Abstract
Using PCR, we screened 105 isolates of poultry-associated Campylobacter jejuni for the presence of class 1 integrons. Of those isolates, 21% (22 of 105) possessed the integrase gene, but only 5 isolates produced an amplicon in a 5′-3′ conserved sequence PCR directed toward amplification of the resistance cassettes. DNA sequencing demonstrated that all five isolates possessed the aminoglycoside resistance gene, aacA4.
Campylobacter jejuni is one of the leading causes of sporadic bacterial enteritis in the United States (6). While most infections are self-limiting, invasive infections and chronic colitis are commonly treated with antibiotics (3). Antibiotic resistance is increasingly a serious problem in some pathogens, and the capability for multidrug resistance is a very real concern. Some authorities regard food animals as the primary source of antibiotic resistance genes present in human food-borne pathogens, whereas others regard imprudent use of antibiotics in humans as the major source of the problem (9). Regardless, it is clear that use of antimicrobials both in animals and in humans can select for resistant bacterial populations.
A genetic element called the integron is potentially a major agent in dissemination of multidrug resistance among gram-negative bacteria. The integron contains an integrase gene and a site-specific integration site where the integrase can link antibiotic resistance gene cassettes in tandem in the integration site when the cassette molecules possess a 59-base element (59-be) (14, 32). Presently there are nine known classes of integrons, based on integrase gene homology (17, 25). Over 60 distinct antibiotic resistance gene cassettes have been characterized within integrons, and as many as 7 of them have been found in a single integron at one time (8, 23). Integrons have a broad distribution among gram-negative fecal bacteria of animal origin, and in fact, many plasmids encoding multiresistance in the Enterobacteriaceae carry a transposon of the Tn21 family, which contains a class 1 integron (12, 13). In one study, 60% of Escherichia coli isolates from poultry possessed a Tn21-like transposon carrying the aadA1 streptomycin-spectinomycin resistance gene cassette (2). Campylobacter species are commonly found among the intestinal flora of chickens, and Gibreel and Skold characterized trimethoprim resistance gene cassettes in Campylobacter that were linked to integrase genes (10, 11). The purpose of this study was to determine whether class 1 integrons are common mediators of antibiotic resistance in C. jejuni associated with poultry, since the microflora of the avian intestinal tract are a rich source of these genetic elements.
C. jejuni was cultured from the nipple drinker waterline and from cecal droppings from three sequential flocks of broiler chickens within a commercial broiler house by the method of Ransom and Rose (29). C. jejuni was similarly cultured from washes of processed broiler carcasses from two different processing plants. Presumptive colonies were confirmed by phase-contrast microscopy and biochemical tests or serological tests per the manufacturer's recommendation (Integrated Diagnostics Inc., Baltimore, Md.). Isolates were screened for possession of the class 1 integrase gene by PCR using a whole-cell template as previously described (2, 26). Positive results were obtained for 22 of the C. jejuni isolates (n = 105); 7 of 40 (17.5%) of the isolates cultured from carcasses obtained from the processing plants, 10 of 58 (17.2%) of the isolates cultured from chicken cecal droppings, and 5 of 7 (71.4%) isolates from the waterline (nipple drinkers) in the grow-out house possessed class 1 integrase. Because the chickens were culture positive in the grow-out house after C. jejuni was cultured from the waterline, we investigated whether the nipple drinkers were the source of the integrase-containing C. jejuni which then colonized the broiler chickens. To test this hypothesis, randomly amplified polymorphic DNA PCR was used to genetically type the isolates, using the method described by Payne et al. (26). Seven distinct randomly amplified polymorphic DNA patterns resulted; however, none of the integrase-containing strains from the waterline were present among the strains isolated from chickens. In fact, class 1 integrase was possessed by three different genetic types isolated from the chickens and two genetic types isolated from the waterline in the flock house.
