Abstract
Mouse mammary tumor virus (MMTV) has been classified as a simple retrovirus with two accessory genes, dut and sag. Cloned MMTV proviruses carrying a trimethoprim (trim) cassette in the envelope gene were defective for Gag protein production and the nuclear export of unspliced gag-pol RNA. Complementation experiments indicated that a trans-acting product was responsible for the Gag defect of such mutants. Analysis of MMTV-infected cells revealed the presence of a novel, doubly spliced RNA that encodes a putative product of 301 amino acids. Overexpression of cDNA from this RNA increased Gag levels from env mutant proviruses or reporter gene expression from unspliced mRNAs and allowed detection of a 33-kDa protein product, which has been named regulator of export of MMTV mRNA, or Rem. The Rem N terminus has motifs similar to the Rev-like export proteins of complex retroviruses, and mutation of the nuclear localization signal (NLS) abolished RNA export and detection within the nucleus. The Rem C terminus has few identifiable features, but removal of this domain increased Rem-mediated export, suggesting an autoregulatory function. A reporter vector developed from the 3′ end of the MMTV provirus was Rem responsive and required both the presence of the MMTV env-U3 junction and a functional Crm1 pathway. The identification of a third accessory protein from a doubly spliced transcript suggests that MMTV is the first murine complex retrovirus to be documented. Manipulation of the MMTV genome may provide mouse models for human retroviral diseases, such as AIDS.
The nuclear export of most cellular mRNAs is coupled to the deposition of exon-junction complexes, which also serve to prevent translation of mRNAs with premature stop codons (2, 35). Therefore, retroviruses have evolved mechanisms to allow export of intron-containing mRNAs from the nucleus to the cytoplasm (14). Simple and complex retroviruses appear to have evolved independent mechanisms for such mRNA export (14). RNA export for the simple betaretrovirus Mason-Pfizer monkey virus (MPMV) involves a cis-acting RNA stem-loop structure known as the constitutive transport element (CTE) (9), which was used to identify the cellular binding factor, nuclear export factor 1 (NXF1), or Tap. Tap is the mammalian counterpart of the yeast export factor Mex67p (47) and appears to facilitate export and expression of CTE-containing mRNAs (8). Moreover, Tap forms a heterodimer with NXT1 (p15) to interact with nucleoporins in the nuclear pore complex (52) and acts as the major receptor for cellular mRNA export (14).
The genus Betaretrovirus also includes the human endogenous retroviruses type K (HERV-Ks) (3). Although classified as simple retroviruses, HERV-Ks encode a trans-acting protein, Rec (31), which is similar to the RNA export factors encoded by the complex retroviruses, e.g., the Rev protein of human immunodeficiency virus (HIV) (33). Rev binds to a cis-acting RNA sequence known as the Rev-responsive element in the HIV envelope coding region (22) and recruits the karyopherin export factor, chromosome region maintenance 1/exportin 1 (Crm1/Xpo1) (18). Crm1 mediates the nuclear export of proteins containing a leucine-rich nuclear export signal (NES) as well as several types of cellular RNA, including 5S rRNA, U small nuclear RNAs, and a distinct class of mRNAs (14). These mRNAs include transcripts of specific early response genes, such as c-fos, cox-2 and type I interferons (29). Crm1-mediated export of cellular mRNAs occurs through recruitment of cellular adaptors, such as HuR, which interacts with pp32 and APRIL, two leucine-rich NES-containing proteins (10). The HIV protein, Rev, also contains a leucine-rich NES and acts as an adaptor for HIV RNA export (51). Rev-mediated RNA export is inhibited by leptomycin B, a drug that binds covalently to Crm1 to block interaction with leucine-rich NESs (53). Rev and Rev-like proteins also appear to shuttle between the nucleus and the cytoplasm (37).
Mouse mammary tumor virus (MMTV) is another betaretrovirus that requires nuclear export of unspliced and partially spliced mRNA for production of virion proteins (Fig. 1A) (15). Experiments from our lab and others suggest that MMTV encodes a Rev-like protein (19, 27; M. Mann, J. A. Mertz, S. Payne, and J. Dudley, Abstr. Cold Spring Harbor Retrovir. Meet., abstr. 314, 2004). In this report, we provide definitive evidence that the MMTV genome expresses a 301-amino-acid protein with RNA export activity. This protein is expressed from a doubly spliced transcript and has been named regulator of export of MMTV mRNA (Rem). Disruption of the rem coding sequence by transposon mutagenesis leads to reduction of unspliced gag-pol mRNA and Gag protein production in the cytoplasm, and this defect can be complemented with an infectious MMTV provirus or the cloned rem cDNA. The Rem N terminus contains the sequence motifs observed in other retroviral export proteins (6, 40). The N-terminal 98 amino acids are sufficient to mediate RNA export activity as determined by the development of a quantitative reporter assay. This assay revealed that the Rem-responsive element (RmRE) includes the envelope-U3 border and that export activity is dependent on Crm1/Xpo1. Surprisingly, the removal of the C terminus increased RNA export, suggesting that this region is an autoregulatory domain. Together, our results indicate that MMTV is a complex retrovirus that encodes a self-regulated RNA export protein.
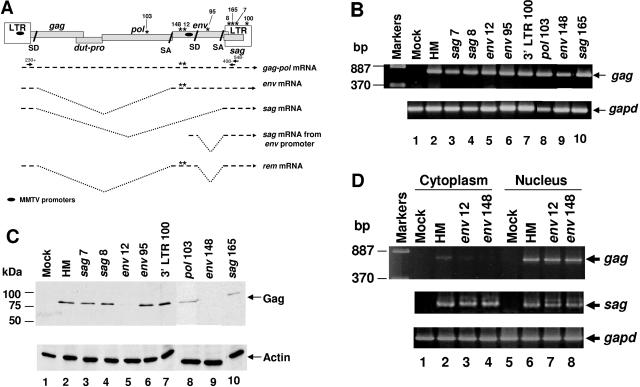
FIG. 1.
trim cassette mutants in the env gene of an infectious MMTV provirus have a defect in nuclear export of unspliced viral RNA. (A) Structure of the MMTV provirus and mRNAs. Asterisks represent the locations of trim insertions within the MMTV provirus. Mutations in two proviruses, env12 and env148, which interrupt the env/rem genes, also are shown within viral mRNAs. Introns are shown by V-shaped dotted lines. Ovals represent MMTV promoters. Arrows and numbers indicate the primers used to identify the novel mRNA. (B) RT-PCR for total MMTV RNA expression. RNA from XC cells transfected with wild-type or mutant MMTV proviral constructs was used in RT-PCRs to detect gag sequences (upper panel) using glyceraldehyde 3-phosphate dehydrogenase (gapd) expression (lower panel) to verify equivalent loading and RNA integrity. (C) Western blotting for MMTV Gag expression. Whole-cell extracts were analyzed by Western blotting with MMTV Gag-specific (upper panel) or actin-specific antibodies (lower panel). (D) RT-PCRs with cytoplasmic and nuclear RNA fractions from transfected XC cells. After transfection with wild-type or mutant proviral constructs, RNA fractions were used for RT-PCRs to detect unspliced gag sequences (upper panel), completely spliced sag sequences (middle panel), or gapd sequences (lower panel).
MATERIALS AND METHODS
Plasmids and in vitro mutagenesis.
