Abstract
Resistance determinants that interfere with normal physiological processes in the bacterial cell usually cause a reduction in biological fitness. Fitness assays revealed that 17 of 18 in vitro-selected chromosomal mutations within the rpoB gene accounting for rifampin resistance in Staphylococcus aureus were associated with a reduction in the level of fitness. There was no obvious correlation between the level of resistance to rifampin and the level of fitness loss caused by rpoB mutations. Among 23 clinical rifampin-resistant S. aureus isolates from six countries, only seven different rpoB genotypes could be identified, whereby the mutation 481His→Asn was present in 21 (91%) of these 23 isolates. The mutation 481His→Asn, in turn, which confers low-level rifampin resistance on its own, was not shown to be associated with a cost of resistance in vitro. The restriction to distinct mutations that confer rifampin resistance in vivo, as demonstrated here, appears to be determined by the Darwinian fitness of the organisms.
The selective pressure of antibiotics in general results in the emergence of resistance mechanisms on the part of bacteria (12). Hence, a direct correlation often exists between antibiotic use and the development of bacterial resistance to antibiotics over time (3). Conversely, the incidence of antibiotic resistance can be reduced by the restriction of antibiotic usage (22). This interrelation between antibiotic usage and resistance development is partly determined by the so-called biological cost of antibiotic resistance (11, 16). Since antibiotic resistance often is characterized by detriments in biological fitness, resistant bacteria can be decimated selectively by a decreased use of antibiotics (10). It appears, however, that resistance never completely disappears. This feature may be owing to the fact that the loss of biological (Darwinian) fitness on the part of resistant bacteria can easily be overcome by the acquisition of compensatory mutations, thereby stabilizing the resistant bacteria within a given population (8, 16, 21).
Antibiotic resistance due to chromosomal mutations that cause structural modifications in the cellular target of the drug, such as rifampin resistance in Staphylococcus aureus, can be associated with a fitness burden. The present study was aimed at analyzing (i) the biological cost of RNA polymerase (rpoB) mutations conferring rifampin resistance on S. aureus, (ii) the relationship between the cost of rpoB mutations and the level of resistance to rifampin, and (iii) the distribution of rpoB mutations in vivo in view of the possible fitness burden associated with resistance.
MATERIALS AND METHODS
Bacteria.
Fifty-four S. aureus isolates were analyzed, including 23 isolates from six countries that were rifampin resistant (Rifr) in vivo as well as 30 mutants derived from a single susceptible isogenic reference strain that were Rifr in vitro. Twenty-six of the Rifr in vitro mutants were selected by culturing a rifampin-susceptible (Rifs) strain on agar containing 0.25 μg of rifampin per ml. The other four Rifr in vitro mutants were obtained by culturing mutant 1 on agar containing 10 μg of rifampin per ml.
Genotyping.
All clinical MRSA isolates were analyzed by pulsed-field gel electrophoresis (PFGE) as described previously (24). Mutations conferring rifampin resistance were determined by partial DNA sequencing of the rpoB gene, including resistance clusters I and II, as described previously (25).
Fitness assay.
The relative fitnesses of the Rifs parental strain and the Rifr in vitro mutants that evolved were determined by paired competition experiments. The parental strain and the Rifr mutant strain were grown to an optical density at 600 nm of 0.6; 1 ml of each culture was mixed and diluted 10−5 in 5 ml of Luria-Bertani medium to create a mixed culture of Rifr and Rifs bacteria that was incubated at 37°C for 20 h. The number of viable Rifr and Rifs cells was determined at the beginning and end of the experiment by plating aliquots of the culture on medium containing either 0.125 μg of rifampin per ml or no drug. The number of generations (G) of the mutant and isogenic parental strain that occurred in the mixed broth was calculated as described by Billington et al. (4): G = (log B − log A)/log 2, where A is the number of CFU per milliliter at time zero and B is the number of CFU per milliliter at the end of the culture period. The relative fitness of each strain was determined from the ratio of the number of generations from Rifr to Rifs strain.