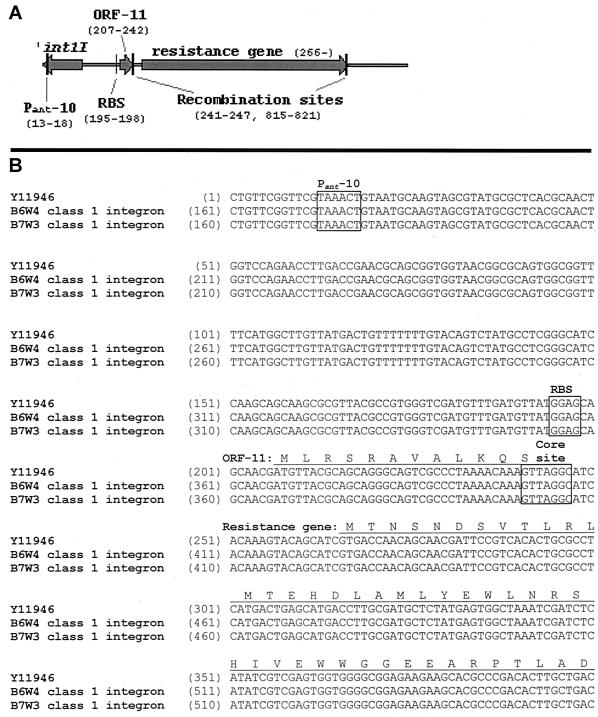
Isolates containing class 1 integrases were assayed for the presence of contiguous resistance gene cassettes, using 5′-3′ conserved sequence (CS) PCR with primers targeted to the 5′ and 3′ conserved regions of the class 1 integron sequences (21). When integrase-positive isolates were assayed, only the C. jejuni strains isolated from the waterline produced PCR amplicons visible on agarose gels. All five of the waterline isolates produced a 900-bp amplicon, suggesting that they possessed a single cassette in the integron. Amplicons from two waterline isolates were gel purified and DNA sequenced at the campus core facility, using an ABI 9600 automated sequencer. Analysis of the DNA sequences, using the online BLAST algorithm at the National Center for Biotechnology Information web server (www.ncbi.nlm.nih.gov), revealed that they were nearly identical to the nucleotide sequence for the aacA4 gene. To determine the identity of upstream sequences which could influence expression of the cassette, PCR was performed using primers int1PS (5′-GAA CAG CAA GGC CGC CAA T-3′) and aacA4R (5′-TTC TTC TCC GCC CCA CCA CT-3′), which were designed to amplify an area spanning the 3′ region of the integrase gene and the 5′ region of the aacA4 gene which would include the putative cassette promoter(s) and 59-be. The primers were based on the sequence submitted to GenBank under accession number AY06966, which also contained aacA4 as the first cassette in the integron and was predicted to produce a 538-bp product. PCR was done using a 10-μl reaction mixture which consisted of 0.2 mM deoxynucleoside triphosphates, 2.0 mM MgCl2, 1× PCR buffer (50 mM Tris [pH 7.4]), bovine serum albumin (0.25 mg/ml), 50 pmol (each) of forward and reverse PCR primers, and 0.5 U of Taq DNA polymerase (Boehringer Mannheim, Indianapolis, Ind.). The program parameters for the Idaho Technology Rapidcycler (Idaho Falls, Idaho) included an incubation at 95°C for 2 min followed by (i) 95°C for 15 s, (ii) 54°C for 15 s, and (iii) 72°C for 30 s for 40 cycles. The PCR product of approximately 550 bp was gel purified, DNA sequenced, and analyzed as described above. Figure 1 shows the sequence homology among the Campylobacter integrons and an integron isolated from an Enterobacter cloacae (5, 16).
FIG. 1.
Comparison of the sequence of Campylobacter integron-associated aacA4 to the sequence submitted to GenBank under accession number Y11946. Panel A shows the schematic annotation of Y11946, which is based on the findings of Hanau-Bercot et al. (16), who demonstrated that expression of the aacA4 was dependent upon the ribosome binding site (RBS) preceding ORF-11. Panel B shows the homology among the Campylobacter integron sequences and Y11946. The locations of the −10 sequence of the Pant promoter (7), the ribosome binding site, and the recombination core site of the 59-be (15) are marked with boxes. The derived translation of the ORF-11 and N terminus of AAC(6′)-Ib7 are shown underlined above their respective DNA sequences.