The pHYB-MTV plasmid has been described previously (48). Transposon mutagenesis was performed using the EZ:TN <DHFR-1> insertion kit (EPICENTRE) as suggested by the manufacturer. A portion of the in vitro reaction mixture was used to electroporate Escherichia coli DH5α using a Bio-Rad Gene Pulser, and transformants were grown on plates containing trimethoprim and ampicillin to select for trim insertions that did not inactivate the ampicillin gene within the pHYB-MTV plasmid backbone. Colony PCR was performed after in vitro mutagenesis of pHYB-MTV to localize transposon integration sites. Selected colonies were used to prepare purified plasmids that were sequenced to verify the exact site of trim cassettes. A bacterial clone was inoculated into five tubes, each containing 5 μl H2O, and used for colony PCR with five different primer pairs and KlenTaq1 polymerase (AB Peptides, St. Louis, MO) in a 20-μl reaction mixture. Primer pairs used for each region were as follows: C3HLTR1+ and Gag620-; C3H230+ and Gag620-; Pol4325+ and Env6337-; Pol 5835+ and Env2251-; and TBLVenv8509+ and pBR3′-. The PCR conditions were denaturation at 94°C for 3 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1.5 min, and a final extension step at 72°C for 10 min.
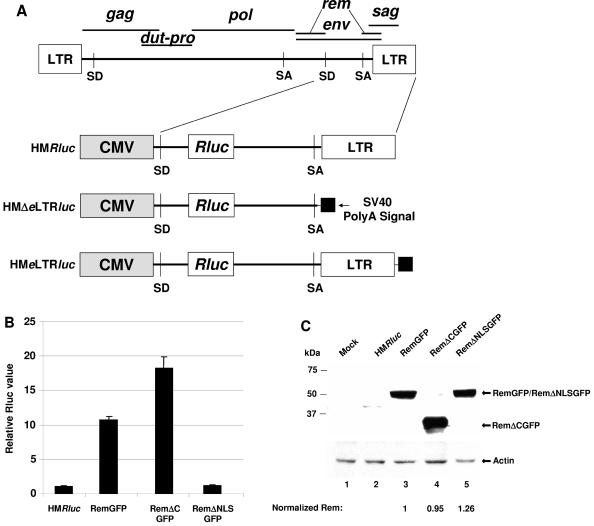
Fusions of gfp to either end of the rem gene were functional in reporter assays with HMRluc and have been used interchangeably. The RemGFP-expression plasmid was generated by PCR amplification of the rem cDNA using RemcKRI+ and RemFLBamHI- primers. The PCR product was digested with EcoRI and BamHI and ligated with EGFPN3 (Clontech) that had been digested with EcoRI and BamHI to fuse the gfp gene in frame to the 3′ end of rem sequences. The RemcKRI+ primer contains a consensus Kozak sequence upstream of the ATG codon. RemΔCGFP was generated in the same manner using a different minus-strand primer, cRemBamHI-. The RemΔNLSGFP construct was produced by deleting the sequences coding for the NLS/ARM using recombinant PCR (36). Plasmids encoding GFPRem and GFPRemΔC were cloned by insertion of the respective cDNA sequences in frame with the green fluorescent protein (GFP) coding sequences in the EGFPC1 vector (Clontech). The pHMRluc plasmid consists of the HincII-to-HindIII fragment of the HYB-MTV proviral construct inserted into pEGFPN3 at the AfeI and HindIII sites in the multiple cloning site. The Renilla luciferase gene from pRL-TK (Promega) was excised by digestion with NheI and XbaI, treated with Klenow to produce blunt ends, and cloned into the XmnI site of pHYB-MTV (245 bp downstream of the splice donor site and 907 bp upstream of the splice acceptor site). The plasmid HMΔeLTRluc was prepared by substitution of the entire long terminal repeat (LTR) and a small portion of the env sequences for the simian virus 40 (SV40) poly(A) signal. The BglII-to-SpeI sequences of pHYB-MTV were excised, treated with Klenow polymerase, and used for blunt-end ligation to an SV40 poly(A) signal cassette. The pHMeLTRluc plasmid was generated by insertion of the BglII-SpeI fragment from pHYB-MTV into an engineered ScaI site downstream of the splice acceptor site and upstream of the SV40 poly(A) signal. The Crm1 (pcΔCAN) and Tap (pcTapA17) dominant-negative expression plasmids were kindly provided by Bryan Cullen (28).
Primers.
Sequences of primers were as follows: C3HLTR1+ (5′ ATG CCG CGC CTG CAG CAG AAA TGG T 3′), gag620- (5′ CCT CCA AAT CAT CCC AAT CCT C 3′), C3H230+ (5′ CAT CAC AAG AGC GGA ACG GAC 3′), pol4235+ (5′ GAA GAG AGC AAT AGC CCT TG 3′), env6337- (5′ GGG CCC CTT TTG GAG AAA ATG AGA GT 3′), pol5835+ (5′ GCC ACG CAC TAC ATC ATC 3′), Env2251- (5′ CGT TAA GAT CTG ACT GCA CTT GG 3′), TBLVenv8509+ (5′ AGC CTT GAC CAA GTG CAG TCA GAT CTT AAC GTG 3′), pBR3′- (5′ CAC CCT GTA TAT GAG TTC CC 3′), C3Hpol6419+ (5′ CGA AAG ACA TTG GGG ACC 3′), Gapd427+ (5′ CAT GTT TGT GAT GGG TGT GAA CCA 3′), Gapd983- (5′ GTT GCT GTA GCC GTA TTC ATT GTC 3′), C3Henv7255+ (5′ ATC GCC TTT AAG AAG GAC GCC TTC T 3′), C3HLTR548- (5′ TAC TTC TAG GCC TGT GGT CA 3′), DHFR-1 FP-1 (5′ GGC GGA AAC ATT GGA TGC GG 3′), C3HLTR408- (5′ TCT ACC TAT TGG ATT GGT CTT ATT GG 3′), RemcKRI+ (5′ GAA TTC GCC ACC ATG CCG AAT CAC CAA TCT GGG 3′), cRemBamHI- (5′ GGA TCC CCC GGT CAC AGG CGG G 3′), RemFLBamHI- (5′ GGA TCC GGT GTA GGA CAC TCT CGG 3′).
Cell lines and transfections.
Growth conditions for rat XC, human Jurkat, and mouse mammary HC11 cells have been described previously (36, 57). Jurkat and HC11 cells were transfected by electroporation (36, 56), whereas XC cells were transfected using DMRIE-C (Invitrogen) (57). For stable transfections, XC cells were cotransfected with pHYB-MTV or penv148 and a neomycin-resistance plasmid (pcDNA3; Invitrogen) at a ratio of 100:1 and cultured in growth medium containing 700 μg G418/ml for 3 weeks before use of pooled transfectants for further experiments.
Western blotting and antibodies.