Statistics.
Statistical analysis was performed by the one-sample t test.
RESULTS
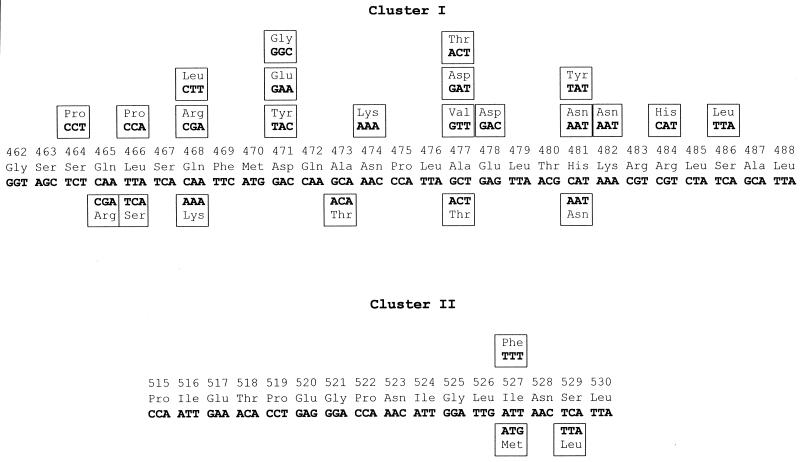
Eighteen different rpoB genotypes could be identified with respect to the in vitro mutations in S. aureus conferring rifampin resistance (Fig. 1). Only one mutational change, 481His→Asn, which was capable of conferring low-level resistance, was not associated with a reduction in the level of fitness (Table 1). In contrast, all other mutants responded to the burden of resistance by means of a distinct decrease in Darwinian fitness. In fact, the biological cost of resistance differed markedly among these mutants, ranging from little loss of fitness (mutant 17) to an immense loss of fitness (mutant 7) (Table 1).
FIG. 1.
Amino acid substitutions in RpoB of rifampin-resistant S. aureus strains. The codon numbering is that of the S. aureus rpoB sequence (1). Mutations identified in vivo and in vitro are shown below and above the wild-type sequence, respectively.
TABLE 1.
Relative fitness of rifampin-resistant S. aureus in vitro mutants
| Strain | No. of independent isolates | rifampin MIC (μg/ml) | Amino acid change | Ratio of wild type/mutant
|
Relative fitness (mean ± SEM)a | |
|---|---|---|---|---|---|---|
| Initialb | Finalb | |||||
| Wild type | ≤0.008 | 1 | ||||
| Mutant 1 | 1 | 4 | 481His→Asn | 1.1 | 1.1 | 1 ± 0.004 |
| Mutant 2 | 1 | 4 | 471Asp→Tyr | 0.8 | 6.0 | 0.88 ± 0.012d |
| Mutant 3 | 1 | 0.5 | 471Asp→Glu | 1.2 | 2.1 | 0.96 ± 0.008d |
| Mutant 4 | 2 | 0.5 | 471Asp→Gly | 0.9 | 7.9 | 0.87 ± 0.022d |
| Mutant 5 | 3 | 4 | 527Ile→Phe | 0.7 | 2.2 | 0.93 ± 0.016d |
| Mutant 6 | 1 | 4 | 464Ser→Pro | 0.7 | 2.0 | 0.93 ± 0.008d |
| Mutant 7 | 2 | 8 | 474Asn→Lys | 0.7 | 298.0 | 0.60 ± 0.040d |
| Mutant 8 | 1 | 1 | 477Ala→Val | 0.7 | 5.0 | 0.88 ± 0.023d |
| Mutant 9 | 1 | 256 | 477Ala→Asp | 0.8 | 3.8 | 0.91 ± 0.013d |
| Mutant 10 | 3 | 256 | 484Arg→His | 0.5 | 34.0 | 0.75 ± 0.019d |