To determine their identities, the 900-bp amplicons from the other three waterline isolates were gel isolated and used as the template in a PCR-enzyme-linked immunosorbent assay (ELISA) as previously described (24). This consisted of the 5′-3′ CS PCR using digoxigenin-labeled nucleotides (Roche Molecular Biochemicals, Indianapolis, Ind.) and a biotinylated aacA4-specific probe. The 5′-3′ CS PCR was performed as previously described (21), except that the reaction mixture (50 mM Tris [pH 8.3], 3 mM MgCl2, 0.25 mg of bovine serum albumin/ml, 0.5% Ficoll) contained 0.1 fM of the 3′ biotinylated probe 5′-GGCATCCAAGCAGCAAG-3′. After 30 cycles, the amplicons were denatured by incubation for 1 min at 96°C, with probe annealing for 15 min at 50°C. Probe-amplicon hybrids were captured in streptavidin-coated wells and detected using digoxigenin antibody conjugate as described by the manufacturer (Roche Molecular Biochemicals). C. jejuni isolates B6W4 and B7W3 were used as the positive controls; Salmonella enterica serotype Typhimurium DT104 was used as the negative control (4). All of the intI1+ waterline isolates contained aacA4, as determined by DNA-DNA hybridization in the PCR-ELISA. Four of the aacA4-containing water isolates (B4W4, B6W2, B6W4, and B7W3) representing one genetic type were cultured during the fourth, sixth, or seventh week of the 49-day grow-out period of the flock, while isolate B7W1, representing a unique strain, was cultured only during the seventh week. Nipples on drinkers in the grow-out house were randomly chosen for removal and culture; therefore, these strains were not acquired from repeated cultures of the same nipple drinker. Since the two different strains were isolated from physically separate nipple drinkers, it is likely that they independently acquired the integron from a common source that colonized the biofilms of the waterline in the broiler house. However, 5′-3′ CS PCR coupled with PCR-ELISA did not detect the cassette among the community DNAs of water samples from other nipple drinkers in the house.
The aminoglycoside resistance gene, aacA4, has been found in integrons from multiple species of bacteria and in class 1 and class 3 integrons (1, 22, 27, 28, 33). aacA4 encodes an aminoglycoside 6′-N-acetyltransferase [AAC(6′)-I or -II] which confers resistance to aminoglycosides containing the purpurosamine ring, including tobramycin, kanamycin, and neomycin (30, 33). Point mutations resulting in a threonine-to-serine substitution (amino acid position 102 in the sequence submitted to GenBank under accession number AAA25688) determine whether the gene also confers resistance to amikacin [AAC(6′)-I] or gentamicin [AAC(6′)-II] (30). Figure 2 shows that the derived amino acid sequence of the aacA4 allele from Campylobacter demonstrated 100% homology to the AAC(6′)-Ib7 enzyme, which has been shown to confer characteristics resulting in higher kanamycin and tobramycin MICs but only slightly increased resistance to gentamicin (5). To evaluate the phenotype conferred by the AAC(6′)-Ib7 enzyme in C. jejuni, four of the isolates containing aacA4 were assayed for tobramycin and gentamicin resistance, using agar dilution on Mueller-Hinton agar containing 5% washed sheep red blood cells (19). Table 1 shows that for the int+ aacA4+ strains tested, tobramycin MICs were 25 to 50 μg/ml and gentamicin MICs were 10 to 50 μg/ml. Among the int+ aacA4 strains tested, MICs were <2 μg/ml. These results strongly suggest that the AAC(6′) activity is expressed in these C. jejuni strains.
FIG. 2.