Western blot assays were performed essentially as described previously (30). Transfected cells were harvested in radioimmunoprecipitation assay buffer as previously described (30). Alternatively, whole-cell extracts were obtained by addition of one volume of 250 mM Tris-HCl, pH 6.8, 20% glycerol, 2% sodium dodecyl sulfate (SDS), 5% β-mercaptoethanol, and 0.2% bromophenol blue to cells in one volume of phosphate-buffered saline (PBS) followed by boiling for 5 min. Proteins were resolved on 8-to-10% polyacrylamide gels containing 1% SDS and transferred to a nitrocellulose membrane. Membranes were blocked with 5% milk in Tris-buffered saline Tween 20 (TBST; 20 mM Tris-HCl, pH 7.6, 137 mM NaCl, and 0.1% Tween 20) for 1 h. The primary antibody was diluted in TBST containing 1% milk (or 5% milk for SU-specific antibody) and incubated with the membrane for 1 h followed by three washes in TBST for 5 min each. The horseradish peroxidase-conjugated secondary antibody was diluted in TBST containing 1% milk and incubated with the membrane for 45 min prior to three additional 5-min washes. All steps were performed at 25°C with shaking. Western Lightning enhanced chemiluminescent reagent (Perkin-Elmer) was used to detect antibody binding. Monoclonal antibodies specific for actin (Oncogene Research), GFP (Becton Dickinson), MMTV Gag (CA specific), or Env (SU specific) (kindly provided by Tanya Golovkina) (45) were used.
Reporter assays.
Luciferase assays were performed using the dual-luciferase reporter assay system (Promega) to quantitate both Renilla and firefly luciferase activities (36).
RNA fractionations and RT-PCR.
Cytoplasmic and nuclear RNA fractions were obtained as described previously (54). Total RNA was obtained using the guanidine isothiocyanate method (12). RNA was treated with amplification-grade DNase I (Invitrogen) and RNase inhibitor (Invitrogen) prior to semiquantitative reverse transcriptase PCR (RT-PCR) (57). Ten micrograms of cytoplasmic, nuclear, or total RNA from tissue culture cells was treated with 3 U amplification-grade DNase I (Invitrogen) and 5 U RNase inhibitor (Invitrogen) for 1 h at 37°C. The DNase I was inactivated by addition of EDTA to a final concentration of 2.5 mM and incubation at 72°C for 15 min. The DNase I-treated RNA (5 μg) was further processed to make cDNA in an RT reaction. The primer poly(dT)17 and deoxynucleotides were added to the RNA at final concentrations of 2.5 pmol/μl and 1 mM, respectively, boiled for 5 min, and placed on ice for 5 min. The RNA was reverse transcribed using 400 U Moloney murine leukemia virus RT, 5 U RNase inhibitor, and 10 mM dithiothreitol in a 50-μl reaction mixture. A 2.5-μl aliquot of cDNA was used in RT-PCRs with specified primers and polymerases for each mRNA. Taq polymerase was used for PCR products less than 1 kb. PCR conditions were denaturation at 94°C for 3 min, 50 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min, with a final extension step at 72°C for 5 min. Expand Long Template polymerase (Roche, Indianapolis, IN) was used for PCR products larger than 1 kb. PCR conditions were denaturation at 94°C for 2 min, 30 cycles of denaturation at 94°C for 20 s, annealing at 55°C for 30 s, and extension at 68°C for 6 min, with a final extension step at 68°C for 8 min. The following primer pairs were used to amplify each product: gag, C3H230+/gag620-; Gapd, Gapd427+/Gapd983-; sag, C3Henv7255+/C3HLTR548-; and gag-pol-trim, C3Hpol6419+/DHFR FP-1.
Immunofluorescence.
Transfected cells were grown on glass coverslips for 48 h and fixed in 4% paraformaldehyde for 15 min. Cells were then permeabilized by incubation in 0.1% Triton X-100 for 10 min. Cells were washed and blocked in 10% fetal calf serum in PBS for 30 min prior to incubation in anti-nucleophosmin (B23) diluted 1:500 in PBS containing 2% fetal calf serum and 0.1% Tween 20 for 1 h. After three washes in PBS, the cells were incubated for 30 min in secondary antibody (1:400). After three washes in PBS for a total of 1 h in the dark, the cells were stained with 300 nM 4′,6-diamidino-2-phenylindole (DAPI) and then washed with PBS. Coverslips were mounted in Vectashield (Vector Labs) for observation in a Leica SP2 AOBS confocal microscope. Antibody to nucleophosmin was purchased from Sigma and Alexa Fluor 594-conjugated secondary antibody specific for mouse immunoglobulin G was obtained from Molecular Probes.
Nucleotide sequence accession number.
The GenBank accession number for the sequence presented here is DQ223969.
RESULTS
To define MMTV sequences required for RNA export of intron-containing mRNAs, an infectious, wild-type provirus, pHYB-MTV (48), was subjected to mutagenesis with the 887-bp EZ:TN transposon containing the trimethoprim resistance (trim) cassette. After electroporation into bacterial cells, individual colonies were screened by colony PCR using a panel of five primer pairs to identify the approximate position of transposon insertions. Selected plasmids were sequenced to determine the exact insertion site and then transiently transfected into rat XC cells (lacking endogenous MMTVs). After 48 h, total RNA was extracted and used for RT-PCR. All transposon mutants, except several in the gag region, expressed gag-pol mRNA (Fig. 1B and data not shown), yet Western blotting indicated that two mutants, env12 and -148, showed defective production of the Gag precursor Pr77 (Fig. 1C). Therefore, insertions within the envelope region reduced Gag production.
If mutations in the MMTV env gene reduced nuclear export of intron-containing viral mRNAs, we anticipated that proviruses carrying these mutations would have decreased cytoplasmic levels of gag-pol transcripts. Therefore, the two mutants that failed to produce the Gag precursor were transiently transfected into XC cells, and RNA was extracted from cytoplasmic and nuclear fractions. RT-PCR analysis showed that the env12 and -148 mutants failed to export gag-pol mRNA into the cytoplasm at wild-type levels (Fig. 1D). The mutants, env12 and -148, are located approximately 200 bp apart at the 5′ end of the env gene (Fig. 1A). This result suggested that there is an element of at least 200 bp that is required for nuclear export of unspliced MMTV RNA.
Since we were unable to detect CTE activity from the MMTV genome (data not shown), RT-PCRs were performed to detect novel MMTV mRNAs that might encode a viral export protein similar to HIV Rev, human T-cell leukemia virus (HTLV) Rex, or HERV-K Rec (14). Total RNA was extracted from XC rat cells stably transfected with pHYB-MTV and subjected to RT-PCR using a forward primer that originated just upstream of the 5′ splice donor site and a reverse primer that originated within the LTR (Fig. 1A). To determine the virus and cell-type specificity of our results, similar experiments were performed using Jurkat T cells transfected with a second MMTV provirus (pHYB-TBLV), which induces T-cell lymphomas rather than mammary tumors (38). MMTV and TBLV proviruses differ only in the U3 region of the LTR and both produce infectious virus (4) and, therefore, each should require nuclear export of unspliced RNA to allow Gag protein synthesis. As expected, both stably transfected cell lines expressed env and sag mRNAs (Fig. 2A). (The product of the gag-pol unspliced RNA is too large to be detected under these conditions.) However, another strong band was detected between the env and sag transcripts. The novel band obtained from both the XC and Jurkat cells was cloned. Sequence analysis verified that this prominent product was derived from a doubly spliced transcript that used previously identified splice donor and acceptor sites (Fig. 1A).
