Abstract
Polymorphism of five tandem repeats that are monomorphic in Bacillus anthracis was investigated in 230 isolates of the B. cereus group and in 5 sequenced B. cereus genomes in search for markers allowing identification of B. cereus and B. thuringiensis strains most closely related to B. anthracis. Using this multiple-locus variable number of tandem repeat analysis (MLVA), a cluster of 30 strains was selected for further characterization. Eventually, six of these were characterized by multilocus sequence type analysis. One of the strains is only six point mutations (of almost 3,000 bp) away from B. anthracis and was also proposed to be closest to B. anthracis by MLVA analysis. However, this strain remains separated from B. anthracis by a number of significant genetic events observed in B. anthracis, including the loss of the hemolysin activity, the presence of four prophages, and the presence of the two virulence plasmids, pXO1 and pXO2. One particular minisatellite marker provides an efficient assay to identify the subset of B. cereus and B. thuringiensis strains closely related to B. anthracis. Based on these results, a very simple assay is proposed that allows the screening of hundreds of strains from the B. cereus complex, with modest equipment and at a low cost, to eventually fill the gap with B. anthracis and better understand the origin and making of this dangerous pathogen.
Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis are spore-forming gram-positive bacteria belonging to the Bacillus cereus group. B. thuringiensis is an insect pathogen producing plasmid-encoded endotoxins and is widely used as a biopesticide against lepidopteran, dipteran, and coleopteran insect pests (36). B. cereus produces an enterotoxin and an emetic toxin that are responsible for diarrhea and emetic syndromes, characteristic of bacillus-associated gastrointestinal illness. B. cereus is also responsible for a variety of nongastrointestinal diseases (9). B. anthracis causes anthrax, a lethal disease in humans and other mammals. Three types of anthrax infection can occur: inhalational, cutaneous, and gastrointestinal. As a result of its high pathogenicity and the possible scattering of its spores, B. anthracis could be used as a biological weapon (19).
B. anthracis appears to be genetically extremely close to some members of the B. cereus-B. thuringiensis group (14). Previous studies based upon multilocus enzyme electrophoresis (MEE) and sequence analysis (14), sequencing of 16S rRNA (2), and pulsed-field gel electrophoresis/MEE analysis (3) lead to the suggestion that these three bacteria belong to the same species and that some of the key phenotypic properties are conferred usually by plasmids. The loss of these plasmids leads to the inability to differentiate the B. cereus and B. thuringiensis strains (3, 14). Moreover, B. cereus strains can be converted into crystal producers by means of plasmid transfer (12), and some strains of B. thuringiensis were also tested for their ability to produce a diarrhea-causing enterotoxin (7).
The presence of the two plasmids pXO1 and pXO2 is essential to the pathogenicity of B. anthracis. Detection of these plasmids in association with a chromosomal marker is a common method to distinguish B. anthracis from B. cereus and B. thuringiensis (32, 33). However, distinction between species is sometimes difficult, and an illness resembling anthrax was shown to be caused by a B. cereus strain (strain G9241) possessing a plasmid with 99.6% similarity to pXO1 (18). This strain was identified by 16S rRNA analysis and by its phenotype as a B. cereus strain, and its virulence was confirmed in A/J mice. Another B. cereus strain, ZK, whose genome was recently sequenced, was responsible for an anthrax-like illness in a zebra (GenBank accession number NC_006274). The pathogenicity in immunosuppressed mice of the B. thuringiensis 97-27 (subsp. konkukian serotype H34), named here CEB97/27 (originally a Centre d'Etudes du Bouchet strain collection name), was also reported by Hernandez et al. (16). This strain was identified as closely related to B. anthracis by suppression subtractive hybridization (31) and by amplified fragment length polymorphism (AFLP) (17).