Comparison of Campylobacter aacA4-derived amino acid sequence to AAC(6′)-Ib and -II sequences. Campylobacter AAC(6′) demonstrates 100% homology to AAC(6′)-Ib7. The serine residue contributing to gentamicin resistance (GenR>AmiR) is marked in bold (30); other dissimilar residues among the AAC(6′) enzymes are marked in bold lowercase letters.
TABLE 1.
Tobramycin and gentamicin MICs for class 1 integron-containing Campylobacter jejuni isolates carrying the aacA4 aminoglycoside resistance gene cassette
| Isolate | Sourcea | aacA4b | Tobramycin MIC (μg/ml)c | Gentamicin MIC (μg/ml)c |
|---|---|---|---|---|
| B5-8 | Feces | − | <0.5 | <0.5 |
| 2A4 | Carcass | − | <0.5 | 1 |
| B6W2 | Water | + | >50 | >50 |
| B6W4 | Water | + | >50 | >50 |
| B7W1 | Water | + | 25 | 10 |
| B7W3 | Water | + | >50 | >50 |
Campylobacter jejuni was cultured from nipple drinkers in a broiler house (water), cecal droppings of three successive flocks of chickens in the same broiler house (feces), or carcass washes of broilers from two different flocks at the processing plant (carcass). All isolates in this table possess class 1 integrase, as detected by PCR (2).
Presence or absence of aacA4 cassette in a class 1 integron as determined by 5′-3′ CS PCR (21) coupled with DNA sequencing or DNA-DNA hybridization (24). Templates from isolates B5-8 and 2A4 do not produce amplicons in 5′-3′ CS PCR.
Resistance phenotype determined by agar dilution (19).
The sequences affecting the expression of some integron cassettes are still being elucidated. Past studies have shown that there are two sets of −35 and −10 sequences, upstream of the cassettes, which are present within the class 1 integrase gene itself (7, 20). The first set comprises the promoter Pant and is commonly involved in transcription of the cassettes (7). While most class 1 integrons possess this promoter, point changes in the −35 or −10 sequences affect the level of transcription (7). The Pant promoter we detected contained a hybrid version (−35, TGGACA; −10, TAAACT) that has been shown to mediate expression in a manner comparable to that of Ptac (20). The second set of −35 and −10 sequences comprise a putative P2 promoter, but the promoter has been shown to initiate transcription only when the spacing is at least 17 bp (31). The P2 region found in many integrons, including this Campylobacter tobramycin resistance integron, contains a 14-bp spacer only between the −35 and −10 sequences and probably does not promote expression. For some integrons it is unclear how resistance is expressed, since one cassette may not contain a recognizable translation initiation region while others demonstrate canonical ribosome binding sites. The integron in this work demonstrated high homology to the sequence submitted to GenBank under accession number Y11946 (Fig. 1), which also does not demonstrate the presence of an apparent ribosome binding site linked to the AAC(6′)-Ib7 open reading frame (ORF) (5). Hanau-Bercot et al. demonstrated that efficient translation of the AAC(6′)-Ib7 in this integron occurred because it was coupled to translation of an 11-amino-acid ORF (ORF-11) immediately upstream (16). Expression in Campylobacter presumably occurred by the same mechanism.
Although the chickens in this study were not colonized with the same Campylobacter jejuni strains present in the waterline, they were colonized with a variety of genetic types that contained the class 1 integrase gene. If gene exchange occurs readily among members of the intestinal microflora, these integrons should have captured antibiotic resistance genes from cassette-containing genetic elements transferred to Campylobacter. Thus, we expected to find the streptomycin resistance gene aadA1, because it is present at a very high incidence in class 1 integrons found among poultry E. coli (2). However, this gene was not detected among the integrons present in poultry C. jejuni, suggesting either that Campylobacter and E. coli do not readily undergo gene exchange or that the elements are not stably maintained.