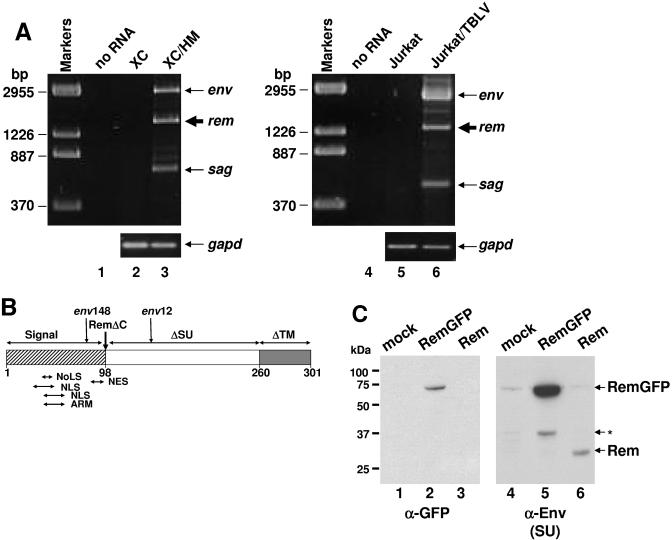
FIG. 2.
The MMTV provirus encodes a novel nuclear protein from a doubly spliced mRNA. (A) Detection of a novel viral mRNA in MMTV-expressing cells. Total RNA was extracted from XC rat fibroblasts stably transfected with the wild-type HYB-MTV (HM) provirus and Jurkat human T cells stably transfected with a thymotropic MMTV provirus (TBLV) and used for RT-PCRs to detect rem mRNA (thick arrow) (lanes 3 and 6). Negative controls contained no RNA (lanes 1 and 4) or RNA from untransfected cells (lanes 2 and 5). Primer pairs used to detect all MMTV or TBLV mRNAs were C3H230+ and C3HLTR548- or C3H230+ and C3HLTR408-, respectively (Fig. 1A). RT-PCR for gapd RNA (lower panel) was used as a control for RNA integrity and loading. (B) Diagram of the Rem protein. Rem contains the entire signal peptide of the MMTV Env protein and part of the Env SU (ΔSU) and TM (ΔTM) sequences. The vertical arrows show the positions of the env12 and -148 mutants and the C-terminal truncation of RemΔC. Locations of predicted sequence motifs are indicated by arrows under the diagram. (C) Western blots showing Rem protein production from the novel MMTV cDNA. Whole-cell lysates from transfected XC cells were analyzed by Western blotting using the indicated antibodies. The band indicated by the asterisk may be a degradation product. NoLS, nucleolar localization signal; NLS, nuclear localization signal; NES, nuclear export signal; ARM, arginine-rich motif.
The open reading frame (ORF) within the novel 1.5-kb cDNA encodes a putative polypeptide of 301 amino acids (Fig. 2B). This protein is in frame with the env coding sequence and consists of the Env signal peptide (98 amino acids), 162 amino acids from the surface (SU) envelope protein, and 41 amino acids from the transmembrane (TM) envelope protein. Predicted protein motifs found using the MotifScan and ExPASy proteomics tools showed glycosylation, myristylation, amidation, and protein kinase C phosphorylation sites. The putative product also contained overlapping nuclear and nucleolar localization signals (NLS and NoLS) and a leucine-rich region that is a potential nuclear export signal (NES), as well as an arginine-rich motif (ARM), an RNA-binding domain found in Rev-like proteins (34). Interestingly, both mutant proviruses (env12 and -148) that have defective export of gag-pol mRNA have insertions of the trim cassette within the predicted ORF. These results and our previous observations suggest that MMTV encodes a trans-acting protein, named Rem for regulator of export of MMTV mRNAs, to facilitate nuclear export of intron-containing viral mRNAs.
To determine if a protein product was produced, rem cDNA and rem cDNA fused to gfp sequences were cloned into an expression vector and transfected into rat XC cells. After 48 h, whole-cell extracts were used for Western blotting and incubated with GFP-specific antibody or a monoclonal antibody specific for Env, which should share amino acid sequences with Rem (Fig. 2C). The full-length Rem protein of ca. 33 kDa was detected with the envelope (SU)-specific antibody (lane 6), but not the GFP-specific antibody (lane 3), consistent with the expected translation product. As anticipated, a RemGFP fusion of 66 kDa was detected with either GFP-specific (lane 2) or SU-specific (lane 5) antibody.
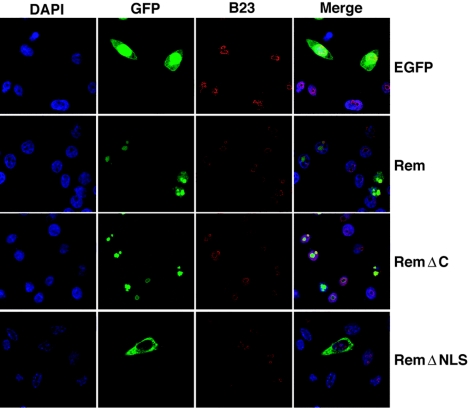
Software predictions suggested that Rem is a nuclear protein. Therefore, XC cells transiently transfected with either the vector control or full-length (Rem) or C-terminally truncated (RemΔC) Rem fusions to GFP were stained with DAPI to identify nuclei and examined by confocal microscopy (Fig. 3). GFP alone was diffusely localized throughout the cell, whereas the full-length Rem localized to the nucleoplasm as well as discrete regions within the nucleus that stained with the nucleolar marker, nucleophosmin (B23) (11). Other Rev-like export proteins also primarily reside in nucleoli (32). RemΔC localized almost entirely within nucleoli, whereas Rem missing the NLS/ARM sequence (RemΔNLS) was excluded from nuclei.
FIG. 3.
Fluorescence localizes GFP-tagged Rem and RemΔC to the nucleus. XC cells were transfected with GFP-expressing constructs and grown on glass coverslips for 48 h prior to processing and incubation with antibodies specific for nucleophosmin (B23). Cells were stained with DAPI and imaged by confocal microscopy.
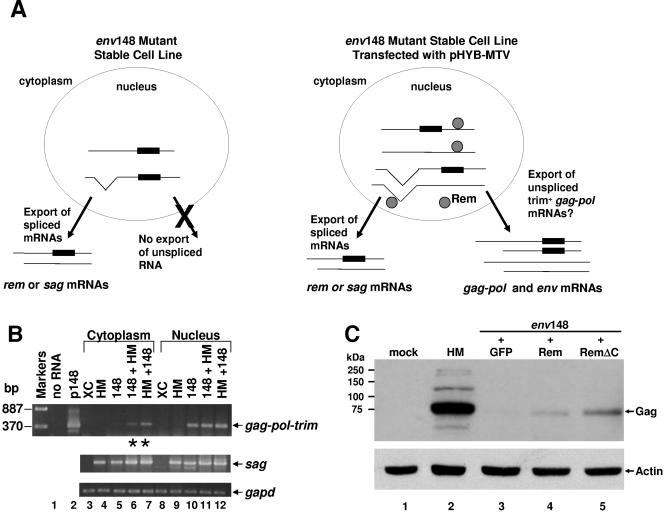
To confirm whether MMTV expresses a trans-acting factor for RNA export, complementation experiments were performed (see the strategy in Fig. 4A). Briefly, cells stably transfected with the mutant provirus env148, which has an insertion between the Rem-encoded NLS/ARM and NES motifs (Fig. 2B), were transiently transfected with wild-type pHYB-MTV. RNA was isolated from cytoplasmic and nuclear fractions of both transfected and untransfected cells and analyzed by semiquantitative RT-PCR (Fig. 4B). Primers in pol and the trim cassette were used to detect only the mutant unspliced mRNA. As expected, the gag-pol mRNA containing trim was detectable in the nuclear fraction of cells transfected with env148 (lane 10), but not in the cytoplasm (lane 5). However, when pHYB-MTV plasmid was transiently transfected into XC cells stably transfected with env148, unspliced RNA containing trim also was observed in the cytoplasmic fractions (lane 6). The reciprocal experiment also verified this result (lane 7). The sag mRNA was used as a control for cytoplasmic export because this RNA is completely spliced and exported by standard cellular mechanisms. These results clearly indicate that MMTV expresses a trans-acting factor that functions in export of intron-containing mRNA. Moreover, the env148 mutation inactivates the export function of this factor.