Several molecular methods have been developed to analyze genetic diversity inside the B. cereus group and to distinguish B. anthracis from the others: AluI restriction of a randomly amplified polymorphic DNA marker specific for the B. cereus complex (6), the restriction site insertion-PCR method (5), sequencing of the long 16S-23S rDNA containing tRNA genes (4), real-time PCR analysis of the rpoB gene alone (30) or associated with the cap gene (24) in a multiplex PCR, characterization of genomic differences that distinguish non-anthrax-causing bacilli from B. anthracis Ames by suppression subtractive hybridization (31), gyrB sequence analysis (25), and partial sequencing of the plcR gene (10, 37).
Multilocus sequence typing (MLST) (15), based on sequencing of 7 essential housekeeping genes, and fluorescent AFLP (17) have proved their efficiency in typing the B. cereus group. Both methods show that the genetic diversity is high inside the B. cereus-B. thuringiensis group, whereas B. anthracis is highly homogeneous and can be considered to be a particularly monomorphic species (all five B. anthracis strains investigated have the same MLST type, whereas more than 50 different MLST types are distinguished among 77 strains from the B. cereus group). However, these methods are laborious and time-consuming when a large number of strains or isolates must be analyzed. Moreover, the AFLP method requires high-quality standards and strict protocols in order to produce data that can be compared between different laboratories. Such analyses will probably be restricted to the most interesting strains as revealed by simpler, higher-throughput investigations.
Multiple-locus variable number of tandem repeat (VNTR) analysis (MLVA) using a collection of polymorphic markers is currently the method of choice to genotype strains inside the B. anthracis species (22, 28). This species is thought to have evolved from a B. cereus-B. thuringiensis strain by acquisition of the pXO plasmids. It is thus interesting to identify the closely related strains in order to better understand the evolutionary origin of this pathogen. In the present study, we show that MLVA can be successfully used for this purpose. This is not the traditional use of this technique. We analyzed the polymorphism of selected VNTRs inside a large collection of B. cereus-B. thuringiensis strains in an attempt to detect strains closely related to B. anthracis. As a result, a two-step assay based on PCR and agarose gel electrophoresis is proposed that represents an interesting approach to rapidly and at a very low cost identify strains of the B. cereus complex that are closest to B. anthracis.
MATERIALS AND METHODS
Bacterial strains.
The microorganisms included in this study are part of the collection maintained by the Centre d'Etudes du Bouchet (CEB). They were obtained from different sources (Tables 1 and 2). A total of 230 strains or isolates were analyzed.
TABLE 1.
Reference strains analyzed in this studya
| Reference | Species | Source | ID | Reference | Species | Source | ID | |
|---|---|---|---|---|---|---|---|---|
| 02/003 | B. thuringiensis | 1 | H1 serovar thuringiensis | 02/069* | B. thuringiensis | 1 | H55 serovar palmanyolensis | |