Among the aminoglycoside antibiotics that are approved for poultry production (34, 35, 36), aacA4 does not confer resistance to streptomycin, although it does confer resistance to gentamicin. Gentamicin is used at the hatchery but not on the broiler farms themselves, and there was no antibiotic selective pressure for this gene in the waterlines of the broiler house, since there was no history of antibiotic usage in the flock house. Since aacA4 was not found among the class 1 integron-containing isolates cultured from chickens, the presence of this gene may reflect human contamination of the farm environment. Studies by Jacobs-Reitsma et al. indicate that flock depopulation is a major risk factor for flock colonization by Campylobacter, presumably as a result of the entrance of farmworkers and equipment into the house (18). In the broiler houses we examined, farmworkers often removed nipple waters for cleaning or replacement between flocks. Contamination with human flora might contribute to the species colonizing the biofilm, ultimately affecting the potential gene pool available for gene transfer.
Nucleotide sequence accession numbers.
The integron-associated aacA4 sequences from the waterline isolates B6W4 and B7W3 were submitted to GenBank under accession numbers AF439785 and AF439786, respectively.
Acknowledgments
Thanks to John Glisson, John Maurer, and Susan Little for critical reading of the manuscript. Thanks to Harold Barnhardt for providing access to isolates for the study and Leeanne Buckner for assistance in producing a whole-cell template for analysis.
REFERENCES
- 1.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass, L., C. A. Liebert, M. D. Lee, D. G. White, A. O. Summers, S. G. Thayer, and J. J. Maurer. 1999. The incidence and characterization of integrons, genetic elements associated with multiple drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43:2925-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser, M. J. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176(Suppl. 2):S103-S105. [DOI] [PubMed] [Google Scholar]
- 4.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casin, I., F. Bordon, P. Bertin, A. Coutrot, I. Podglajen, R. Brasseur, and E. Collatz. 1998. Aminoglycoside 6′-N-acetyltransferase variants of the Ib type with altered substrate profile in clinical isolates of Enterobacter cloacae and Citrobacter freundii. Antimicrob. Agents Chemother. 42:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2001. Preliminary FoodNet data on the incidence of foodborne illnesses—selected sites, United States, 2000. Morb. Mortal. Wkly. Rep. 50:241-246. [PubMed] [Google Scholar]
- 7.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fluit, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 9.Food and Drug Administration. 2001. Hot topics. Antibiotic resistance: a growing threat. [Online.] http://www.fda.gov/oc/opacom/hottopics/anti_resist.html.
- 10.Gibreel, A., and O. Skold. 1998. High-level resistance to trimethoprim in clinical isolates of Campylobacter jejuni by acquisition of foreign genes (dfr1 and dfr9) expressing drug-insensitive dihydrofolate reductases. Antimicrob. Agents Chemother. 42:3059-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibreel, A., and O. Skold. 2000. An integron cassette carrying dfr1 with 90-bp repeat sequences located on the chromosome of trimethoprim-resistant isolates of Campylobacter jejuni. Microb. Drug Resist. 6:91-98. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein, C., M. D. Lee, S. Sanchez, C. Hudson, B. Phillips, B. Register, M. Grady, C. Liebert, A. O. Summers, D. G. White, and J. J. Maurer. 2001. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45:723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grinsted, J., F. de la Cruz, and R. Schmitt. 1990. The Tn21 subgroup of bacterial transposable elements. Plasmid 24:163-189. [DOI] [PubMed] [Google Scholar]
- 14.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 15.Hall, R. M., D. E. Brookes, and H. W. Stokes. 1991. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 5:1941-1959. [DOI] [PubMed] [Google Scholar]