| Mutant 11 | 3 | 512 | 486Ser→Leu | 0.5 | 5.1 | 0.86 ± 0.011d |
| Mutant 12 | 1 | >512 | 468Gln→Arg | 0.8 | 20.1 | 0.80 ± 0.007d |
| Mutant 13 | 1 | 512 | 468Gln→Leu | 1.0 | 2.5 | 0.95 ± 0.016d |
| Mutant 14 | 5 | 512 | 481His→Tyr | 1.0 | 2.7 | 0.93 ± 0.011d |
| Mutant 15c | 1 | >512 | 481His→Asn, 482Lys→Asn | 0.8 | 1.9 | 0.94 ± 0.024d |
| Mutant 16c | 1 | 256 | 481His→Asn, 478Glu→Asp | 0.6 | 2.2 | 0.92 ± 0.019d |
| Mutant 17c | 1 | 256 | 481His→Asn, 477Ala→Thr | 0.7 | 1.2 | 0.98 ± 0.015 |
| Mutant 18c | 1 | >512 | 481His→Asn, 466Leu→Pro | 0.6 | 5.6 | 0.87 ± 0.036d |
Mean values and standard errors of the mean (SEMs) were derived from a total of four different fitness assays.
Mean values were derived from a total of four different fitness assays.
Mutant was obtained by culturing mutant 1 on agar containing 10 μg of rifampin per ml.
The fitness reduction is significant, as determined by the one-sample t test (P < 0.05).
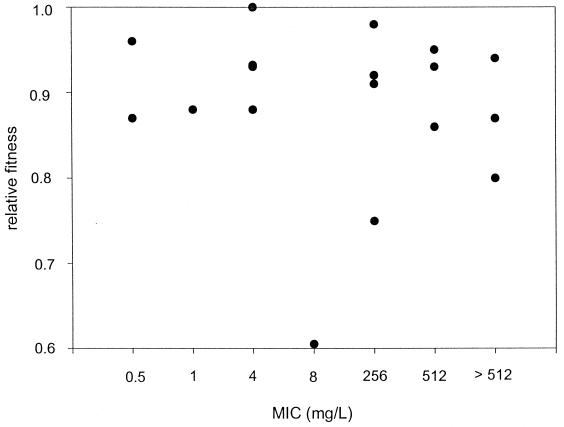
Interestingly, no relationship was detected between the cost of the rpoB mutation and the level of resistance (MIC) to rifampin (Fig. 2).
FIG. 2.
Correlation between rifampin MIC and fitness of S. aureus mutants.
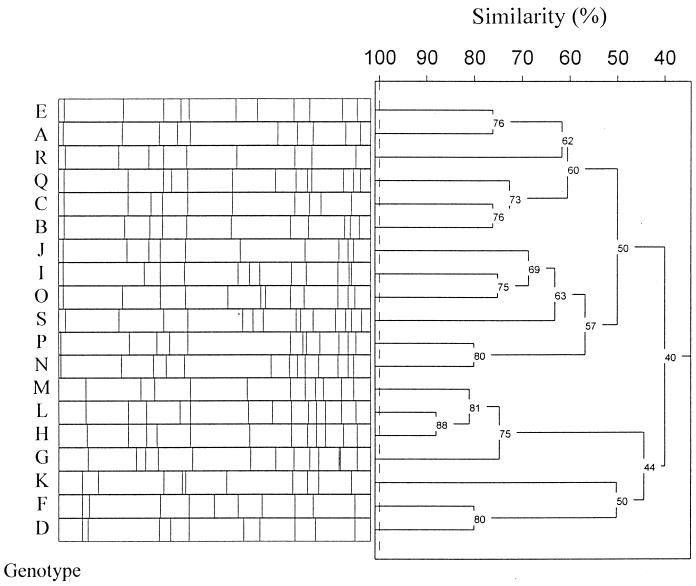
Macrorestriction analysis by PFGE of chromosomal DNA of 23 rifampin-resistant in vivo isolates from six countries revealed 19 different genotypes (Fig. 3). Remarkably, only seven different rpoB genotypes could be demonstrated in vivo, whereby the 481His→Asn mutation, which prevailed in 91% of isolates, was not demonstrably associated with a cost of resistance in vitro (Fig. 1; Table 2). Whereas low-level resistance in vivo is attributable solely to this single mutation at amino acid position 481, high-level resistance in vivo generally is traceable to multiple mutations, including the 481His→Asn mutation, thereby indicating a step-by-step mechanism in resistance development.