| 02/004 | B. thuringiensis | 1 | H2 serovar finitimus | 02/070 | B. thuringiensis | 1 | H56 serovar rongseni | |
| 02/005 | B. thuringiensis | 1 | H3aH3c serovar alesti | 02/071 | B. thuringiensis | 1 | H57 serovar pirenaica | |
| 02/006 | B. thuringiensis | 1 | H3aH3bH3c serovar kurstaki | 02/072 | B. thuringiensis | 1 | H58 serovar argentinensis | |
| 02/007 | B. thuringiensis | 1 | H3aH3d serovar sumiyoshiensis | 02/073 | B. thuringiensis | 1 | H59 serovar iberica | |
| 02/008 | B. thuringiensis | 1 | H3aH3dH3e serovar fukuokaensis | 02/074 | B. thuringiensis | 1 | H60 serovar pingluonsis | |
| 02/009 | B. thuringiensis | 1 | H4aH4b serovar sotto | 02/075 | B. thuringiensis | 1 | H61 serovar sylvestriensis | |
| 02/010 | B. thuringiensis | 1 | H4aH4c serovar kenyae | 02/076 | B. thuringiensis | 1 | H62 serovar zhaodongensis | |
| 02/011 | B. thuringiensis | 1 | H5aH5b serovar galleriae | 02/077 | B. thuringiensis | 1 | H63 serovar bolivia | |
| 02/012 | B. thuringiensis | 1 | H5aH5c serovar canadensis | 02/078 | B. thuringiensis | 1 | H64 serovar azorensis | |
| 02/013 | B. thuringiensis | 1 | H6 serovar entomocidus | 02/079 | B. thuringiensis | 1 | H65 serovar pulsiensis | |
| 02/014* | B. thuringiensis | 1 | H7 serovar aizawai | 02/080 | B. thuringiensis | 1 | H66 serovar graciosensis | |
| 02/015* | B. thuringiensis | 1 | H8aH8b serovar morrisoni | 02/081 | B. thuringiensis | 1 | H67 serovar vazensis | |
| 02/016 | B. thuringiensis | 1 | H8aH8c serovar ostriniae | 02/082 | B. thuringiensis | 1 | H68 serovar thailandensis | |
| 02/017 | B. thuringiensis | 1 | H8bH8d serovar nigeriensis | 02/083 | B. thuringiensis | 1 | H69 serovar pahangi | |
| 02/018 | B. thuringiensis | 1 | H9 serovar tolworthi | 02/084 | B. thuringiensis | 1 | H70 serovar sinensis | |
| 02/019 | B. thuringiensis | 1 | H10aH10b serovar darmstadiensis | 02/350 | B. cereus | 2 | F4430/73 | |
| 02/020 | B. thuringiensis | 1 | H10aH10c serovar londrina | 02/351 | B. cereus | 2 | DSM 4282 | |
| 02/021* | B. thuringiensis | 1 | H11aH11b serovar ioumanoffi | 02/352 | B. cereus | 2 | 98HMPL63 | |
| 02/022 | B. thuringiensis | 1 | H11aH11c serovar kyushuensis | 02/353 | B. cereus | 2 | F2769/77 | |
| 02/023 | B. thuringiensis | 1 | H12 serovar thompsoni | 02/354 | B. cereus | 2 | F284/78 | |
| 02/024 | B. thuringiensis | 1 | H13 serovar pakistani | 02/355 | B. cereus | 2 | F4815/94 | |
| 02/025 | B. thuringiensis | 1 | H14 serovar israelensis | 02/356 | B. cereus | 2 | F3003/73 | |
| 02/026 | B. thuringiensis | 1 | H15 serovar dakota | 02/357 | B. cereus | 2 | F2081A/98 | |
| 02/027 | B. thuringiensis | 1 | H16 serovar indiana | 02/358 | B. cereus | 2 | F2081B/98 | |
| 02/028 | B. thuringiensis | 1 | H17 serovar tohokuensis | 02/359 | B. cereus | 2 | F2085/98 | |
| 02/029* | B. thuringiensis | 1 | H18aH18b serovar kumamotoensis | 02/360 | B. cereus | 2 | DSM 2301 | |
| 02/030 | B. thuringiensis | 1 | H18aH18c serovar yosoo | 02/361 | B. cereus | 2 | LMG 17605 | |
| 02/031 | B. thuringiensis | 1 | H19 serovar tochigiensis | 02/362 | B. cereus | 2 | DSM 4222 | |
| 02/032 | B. thuringiensis | 1 | H20aH20b serovar yunnanensis | 02/363 | B. cereus | 2 | DSM 8438 | |