- 16.Hanau-Bercot, B., I. Podglajen, I. Casin, and E. Collatz. 2002. An intrinsic control element for translational initiation in class 1 integrons. Mol. Microbiol. 44:119-130. [DOI] [PubMed] [Google Scholar]
- 17.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs-Reitsma, W. F., A. W. van de Giessen, N. M. Bolder, and R. W. A. W. Mulder. 1995. Epidemiology of Campylobacter spp. at two Dutch broiler farms. Epidemiol. Infect. 114:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karmali, M. A., S. De Grandis, and P. C. Fleming. 1980. Antimicrobial susceptibility of Campylobacter jejuni and Campylobacter fetus subsp. fetus to eight cephalosporins with special reference to species differentiation. Antimicrob. Agents Chemother. 18:948-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 21.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mugnier, P., I. Podglajen, F. W. Goldstein, and E. Collatz. 1998. Carbapenems as inhibitors of OXA-13, a novel, integron-encoded beta-lactamase in Pseudomonas aeruginosa. Microbiology 144:1021-1031. [DOI] [PubMed] [Google Scholar]
- 23.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nevinny-Stickel, C., and E. D. Albert. 1993. HLA class II typing in a microtitre plate format using digoxigenin-labelled amplified DNA and biotin-labelled oligonucleotide probes. Eur. J. Immunogenet. 20:419-427. [DOI] [PubMed] [Google Scholar]
- 25.Nield, B. S., A. J. Holmes, M. R. Gillings, G. D. Recchia, B. C. Mabbutt, K. M. Nevalainen, and H. W. Stokes. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195:59-65. [DOI] [PubMed] [Google Scholar]
- 26.Payne, R. E., M. D. Lee, D. W. Dreesen, and H. M. Barnhardt. 1999. Molecular epidemiology of Campylobacter jejuni in broiler flocks using randomly amplified polymorphic DNA-PCR and 23S rRNA-PCR and role of litter in its transmission. Appl. Environ. Microbiol. 65:260-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel, L., I. Le Thomas, T. Naas, A. Karim, and P. Nordmann. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preston, K. E., M. A. Kacia, R. J. Limberger, W. A. Archinal, and R. A. Venezia. 1997. The resistance and integrase genes of pACM1, a conjugative multiple-resistance plasmid, from Klebsiella oxytoca. Plasmid 37:105-118. [DOI] [PubMed] [Google Scholar]
- 29.Ransom, G. M., and B. E. Rose. 1998. Isolation, identification, and enumeration of Campylobacter jejuni/coli from meat and poultry products, p. 6.1-6.10. In USDA/FSIS microbiology laboratory guidebook, 3rd ed. U.S. Government Printing Office, Washington, D.C.
- 30.Rather, P. N., H. Munayyer, P. A. Mann, R. S. Hare, G. H. Miller, and K. J. Shaw. 1992. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase Ib and IIa proteins. J. Bacteriol. 174:3196-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt, F. R., E. J. Nucken, and R. B. Henschke. 1988. Nucleotide sequence analysis of 2"-aminoglycoside nucleotidyl-transferase ANT(2") from Tn4000: its relationship with AAD(3") and impact on Tn21 evolution. Mol. Microbiol. 2:709-717. [DOI] [PubMed] [Google Scholar]
- 32.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 33.Tran van Nhieu, G., and E. Collatz. 1987. Primary structure of an aminoglycoside 6′-N-acetyltransferase AAC(6′)-4, fused in vivo with the signal peptide of the Tn3-encoded β-lactamase. J. Bacteriol. 169:5708-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.U.S. Department of Health and Human Services. 2001. Code of Federal regulations, title 21, vol. 6, part 522, revised as of April 1. Implantation or injectable dosage form new animal drugs: gentamicin sulfate injection. Document 21CFR522.1044. U.S. Government Printing Office, Washington, D.C.
- 35.U.S. Department of Health and Human Services. 2001. Code of Federal regulations, title 21, vol. 6, part 520, revised as of April 1. Oral dosage form new animal drugs: streptomycin sulfate oral solution. Document 21CFR520.2158a. U.S. Government Printing Office, Washington, D.C.
- 36.U.S. Department of Health and Human Services. 2001. Code of Federal regulations, title 21, vol. 6, part 520, revised as of April 1. Oral dosage form new animal drugs: neomycin sulfate soluble powder. Document 21CFR520.1484. U.S. Government Printing Office, Washington, D.C.