Abstract
Comparative analysis of capsid protein structures in the eukaryote-infecting herpesviruses (Herpesviridae) and the prokaryote-infecting tailed DNA bacteriophages (Caudovirales) revealed a characteristic fold that is restricted to these two virus lineages and is indicative of common ancestry. This fold not only serves as a major architectural element in capsid stability but also enables the conformational flexibility observed during viral assembly and maturation. On the basis of this and other emerging relationships, it seems increasingly likely that the very diverse collection of extant viruses may have arisen from a relatively small number of primordial progenitors.
There are many unresolved questions concerning the origins of viruses and their subsequent evolutionary histories (10). In the nature of their genomes, replication mechanisms, and particle structures, viruses represent a very diverse group of entities, which seems to imply multiple independent origins. To make order of this diversity, viruses have traditionally been grouped using a wide range of physical and biological properties (18). With the explosive growth in sequence availability, genomic comparison has increasingly been used to supplement and extend other classification criteria. However, in such rapidly evolving organisms as viruses, sequence-based methods are less effective at uncovering deeply rooted evolutionary links. A likely place to find evidence for shared ancestry between highly diverged viruses is in fundamental structures, such as the capsid, that are likely to have been established at a very early stage in the history of the viruses, predating any evolutionary split. In support of this supposition, recent analyses of capsid protein structures have revealed previously unsuspected relationships among apparently distinct virus families (1, 3). Here we propose, based on analysis of their capsid structures, that two lineages of large double-stranded DNA viruses, the Herpesviridae and Caudovirales, are structurally and evolutionarily related.
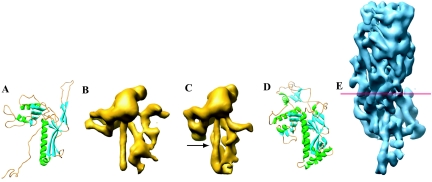
Potential links between the eukaryote-infecting Herpesviridae and the prokaryote-infecting Caudovirales have been suggested previously because of parallels in their capsid assembly pathways and similarities between their portal complexes, through which DNA enters the capsid (2, 16). While only one bacteriophage portal structure has been determined by X-ray crystallography (15), the distinctive 12-fold arrangement of subunits has been reported for several other bacteriophages and also one herpesvirus (17). However, the overall appearance of herpesvirus particles is very different from that of the Caudovirales and sequence comparison has not provided any evidence for common origins in either capsid shell or portal proteins. To investigate whether evidence for potential relationships could be detected at the protein structural level, we compared the capsid structure of herpes simplex virus type 1 (HSV-1) with those of four bacteriophages, P22, Φ29, T4, and HK97, all members of the Caudovirales (7, 13, 14, 19) (Fig. 1). The crystal structure of the HK97 capsid (19) shows that the capsid protein is roughly triangular (Fig. 1A) and contains a fold not found in any other protein in the SCOP database (http://scop.mrc-lmb.cam.ac.uk/scop/). This signature fold consists of three α-helices and two β-sheets. The sub-nanometer-resolution cryoelectron microscopy (cryoEM) structures of P22 (13) (Fig. 1B and C) and Φ29 (14) particles and the X-ray crystal structure of gp24, the T4 pentavalent capsid protein (8) (Fig. 1D), established that the capsid proteins of these bacteriophages follow the same fold design despite having disparate sequences (<15% identity).
FIG. 1.
A gallery of bacteriophage capsid protein structures determined by either X-ray crystallography or cryoEM. HK97 gp5 (A), mature P22 gp5 (B), procapsid P22 gp5 (C), and T4 gp24 (D) are shown in comparison to HSV-1 VP5 (E). VP5, the 145-kDa capsid protein, was segmented from an approximately 8-Å cryoEM map of the HSV-1 capsid. The red line demarcates the boundary between the floor domain and the other two domains of VP5 (upper and middle domains). The N-terminal helix in P22 that has been proposed to undergo refolding is indicated by the arrow in panel C.