FIG. 4.
Defective Gag expression from an env mutant provirus is complemented by an infectious MMTV provirus or Rem cDNA. (A) Complementation assay for gag-pol mRNA export. Rectangles represent trim cassettes, and circles represent Rem protein. Lines, some with V shapes to show spliced regions, represent capped and polyadenylated viral RNAs. For simplicity, the doubly spliced rem RNA is not shown in the nucleus. (B) The Gag production defect of the env148 mutant is complemented by cotransfection of an infectious wild-type MMTV provirus. XC cells stably transfected with penv148 (148) or pHYB-MTV (HM) were transiently transfected with pHYB-MTV or penv148, respectively. After fractionation, cytoplasmic and nuclear RNAs were used in RT-PCRs to detect trim-containing unspliced viral RNA (gag-pol-trim), completely spliced sag RNA, or gapd RNA. Negative controls included reactions lacking RNA (lane 1) or RNA from untransfected XC cells (lanes 3 and 8). The penv148 vector (p148) was used as a size marker for the product of unspliced RNA (lane 2). Asterisks represent lanes where complementation occurred. (C) Complementation of the env148 mutant defect by cotransfection with RemGFP or RemΔCGFP expression plasmids. HC11 cells were transiently transfected for 24 h and then incubated with 10−6 M dexamethasone for an additional 48 h to induce the LTR promoter. Whole-cell lysates were subjected to Western blotting and incubation with Gag-specific (upper panel) or actin-specific antibodies (lower panel).
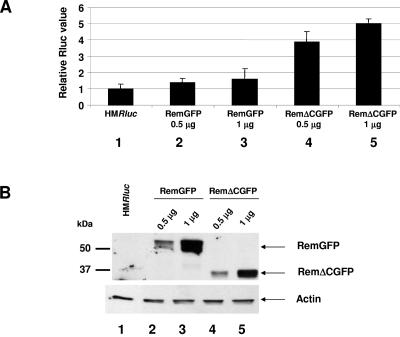
To determine if Rem is a trans-acting RNA export protein, we performed a second type of complementation experiment. The cloned cDNAs expressing full-length Rem or RemΔC fused to GFP or empty vector were transiently cotransfected with the env148 mutant into HC11 mouse mammary cells, which are permissive for MMTV replication. After 72 h, cells were harvested and analyzed by Western blotting using a Gag-specific antibody (Fig. 4C). By Scion Image software, remGFP and remΔCGFP vector-transfected cells showed 11- to 42-fold (lanes 4 and 5) increased Gag expression relative to cells transfected with control vector (lane 3). However, Gag expression levels comparable to that of the wild-type pHYB-MTV vector were not attained (lane 2).
To further quantitate the effects of Rem and RemΔC and to identify a Rem-binding site on MMTV RNA, we developed an MMTV reporter plasmid based on the pDM128 vector used for assays of HIV Rev function (26). This vector (pHMRluc) contains the 3′ end of the MMTV genome with a Renilla luciferase gene downstream of the splice donor in the env gene, thus eliminating Rem and Env expression (Fig. 5A). Because the reporter gene is flanked by splice donor and acceptor sites, splicing of the primary transcript eliminates the luciferase gene and enzyme activity. In the presence of Rem and the RmRE, the unspliced primary transcript should be exported from the nucleus, leading to an increase in reporter activity. Cotransfection of pHMRluc with a vector expressing Rem elevated luciferase activity in HC11 mammary cell extracts (Fig. 5B). This response was eliminated by deletion of the Rem NLS/ARM sequence (RemΔNLS). Surprisingly, deletion of the Rem C terminus (RemΔC) increased export of the reporter RNA (ca. 18-fold relative to vector alone) compared to that achieved with the full-length protein (ca. 11-fold relative to vector alone) (P = 0.001; two-tailed Student's t test). Western blotting confirmed equivalent expression of Rem in each transfection (Fig. 5C). Similar results were obtained in XC rat fibroblasts that lack endogenous MMTVs, although the effect of C-terminal deletion was greater in these cells than in HC11 mouse mammary cells (compare Fig. 5B and 6A). Interestingly, a doublet of Rem protein was more apparent in the less-permissive XC cells, suggesting that Rem function may be regulated by posttranslational modifications (Fig. 6B). These experiments indicate that the Rem C terminus contains a negative regulatory domain that directly or indirectly controls the export function of the N terminus.
FIG. 5.
Rem-responsive vectors localize the RmRE. (A) Diagram of reporter vectors. (B) An MMTV-based reporter plasmid is responsive to rem expression vectors. HC11 cells were transfected and harvested at 48 h for reporter assays. Luciferase values were normalized for DNA uptake, and HMRluc alone was assigned a value of 1. Standard deviations from the averages of triplicate assays are given. This experiment is representative of at least three independent assays. (C) Western blot analysis confirms equal expression of RemGFP and RemΔCGFP. Whole-cell extracts were prepared from transfected HC11 cells and analyzed by incubation of Western blots with GFP-specific (upper panel) or actin-specific antibodies (lower panel). Normalized amounts of Rem expression are indicated after quantitation of GFP and actin levels using Scion Image software. SD, splice donor; SA, splice acceptor (46).
FIG. 6.
Activity of the HMRluc vector in XC cells with RemGFP and RemΔCGFP. (A) XC rat fibroblasts were transfected with HMRluc alone (lane 1) or cotransfected with HMRluc and plasmids expressing RemGFP (lanes 2 and 3) or RemΔCGFP (lanes 4 and 5). Cells were harvested at 36 h for reporter assays. Luciferase values were normalized for DNA uptake, and HMRluc alone was assigned a value of 1. Standard deviations from the averages of triplicate assays are given. This experiment is representative of at least four independent assays. (B) Western blot analysis confirmed expression of RemGFP and RemΔCGFP. Whole-cell extracts were prepared from transfected XC cells (shown in panel A) and analyzed by incubation of Western blots with GFP-specific (upper panel) or actin-specific antibodies (lower panel).