FIG. 3.
SmaI macrorestriction patterns and corresponding dendrogram from representatives of the S. aureus PFGE genotypes (indicated by capital letters).
TABLE 2.
In vivo mutations conferring rifampin resistance on S. aureus isolates of various PFGE genotypes
| Origin | PFGE genotypea | Rifampin MIC (μg/ml) | Amino acid change(s) |
|---|---|---|---|
| Germany | A | 2 | 481His→Asn |
| Germany | B | 2 | 481His→Asn |
| Germany | N | 2 | 481His→Asn |
| Germany | P | 2 | 481His→Asn |
| United States | K | 2 | 481His→Asn |
| Germany | D | 2 | 481His→Asn |
| Germany | O | 256 | 481His→Asn, 529Ser→Leu |
| France | C | 256 | 481His→Asn, 529Ser→Leu |
| France | B | 256 | 481His→Asn, 529Ser→Leu |
| Slovenia | I | 256 | 481His→Asn, 529Ser→Leu |
| Poland | J | 256 | 481His→Asn, 529Ser→Leu |
| Italy | S | 256 | 481His→Asn, 529Ser→Leu |
| Italy | Q | 256 | 481His→Asn, 529Ser→Leu |
| Germany | B | 512 | 481His→Asn, 465Gln→Arg, 529Ser→Leu |
| Germany | E | >512 | 481His→Asn, 473Ala→Thr, 477Ala→Thr |
| Germany | L | 512 | 481His→Asn, 466Leu→Ser |
| Germany | M | 512 | 481His→Asn, 466Leu→Ser |
| Germany | H | 512 | 481His→Asn, 466Leu→Ser |
| Germany | G | 512 | 481His→Asn, 466Leu→Ser |
| Italy | S | 512 | 481His→Asn, 527Ile→Met |
| Italy | R | 512 | 481His→Asn, 527Ile→Met |
| Germany | D | 512 | 468Gln→Lys |
| Germany | F | 512 | 468Gln→Lys |
S. aureus strains exhibiting the same macrorestriction pattern among PFGE analyses were assigned to the same PFGE genotype. The genotypes were arbitrarily denoted by capital letters.
DISCUSSION
From the perspective of the human population, a biological cost is associated with antibiotic resistance insofar as, first, infections due to resistant pathogens constitute increasing causes of morbidity and mortality worldwide and, second, the financial burden for the development of new and potent antibiotics must be borne by society (3, 14). In addition, however, there is the price associated with antibiotic resistance from the perspective of the bacterium itself, to the extent that resistance determinants may interfere with the normal physiological process in the cell and hence cause a reduction in the level of biological fitness (2, 16, 18).
In point of fact, several recent studies have shown that isolates with resistance genotypes are less fit than their sensitive counterparts in the absence of antibiotic selective pressure, thereby indicating a considerable cost of resistance (5, 7, 9, 23). The biological cost of antibiotic resistance, however, can be considerably diminished and even compensated for by evolutionary changes within the bacterial genome (6, 13, 15, 19, 20). Hence, bacteria often do pay a metabolic price, such as reduced growth rate, reduced invasiveness, or loss of virulence, for the acquisition of drug resistance in the short term; but their adaptation to the physiological cost is likely to foster the stable maintenance of resistance in the long term.
