIVagm variant from a chacma baboon. J. Gen. Virol. 79:1809-1814. [DOI] [PubMed] [Google Scholar]
- 24.Yang, Z. 1997. PALM: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biol. Sci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 25.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]