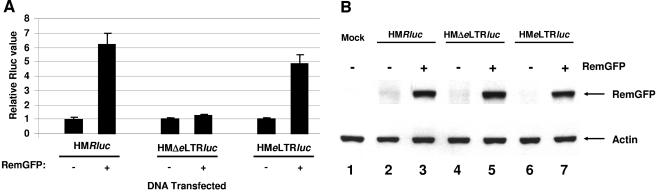
To determine the sequences necessary for the response to Rem, additional reporter vectors were constructed (Fig. 5A). Rem responsiveness was dependent on the presence of a downstream LTR, since the use of the SV40 polyadenylation signal allowed basal reporter activity (pHMΔeLTRluc) but abolished the Rem response (Fig. 7A). Reconstitution of Rem responsiveness was achieved by addition of a fragment containing the MMTV LTR and the 3′ end of the env gene (pHMeLTRluc). Western blot assays confirmed Rem expression, as expected (Fig. 7B). These data are consistent with localization of the RmRE near the 3′ end of the MMTV genome.
FIG. 7.
The RmRE includes the env-U3 border. (A) HC11 cells were transiently transfected and harvested after 48 h. Reporter assays were performed as described in the legend for Fig. 5B. Each independent reporter vector in the absence of Rem was assigned a relative value of 1. (B) Western blot analysis confirmed Rem expression. Whole-cell extracts were prepared from transfected HC11 cells used in panel A and analyzed by Western blotting after incubation with GFP-specific (upper panel) or actin-specific (lower panel) antibodies.
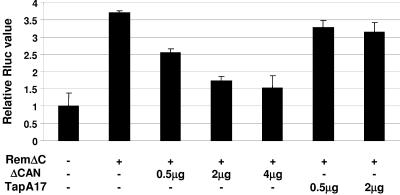
Other retroviral export proteins have leucine-rich NESs that are dependent on the Crm1 export pathway (14). Based on the observation that Rem has a leucine-rich region within the N-terminal 98 amino acids (Fig. 2B), pHMRluc was cotransfected with RemΔCGFP in the presence or absence of a dominant-negative inhibitor of Crm1 (ΔCAN) (Fig. 8). Rem responsiveness was largely abolished in a dose-dependent manner by expression of ΔCAN, in agreement with the idea that Crm1 mediates export of intron-containing MMTV RNAs. As a control for nonspecific effects on RNA metabolism, cotransfection of two different amounts of a dominant-negative inhibitor of NXF1/Tap (TapA17) had no significant effect on luciferase activity in the presence of Rem.
FIG. 8.
Crm1 dependence of Rem activity. XC fibroblasts were transfected with HMRluc in the absence or presence of RemΔCGFP as indicated. Increasing amounts of dominant-negative expression plasmids (ΔCAN or TapA17) also were transfected. Cells were harvested at 48 h, and extracts were assayed for luciferase activity as described in the legend for Fig. 5. The reporter vector in the absence of Rem was assigned a relative value of 1.
DISCUSSION
Preliminary data from our laboratory revealed that MMTV expresses a doubly spliced mRNA encoding a protein of 301 amino acids within the same reading frame as the envelope protein. This protein, which we named Rem, had Rev-like RNA export motifs (Fig. 2B), and the rem cDNA could complement defective Gag production from an MMTV provirus carrying an insertion mutation within the envelope gene (19). A report by Indik et al. confirmed the presence of the Rem protein, but no evidence for RNA export activity was provided (27). In this study, we have demonstrated that infectious MMTV proviruses express the same doubly spliced RNA encoding Rem protein from human Jurkat T cells and XC rat fibroblasts, which lack endogenous MMTVs (Fig. 2A). Furthermore, we have shown that the cloned rem cDNA was expressed both as a GFP-fusion protein and in the untagged form (Fig. 2C). Both the infectious HYB-MTV provirus and cloned rem cDNA separately could complement the defective export of unspliced gag-pol mRNA and Gag production of a mutant MMTV provirus (env148) carrying a transposon insertion within the rem open reading frame (Fig. 4). Complementation of the Gag defect of the env148 provirus was up to 42-fold; however, wild-type levels of Gag protein were not achieved. The explanation for this effect is unclear but may be due to additional proviral defects caused by trimethoprim cassette insertion. Nevertheless, our experiments definitively show that MMTV encodes an RNA export protein from a doubly spliced mRNA.
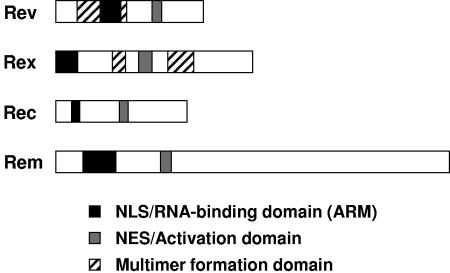
All retroviral export proteins contain a nuclear localization signal, an RNA-binding motif, and a leucine-rich domain containing an NES (25, 32) (Fig. 9). Rem contains an NLS/NoLS with an overlapping arginine-rich RNA-binding domain. As expected, expression of the GFP-tagged Rem was largely nucleolar, and localization was dependent on the N-terminal NLS/ARM (Fig. 3). The Rem protein also has a leucine-rich domain, which is typical of RNA export proteins that use the Crm1 export pathway (13). Deletion of the NLS/ARM (Fig. 5B) or expression of a dominant-negative mutant of Crm1/Xpo1 (Fig. 8) is sufficient to inactivate Rem function in RNA export. Previous results have reported the leptomycin B sensitivity of the nuclear export of unspliced gag-pol, but not partially spliced env transcripts in MMTV-infected cells (27). These experiments indicate that Rem, like other retroviral export proteins, uses Crm1 to mediate export of unspliced MMTV RNAs.
FIG. 9.
Domain structure of retroviral RNA export proteins.
Unlike other retroviral export proteins, except HERV-K Rec, the major motifs needed for Rem export function are localized to the signal peptide used for the envelope protein precursor (Fig. 2B). Interestingly, in nonprimate lentiviruses, e.g., equine infectious anemia virus (5) and caprine arthritis-encephalitis virus (46), rev-coding sequences are derived from part of the signal peptide-coding region and then joined to the TM region of the env gene in a different frame. A similar strategy is used for the HERV-K Rec protein (31). In MMTV Rem, the entire signal peptide and part of the SU sequence are joined by splicing to part of TM sequences, but in the same frame.
It is somewhat surprising that the HERV-K Rec and MMTV Rem proteins use the Env signal peptides to direct their export proteins to the nucleus (31). The majority of such signal sequences are recognized as nascent peptides by signal recognition particles (SRPs) and then directed to the endoplasmic reticulum for cotranslational transfer across the endoplasmic reticulum membrane (49). However, it is apparent that the efficiency of SRP recognition is affected by signal sequence hydrophobicity and length (16) and that there is a limited concentration of SRP relative to ribosomes (1). Both HERV-K and MMTV have envelope proteins with long signal peptides (31).
In this study, we also developed a vector that quantitatively measures Rem export function (Fig. 5). Expression of the 3′ end of the MMTV provirus from a cytomegalovirus promoter allowed Rem-responsive production of luciferase activity. Further manipulation of this vector allowed the mapping of the RmRE to a region including the env-U3 junction. Preliminary data suggest that removal of 3′ end of the U3 region also inactivates the RmRE (data not shown). These results are consistent with localization of the HIV Rev-responsive element within the envelope gene and the HERV-K Rec-responsive element and HTLV Rex-responsive elements to the LTR (23).