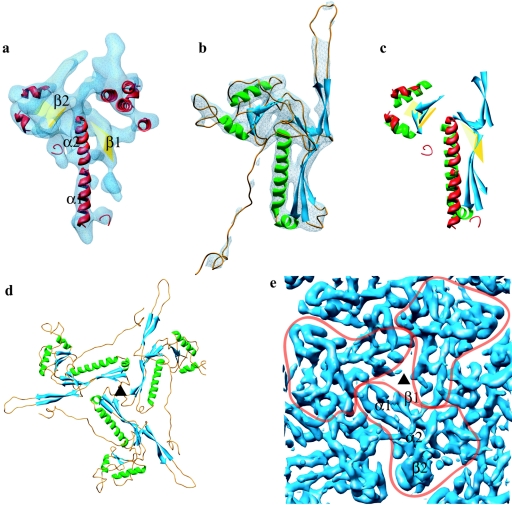
Our analysis of an improved (approximately 8-Å) cryoEM map of the HSV-1 capsid obtained with a larger data set than that published previously (20) now reveals that the major capsid protein, VP5, has the same structural organization in its floor (Fig. 1E). Although VP5 is much larger than the bacteriophage capsid proteins (Fig. 1), the size disparity is almost entirely accounted for by the middle and upper domains of VP5, which form the large penton and hexon towers. The VP5 floor domain has very similar dimensions and capsomere spacing to the HK97 capsid protein, gp5. Both the overall shape of the HK97 gp5 protein and the disposition of its secondary structural elements are preserved in the VP5 floor domain (Fig. 2a and b). This is shown by the positional match of the α-helix centroids with <2.5-Å root mean square deviation (Fig. 2c) and by the close match in the locations of the β-sheets. Additionally, the relative orientations and molecular interfaces of the subunits are retained, with the α-helices and β-sheets from the three subunits giving rise to a common architecture at the three-fold axes in the floors of HK97 and HSV-1 (Fig. 2d and e). Thus, the exhibited fold can be considered a structural signature for these viruses, which is analogous to the well-known β-sandwich fold of many RNA virus particles (9) or the double β-barrel of some DNA virus capsids (1).
FIG. 2.
Match of the secondary structure elements of HSV-1 capsid VP5 and HK97 phage gp5 and their molecular interactions in the capsids. (a) The isolated VP5 floor domain, in blue, viewed from outside the capsid. SSEhunter identified two long α-helices (red; α1 and α2) adjacent to a large β-sheet (yellow; β1) in the floor domain, as well as a second β-sheet (yellow; β2) and several smaller helices flanking α1 and α2. (b) The HK97 capsid protein (gp5) shown in the same view reveals a similar structural motif. A simulated density map for gp5 at approximately 8-Å resolution is shown in pale blue. (c) Alignment of the secondary structure elements by use of Foldhunter (12) demonstrated a clear match between the floor domain of VP5 and the core structure of gp5. (d) Arrangement of gp5 subunits around a local three-fold axis (▴) in the HK97 capsid as viewed from inside the capsid. (e) Organization of the HSV-1 capsid floor as shown in the same view in panel d. Individual VP5 subunit floor domains are demarcated with the long α-helices (α1, α2) and associated β-sheets (β1, β2) annotated in one subunit.
CryoEM “snapshots” of HSV-1 capsids undergoing angularization during maturation show extensive structural rearrangements in the floor domain (11). Similarly, P22 capsid proteins undergo large movements during maturation, including rotation of the β-sheet about the long α-helix and refolding of another α-helix, resulting in capsid expansion by over 100 Å in external diameter (13). The similar dispositions of secondary structural elements in P22 and HSV-1 raise the possibility that an equivalent rotation produces the changes in the floor domain seen during HSV-1 maturation. Since the signature capsid protein fold has been found in all of the sufficiently well-studied capsids that undergo reconfiguration, it is likely that its development was an important factor in meeting the potentially conflicting demands imposed by the need to maintain capsid stability while allowing for conformational changes associated with virus maturation.
The observation that herpesviruses and tailed bacteriophages are related through their capsid protein structures triggers the need for a re-evaluation of evolutionary evidence from sequence comparisons. The divergence between the evolutionarily distant fish, mollusk, and mammalian herpesviruses is so great that their common ancestry cannot be deduced from sequence similarity (4, 6). The best candidate for a protein that is specific to herpesviruses is a packaging protein, the putative terminase encoded by HSV-1 gene UL15. In the past it has not been considered diagnostic for Herpesviridae, as it is distantly related to an equivalent function identified in certain Caudovirales (5). However, in light of the evolutionary link demonstrated through their capsid protein folds, we can now interpret the occurrence of related packaging proteins as independent evidence for common ancestry of these two viral lineages.