Our study aimed to investigate the intrinsic detriments in biological fitness associated with RNA polymerase (rpoB) mutations that confer rifampin resistance in S. aureus. Three principal findings emerged. First, the competition assays of in vitro-selected Rifr mutants with their Rifs isogenic counterpart revealed that only one rpoB genotype displayed no fitness burden, whereas the other mutations were associated in some cases with a considerable loss of fitness. It must be kept in mind, however, that growth defects in vitro do not always necessarily go along with growth defects in vivo. Nonetheless, Moorman and Mandell (17) also showed that rifampin-resistant strains of S. aureus display growth defects in vitro and in some cases display reduced virulence in an animal model. Second, no relationship between the magnitude of the biological cost and the level of resistance to rifampin could be detected. Third, the variation in frequency of rpoB mutations conferring resistance to rifampin in S. aureus in vivo appears to be a function of the Darwinian fitness of the organism. Indeed, the 481His→Asn mutation, which was not associated with a loss of fitness in vitro, was shown to be prevalent in 91% of the Rifr clinical S. aureus isolates tested. Moreover, since the in vivo isolates that display the 481His→Asn mutation were obtained from six countries and revealed different genetic backgrounds by macrorestriction analysis, it seems very unlikely that the high prevalence of this “no-cost” mutation can be explained by the clonal spread of a rifampin-resistant ancestral strain. As such, it seems likely that, in vivo, a functional restriction on RNA polymerase and subsequent bacterial fitness limit the expression of the full range of theoretical mutants defined in vitro. Interestingly, high-level resistance within clinical isolates was mainly attributable to double mutations within rpoB, including the mutational change 481His→Asn. This in turn is indicative of a resistance-mediating function on the part of the additional mutation, since the 481His→Asn mutational change on its own confers only low-level resistance. Regarding the two isolates that exhibited three mutational changes within rpoB in vivo, however, it still remains a matter of speculation as to whether both of the additional mutations, besides the 481His→Asn mutation, contribute to resistance.
These findings demonstrate that the ability of resistance determinants to survive in bacteria is due not only to compensatory mutations that substantially diminish the biological cost of antibiotic resistance, as shown by others (2, 6, 15), but also to the selection among resistance alleles favoring those that impose the lowest (if any) biological costs, as proposed here.
Another example of a no-cost resistance mutation has been elucidated in vitro for the rpsL gene, which is responsible for resistance to high concentrations of streptomycin in Salmonella enterica subsp. enterica serovar Typhimurium (5). Moreover, rpsL mutations responsible for streptomycin resistance in clinical isolates of Mycobacterium tuberculosis were shown to be the same as those with no cost in experiments performed with Salmonella serovar Typhimurium (8).
Extrapolation of these findings makes it tempting to speculate that resistance will never disappear completely because there is no evolutionary disadvantage to being resistant once adaptation has taken place, i.e., by acquisition of compensatory mutations, or when fitness has not been diminished, i.e., by selection of no-cost mutations. Consequently, policies of decreased antibiotic usage intended to reduce resistance might not be as successful as originally anticipated. Finally, strict hygienic and antibiotic treatment regimens for prevention of the spread of resistance together with continued drug development are required if the human population wishes to stay one step ahead of antibiotic resistance.
Acknowledgments
We thank Denia Franck and Marita Parschau for technical assistance.
This work was supported by grants from the Marie Christine Held and Erika Hecker Foundation.