The sizes of different retroviral export proteins range from 12 to 21 kDa (25). However, the 33-kDa Rem protein is considerably larger. Interestingly, all motifs necessary for RNA export are localized within the first 98 amino acids of Rem (RemΔC), making it similar in structure to the related HERV-K Rec protein. Our results showed that RemΔC is sufficient for export of unspliced RNAs containing the RmRE. On the other hand, the full-length Rem protein induced significantly less export as determined by Gag expression from a rem-defective provirus (Fig. 4C) and by luciferase activity expressed from unspliced transcripts of the reporter vector in both HC11 mouse mammary cells (Fig. 5) and in XC rat fibroblasts (Fig. 6). Such experiments indicate that the Rem C terminus is likely to be a negative regulatory domain.
Recently, the p28 and p30 proteins of HTLV-1 and -2 have been reported to posttranscriptionally suppress the expression of the positive regulators, Tax and Rex, by retaining the doubly spliced mRNA for these proteins in the nucleus (41, 55). This regulatory strategy has been proposed to promote HTLV survival by minimizing the expression of highly immunogenic proteins, such as Tax (41). Examination of the Rem amino acid sequence using available software provides little information about the potential function(s) of the C terminus. Rev is considerably smaller than Rem but has been shown to regulate splicing (44). Moreover, use of the CTE/Tap pathway, rather than the Rev/Crm1 pathway, for HIV gag-pol RNA export restores viral budding in mouse cells (50). Although the exact role of the Rem C terminus is currently unknown, our previous work has confirmed the importance of negative regulation of transcription in the virus life cycle (6a, 7, 30, 56, 57). Suppression of MMTV expression, both in lymphoid cells and in early stages of mammary gland development, allows maximum virus production only for short periods of time during lactation (57). We suggest that modification of Rem activity at various times during mammary differentiation or in different cell types may allow posttranscriptional, as well as transcriptional, control of MMTV production.
Betaretroviruses have been shown to use two different mechanisms for export of intron-containing mRNAs to the cytoplasm. MPMV uses a CTE to recruit NXF1/Tap for nuclear export of viral RNA, whereas the HERV-Ks and MMTV employ Rev-like proteins as adaptors for recruitment of Crm1/Xpo1 for RNA export (14, 32). Moreover, MPMV encodes only a single accessory gene, dut, whereas MMTV has previously been shown to encode two accessory genes, sag and dut. Identification of the rem gene indicates that MMTV has at least three accessory genes. These differences suggest that MPMV and MMTV/HERV-K should be classified in different genera. Moreover, discovery of the RNA export strategy of another betaretrovirus, Jaagsiekte sheep retrovirus (42), will be intriguing and relevant for classification.
Complex retroviruses have multiple accessory genes, at least one of which is encoded by a multiply spliced RNA. The MMTV superantigen (Sag) is translated from a singly spliced RNA to give expression at the surface of antigen-presenting cells in conjunction with major histocompatibility complex class II protein (39). Sag is required for the amplification of viral expression in B and T cells and transmission of the infection among mammary gland cells (20, 21). The dut gene encodes a dUTPase (DU) from unspliced mRNA. DU is believed to be involved in the replication of a variety of lentiviruses, including feline immunodeficiency virus and equine infectious anemia virus, in nondividing cells (17). Feline immunodeficiency virus mutants that lack DU have reduced replication in macrophages and an increase in G-to-A transitions in proviruses integrated in these cells (43). Although simian immunodeficiency virus and HIV lack a dUTPase, the ability of Vif to counteract the action of APOBECs clearly serves the same function (24). The expression of an RNA export protein from a doubly spliced mRNA indicates that MMTV is the first complex murine virus to be described. These data also suggest that MMTV might be adapted by the insertion of novel accessory genes to develop mouse models of diseases, such as AIDS, induced by human complex retroviruses.
Acknowledgments
We thank members of the Dudley and Payne laboratories as well as Tom Hope and Jon Huibregtse for helpful suggestions and reagents.
This work was supported by the Texas Advanced Technology and Research Program and the Foundation for Research.
REFERENCES
- 1.Abell, B. M., M. R. Pool, O. Schlenker, I. Sinning, and S. High. 2004. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 23:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, C. R. 2005. Nonsense-mediated RNA decay: a molecular system micromanaging individual gene activities and suppressing genomic noise. Bioessays 27:463-466. [DOI] [PubMed] [Google Scholar]
- 3.Baillie, G. J., L. N. van de Lagemaat, C. Baust, and D. L. Mager. 2004. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J. Virol. 78:5784-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, J. K., H. Diggelmann, G. A. Dekaban, G. F. Grossi, R. Semmler, P. A. Waight, and R. F. Fletcher. 1988. Alterations in the U3 region of the long terminal repeat of an infectious thymotropic type B retrovirus. J. Virol. 62:2985-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beisel, C. E., J. F. Edwards, L. L. Dunn, and N. R. Rice. 1993. Analysis of multiple mRNAs from pathogenic equine infectious anemia virus (EIAV) in an acutely infected horse reveals a novel protein, Ttm, derived from the carboxy terminus of the EIAV transmembrane protein. J. Virol. 67:832-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, J., C. Aepinus, M. Dobrovnik, B. Fleckenstein, J. Hauber, and E. Bohnlein. 1991. Mutational analysis of functional domains in the HIV-1 Rev trans-regulatory protein. Virology 183:630-635. [DOI] [PubMed] [Google Scholar]
- 6a.Bhadra, S., M. M. Lozano, and J. P. Dudley. 2005. Conversion of mouse mammary tumor virus to a lymphomagenic virus. J. Virol. 79:12592-12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bramblett, D., C. L. Hsu, M. Lozano, K. Earnest, C. Fabritius, and J. Dudley. 1995. A redundant nuclear protein binding site contributes to negative regulation of the mouse mammary tumor virus long terminal repeat. J. Virol. 69:7868-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, I. C., E. Rohrbach, C. Schmitt, and E. Izaurralde. 1999. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 18:1953-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray, M., S. Prasad, J. W. Dubay, E. Hunter, K. T. Jeang, D. Rekosh, and M. L. Hammarskjold. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. USA 91:1256-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, W. Y., Q. R. Liu, J. Borjigin, H. Busch, O. M. Rennert, L. A. Tease, and P. K. Chan. 1989. Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry 28:1033-1039. [DOI] [PubMed] [Google Scholar]
- 12.Chirgwin, J. M., A. E. Przybyla, R. J. MacDonald, and W. J. Rutter. 1979. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18:5294-5299. [DOI] [PubMed] [Google Scholar]
- 13.Cullen, B. R. 2002. Using retroviruses to study the nuclear export of mRNA. Results Probl. Cell Differ. 35:151-168. [DOI] [PubMed] [Google Scholar]
- 14.Cullen, B. R. 2003. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28:419-424. [DOI] [PubMed] [Google Scholar]
- 15.Dudley, J. P. 1999. Mouse mammary tumor virus, p. 965-972. In R. G. Webster and A. Granoff (ed.), Encyclopedia of virology. Academic Press Ltd., London, United Kingdom.