The arrangement of subunit proteins in the portal and the characteristic folds of the capsid proteins are sufficiently distinctive to suggest that each evolved only once. Both are fundamental components of the capsid structure, which represents one of the defining features of any virus. When considered together with the retained homology in the terminase protein, they provide a compelling molecular evidence-based argument in support of a common origin for the particle-packaging complex in the Caudovirales and Herpesviridae. This linkage between the most abundant set of bacteriophages and a major family of eukaryotic viruses highlights the growing evidence of relationships across these major biological divides. In light of this and other recent studies (1, 3), it is becoming increasingly plausible that extant viruses may have arisen from a relatively small number of primordial progenitors.
It is not clear whether the existence of related viruses infecting cells from different domains of life reflects the presence of a common ancestor that predates the separation of the domains or is a result of later adaptation to a new host cell. Superficially, the strategy adopted by incoming herpesvirus capsids to target and release DNA into eukaryotic nuclei appears analogous to that employed by tailed bacteriophages to infect bacterial cells. Although this resemblance may be coincidental, it is conceivable that if eukaryotic cells are the products of symbiosis, as has been postulated, it actually reflects the retention of an ancient pathway of infection. In this case, the common ancestor of the Caudovirales and Herpesviridae would predate the incorporation of the prokaryotic derived nucleus into the eukaryotic cell.
Acknowledgments
This investigation was stimulated by Roger Hendrix of Pittsburgh University. We thank Duncan McGeoch for critically reading the manuscript.
Research has been supported by the National Institutes of Health and National Science Foundation and by the UK Medical Research Council and the UK-Texas Bioscience Initiative of the Department of Trade and Industry.
REFERENCES
- 1.Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673-685. [DOI] [PubMed] [Google Scholar]
- 2.Casjens, S., and J. King. 1975. Virus assembly. Annu. Rev. Biochem. 44:555-611. [DOI] [PubMed] [Google Scholar]
- 3.Coulibaly, F., C. Chevalier, I. Gutsche, J. Pous, J. Navaza, S. Bressanelli, B. Delmas, and F. A. Rey. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761-772. [DOI] [PubMed] [Google Scholar]
- 4.Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 5.Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 6.Davison, A. J., B. L. Trus, N. Q. Cheng, A. C. Steven, M. S. Watson, C. Cunningham, R. M. Le Deuff, and T. Renault. 2005. A novel class of herpesvirus with bivalve hosts. J. Gen. Virol. 86:41-53. [DOI] [PubMed] [Google Scholar]
- 7.Fokine, A., P. R. Chipman, P. G. Leiman, V. V. Mesyanzhinov, V. B. Rao, and M. G. Rossmann. 2004. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. USA 101:6003-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fokine, A., P. G. Leiman, M. M. Shneider, B. Ahvazi, K. M. Boeshans, A. C. Steven, L. W. Black, V. V. Mesyanzhinov, and M. G. Rossmann. 2005. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. USA 102:7163-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison, S. C. 2001. Principles of virus structure, p. 53-85. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strauss (ed.), Fields virology, 4th ed., vol. 1. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 10.Hendrix, R. W. 1999. Evolution: the long evolutionary reach of viruses. Curr. Biol. 9:R914-R917. [DOI] [PubMed] [Google Scholar]
- 11.Heymann, J. B., N. Q. Cheng, W. W. Newcomb, B. L. Trus, J. C. Brown, and A. C. Steven. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10:334-341. [DOI] [PubMed] [Google Scholar]
- 12.Jiang, W., M. L. Baker, S. J. Ludtke, and W. Chiu. 2001. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 308:1033-1044. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, W., Z. L. Li, Z. X. Zhang, M. L. Baker, P. E. Prevelige, and W. Chiu. 2003. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 10:131-135. [DOI] [PubMed] [Google Scholar]
- 14.Morais, M. C., K. H. Choi, J. S. Koti, P. R. Chipman, D. L. Anderson, and M. G. Rossmann. 2005. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of Φ29. Mol. Cell 18:149-159. [DOI] [PubMed] [Google Scholar]
- 15.Simpson, A. A., Y. Z. Tao, P. G. Leiman, M. O. Badasso, Y. N. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes, D. L. Anderson, T. S. Baker, and M. G. Rossmann. 2000. Structure of the bacteriophage φ29 DNA packaging motor. Nature 408:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 17.Trus, B. L., N. Q. Chen, W. W. Newcomb, F. L. Homa, J. C. Brown, and A. C. Steven. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.). 2000. Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 19.Wikoff, W. R., L. Liljas, R. L. Duda, H. Tsuruta, R. W. Hendrix, and J. E. Johnson. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289:2129-2133. [DOI] [PubMed] [Google Scholar]
- 20.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 Å. Science 288:877-880. [DOI] [PubMed] [Google Scholar]