REFERENCES
- 1.Aboshkiwa, M., G. Rowland, and G. Coleman. 1995. Nucleotide sequence of the Staphylococcus aureus RNA polymerase rpoB gene and comparison of its predicted amino acid sequence with those of other bacteria. Biochem. Biophys. Acta 1262:73-78. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 3.Austin, D. J., K. G. Kristinsson, and R. M. Anderson. 1999. The relationship between the volume of antimicrobial consumption in human communities and the frequency of resistance. Proc. Natl. Acad. Sci. USA 96:1152-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billington, O. J., T. D. McHugh, and S. H. Gillespie. 1999. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:1866-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Björkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Björkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479-1482. [DOI] [PubMed] [Google Scholar]
- 7.Björkman, J., P. Samuelsson, D. I. Andersson, and D. Hughes. 1999. Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol. Microbiol. 31:53-58. [DOI] [PubMed] [Google Scholar]
- 8.Böttger, E. C., B. Springer, M. Pletschette, and P. Sander. 1998. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat. Med. 4:1343-1344. [DOI] [PubMed] [Google Scholar]
- 9.Davies, A. P., O. J. Billington, B. A. Bannister, W. R. C. Weir, T. D. McHugh, and S. H. Gillespie. 2000. Comparison of fitness of two isolates of Mycobacterium tuberculosis, one of which had developed multi-drug resistance during the course of treatment. J. Infect. 41:184-187. [DOI] [PubMed] [Google Scholar]
- 10.Gillespie, S. H. 2001. Antibiotic resistance in the absence of selective pressure. Int. J. Antimicrob. Agents 17:171-176. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie, S. H., and T. D. McHugh. 1997. The biological cost of antimicrobial resistance. Trends Microbiol. 5:337-339. [DOI] [PubMed] [Google Scholar]
- 12.Guillemot, D. 1999. Antibiotic use in humans and bacterial resistance. Curr. Opin. Microbiol. 2:494-498. [DOI] [PubMed] [Google Scholar]
- 13.Laurent, F., H. Lelievre, M. Cornu, F. Vandenesch, G. Carret, J. Etienne, and J. P. Flandrois. 2001. Fitness and competitive growth advantage of new gentamicin-susceptible MRSA clones spreading in French hospitals. J. Antimicrob. Agents 47:277-283. [DOI] [PubMed] [Google Scholar]
- 14.Lenski, R. E. 1998. Bacterial evolution and the cost of antibiotic resistance. Int. Microbiol. 1:265-270. [PubMed] [Google Scholar]
- 15.Levin, B. R., V. Perrot, and N. Walker. 2000. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154:985-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipsitch, M. 2001. The rise and fall of antimicrobial resistance. Trends Microbiol. 9:438-444. [DOI] [PubMed] [Google Scholar]
- 17.Moorman, D. R., and G. L. Mandell. 1981. Characteristics of rifampin-resistant variants obtained from clinical isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 203:709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris, A., J. D. Kellner, and D. E. Low. 1998. The superbugs: evolution, dissemination and fitness. Curr. Opin. Microbiol. 1:524-529. [DOI] [PubMed] [Google Scholar]
- 19.Nagaev, I., J. Björkman, D. I. Andersson, and D. Hughes. 2001. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 40:433-439. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds, M. G. 2000. Compensatory evolution in rifampicin-resistant Escherichia coli. Genetics 156:1471-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salyers, A. A., and C. F. Amabile-Cuevas. 1997. Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 41:2321-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seppala, H., T. Klaukka, J. Vuopio-Varkila, A. Muotiala, H. Helenius, K. Lager, and P. Huovinen. 1997. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. N. Engl. J. Med. 337:441-446. [DOI] [PubMed] [Google Scholar]
- 23.Wang, G., M. Sayeedur Rahmann, M. Zafri Humayun, and D. E. Taylor. 1999. Multiplex sequence analysis demonstrates the competitive growth advantage of the A-to-G mutants of clarithromycin-resistant Helicobacter pylori. Antimicrob. Agents Chemother. 43:683-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wichelhaus, T. A., J. Schulze, K. P. Hunfeld, V. Schäfer, and V. Brade. 1997. Clonal heterogeneity, distribution, and pathogenicity of methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 16:893-897. [DOI] [PubMed] [Google Scholar]
- 25.Wichelhaus, T. A., V. Schäfer, V. Brade, and B. Böddinghaus. 1999. Molecular characterization of rpoB mutations conferring cross-resistance to rifamycins on methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2813-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]