- 16.Egea, P. F., R. M. Stroud, and P. Walter. 2005. Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biol. 15:213-220. [DOI] [PubMed] [Google Scholar]
- 17.Elder, J. H., D. L. Lerner, C. S. Hasselkus-Light, D. J. Fontenot, E. Hunter, P. A. Luciw, R. C. Montelaro, and T. R. Phillips. 1992. Distinct subsets of retroviruses encode dUTPase. J. Virol. 66:1791-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 19.Freed, E. O., and S. R. Ross. 2004. Retroviruses 2004: review of the 2004 Cold Spring Harbor Retroviruses Conference. Retrovirology 1:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golovkina, T. V., A. Chervonsky, J. P. Dudley, and S. R. Ross. 1992. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell 69:637-645. [DOI] [PubMed] [Google Scholar]
- 21.Golovkina, T. V., J. P. Dudley, and S. R. Ross. 1998. B and T cells are required for mouse mammary tumor virus spread within the mammary gland. J. Immunol. 161:2375-2382. [PubMed] [Google Scholar]
- 22.Hadzopoulou-Cladaras, M., B. K. Felber, C. Cladaras, A. Athanassopoulos, A. Tse, and G. N. Pavlakis. 1989. The Rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J. Virol. 63:1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanly, S. M., L. T. Rimsky, M. H. Malim, J. H. Kim, J. Hauber, D. M. Duc, S. Y. Le, J. V. Maizel, B. R. Cullen, and W. C. Greene. 1989. Comparative analysis of the HTLV-I Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes Dev. 3:1534-1544. [DOI] [PubMed] [Google Scholar]
- 24.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 25.Hauber, J. 2001. Nuclear export mediated by the Rev/Rex class of retroviral trans-activator proteins. Curr. Top. Microbiol. Immunol. 259:55-76. [DOI] [PubMed] [Google Scholar]
- 26.Hope, T. J., X. J. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Indik, S., W. H. Gunzburg, B. Salmons, and F. Rouault. 2005. A novel, mouse mammary tumor virus encoded protein with Rev-like properties. Virology 337:1-6. [DOI] [PubMed] [Google Scholar]
- 28.Kang, Y., and B. R. Cullen. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 13:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura, T., I. Hashimoto, T. Nagase, and J. Fujisawa. 2004. CRM1-dependent, but not ARE-mediated, nuclear export of IFN-α1 mRNA. J. Cell Sci. 117:2259-2270. [DOI] [PubMed] [Google Scholar]
- 30.Liu, J., D. Bramblett, Q. Zhu, M. Lozano, R. Kobayashi, S. R. Ross, and J. P. Dudley. 1997. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol. Cell. Biol. 17:5275-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lower, R., R. R. Tonjes, C. Korbmacher, R. Kurth, and J. Lower. 1995. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magin, C., J. Hesse, J. Lower, and R. Lower. 2000. Corf, the Rev/Rex homologue of HTDV/HERV-K, encodes an arginine-rich nuclear localization signal that exerts a trans-dominant phenotype when mutated. Virology 274:11-16. [DOI] [PubMed] [Google Scholar]
- 33.Malim, M. H., J. Hauber, R. Fenrick, and B. R. Cullen. 1988. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335:181-183. [DOI] [PubMed] [Google Scholar]
- 34.Malim, M. H., L. S. Tiley, D. F. McCarn, J. R. Rusche, J. Hauber, and B. R. Cullen. 1990. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 60:675-683. [DOI] [PubMed] [Google Scholar]
- 35.Matlin, A. J., F. Clark, and C. W. Smith. 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6:386-398. [DOI] [PubMed] [Google Scholar]
- 36.Mertz, J. A., F. Mustafa, S. Meyers, and J. P. Dudley. 2001. Type B leukemogenic virus has a T-cell-specific enhancer that binds AML-1. J. Virol. 75:2174-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 38.Mustafa, F., S. Bhadra, D. Johnston, M. Lozano, and J. P. Dudley. 2003. The type B leukemogenic virus truncated superantigen is dispensable for T-cell lymphomagenesis. J. Virol. 77:3866-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mustafa, F., M. Lozano, and J. P. Dudley. 2000. C3H mouse mammary tumor virus superantigen function requires a splice donor site in the envelope gene. J. Virol. 74:9431-9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narayan, M., I. Younis, D. M. D'Agostino, and P. L. Green. 2003. Functional domain structure of human T-cell leukemia virus type 2 rex. J. Virol. 77:12829-12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicot, C., M. Dundr, J. M. Johnson, J. R. Fullen, N. Alonzo, R. Fukumoto, G. L. Princler, D. Derse, T. Misteli, and G. Franchini. 2004. HTLV-1-encoded p30II is a post-transcriptional negative regulator of viral replication. Nat. Med. 10:197-201. [DOI] [PubMed] [Google Scholar]
- 42.Palmarini, M., and H. Fan. 2003. Molecular biology of Jaagsiekte sheep retrovirus. Curr. Top. Microbiol. Immunol. 275:81-115. [DOI] [PubMed] [Google Scholar]
- 43.Payne, S. L., and J. H. Elder. 2001. The role of retroviral dUTPases in replication and virulence. Curr. Protein Peptide Sci. 2:381-388. [DOI] [PubMed] [Google Scholar]
- 44.Powell, D. M., M. C. Amaral, J. Y. Wu, T. Maniatis, and W. C. Greene. 1997. HIV Rev-dependent binding of SF2/ASF to the Rev response element: possible role in Rev-mediated inhibition of HIV RNA splicing. Proc. Natl. Acad. Sci. USA 94:973-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purdy, A., L. Case, M. Duvall, M. Overstrom-Coleman, N. Monnier, A. Chervonsky, and T. Golovkina. 2003. Unique resistance of I/LnJ mice to a retrovirus is due to sustained interferon gamma-dependent production of virus-neutralizing antibodies. J. Exp. Med. 197:233-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saltarelli, M., G. Querat, D. A. Konings, R. Vigne, and J. E. Clements. 1990. Nucleotide sequence and transcriptional analysis of molecular clones of CAEV which generate infectious virus. Virology 179:347-364. [DOI] [PubMed] [Google Scholar]
- 47.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shackleford, G. M., and H. E. Varmus. 1988. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc. Natl. Acad. Sci. USA 85:9655-9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shan, S. O., and P. Walter. 2005. Co-translational protein targeting by the signal recognition particle. FEBS Lett. 579:921-926. [DOI] [PubMed] [Google Scholar]
- 50.Swanson, C. M., B. A. Puffer, K. M. Ahmad, R. W. Doms, and M. H. Malim. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 23:2632-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 52.Wiegand, H. L., G. A. Coburn, Y. Zeng, Y. Kang, H. P. Bogerd, and B. R. Cullen. 2002. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 22:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 54.Yang, J. N., and J. Dudley. 1992. Endogenous Mtv-8 or a closely linked sequence stimulates rearrangement of the downstream Vκ9 gene. J. Immunol. 149:1242-1251. [PubMed] [Google Scholar]
- 55.Younis, I., L. Khair, M. Dundr, M. D. Lairmore, G. Franchini, and P. L. Green. 2004. Repression of human T-cell leukemia virus type 1 and type 2 replication by a viral mRNA-encoded posttranscriptional regulator. J. Virol. 78:11077-11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, Q., K. Gregg, M. Lozano, J. Liu, and J. P. Dudley. 2000. CDP is a repressor of mouse mammary tumor virus expression in the mammary gland. J. Virol. 74:6348-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, Q., U. Maitra, D. Johnston, M. Lozano, and J. P. Dudley. 2004. The homeodomain protein CDP regulates mammary-specific gene transcription and tumorigenesis. Mol. Cell. Biol. 24:4810-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]