| 02/033 | B. thuringiensis | 1 | H20aH20c serovar pondicheriensis | 02/364 | B. cereus | 2 | ATCC 4342 | |
| 02/034 | B. thuringiensis | 1 | H21 serovar colmeri | 02/367 | B. cereus | 2 | LMG 6923T | |
| 02/035 | B. thuringiensis | 1 | H22 serovar shandongiensis | 02/368 | B. thuringiensis | 2 | CIP 53.137T | |
| 02/036 | B. thuringiensis | 1 | H23 serovar japonensis | 02/369 | B. thuringiensis | 2 | T03A001 | |
| 02/037 | B. thuringiensis | 1 | H24aH24b serovar neoleonensis | 02/370 | B. thuringiensis | 2 | T14007 | |
| 02/038 | B. thuringiensis | 1 | H25 serovar coreanensis | 02/371 | B. thuringiensis | 2 | T01016 | |
| 02/039 | B. thuringiensis | 1 | H26 serovar silo | 02/373 | B. cereus | 2 | CIP 78.3 | |
| 02/040 | B. thuringiensis | 1 | H27 serovar mexicanensis | 02/374 | B. cereus | 2 | CIP 51.27 | |
| 02/041* | B. thuringiensis | 1 | H28aH28b serovar monterrey | 02/375 | B. cereus | 2 | F4635A/90 | |
| 02/042 | B. thuringiensis | 1 | H28aH28c serovar jegathesan | 02/376 | B. cereus | 2 | F4620/90 | |
| 02/043* | B. thuringiensis | 1 | H29 serovar amagiensis | 02/379 | B. cereus | 2 | CIP 58.32 | |
| 02/044 | B. thuringiensis | 1 | H30 serovar medellin | 02/381 | B. cereus | 2 | P1-1 | |
| 02/045 | B. thuringiensis | 1 | H31 serovar toguchini | 02/382 | B. cereus | 2 | P2-3 | |
| 02/046 | B. thuringiensis | 1 | H32 serovar cameroun | 02/383 | B. cereus | 2 | P14-1 | |
| 02/047 | B. thuringiensis | 1 | H33 serovar leesis | 02/384 | B. cereus | 2 | C413 | |
| 02/048 | B. thuringiensis | 1 | H34 serovar konkukian | 02/385 | B. cereus | 2 | TC414 | |
| 02/049 | B. thuringiensis | 1 | H35 serovar seoulensis | 02/386 | B. cereus | 2 | Z421 | |
| 02/050 | B. thuringiensis | 1 | H36 serovar malaysiensis | 02/387 | B. cereus | 2 | TL811 | |
| 02/051* | B. thuringiensis | 1 | H37 serovar andaluciensis | 02/388 | B. cereus | 2 | B2114 | |
| 02/052 | B. thuringiensis | 1 | H38 serovar oswaldocruzi | 02/389 | B. cereus | 2 | L2103 | |
| 02/053 | B. thuringiensis | 1 | H39 serovar brasiliensis | 02/391 | B. cereus | 2 | P2103 | |
| 02/054 | B. thuringiensis | 1 | H40 serovar huazhongensis | 02/392 | B. cereus | 2 | P15-2 | |
| 02/055 | B. thuringiensis | 1 | H41 serovar sooncheon | 02/393 | B. cereus | 2 | P22-4 | |
| 02/056 | B. thuringiensis | 1 | H42 serovar jinghongiensis | 02/394 | B. cereus | 2 | P24-1 | |
| 02/057 | B. thuringiensis | 1 | H43 serovar guiyangiensis | 02/395 | B. cereus | 2 | TZ415 | |
| 02/058 | B. thuringiensis | 1 | H44 serovar higo | 02/396 | B. cereus | 2 | TZ427 | |
| 02/059 | B. thuringiensis | 1 | H45 serovar roskildiensis | 02/397 | B. cereus | 2 | C2109 | |
| 02/060 | B. thuringiensis | 1 | H46 serovar chanpaisis | 02/398 | B. cereus | 2 | Z4222 | |
| 02/061 | B. thuringiensis | 1 | H47 serovar wratislaviensis | 02/400 | B. cereus | 2 | K1231 | |
| 02/062 | B. thuringiensis | 1 | H48 serovar balearica | 02/402 | B. cereus | 2 | C2104 | |
| 02/063 | B. thuringiensis | 1 | H49 serovar muju | 96/011 | B. cereus | ATCC 14579 | ||
| 02/064 | B. thuringiensis | 1 | H50 serovar navarrensis | 97/027 | B. thuringiensis | 3 | H34 serovar konkukian | |
| 02/065 | B. thuringiensis | 1 | H51 serovar xiaguangiensis | 98/020 | B. thuringiensis | 4 | H3aH3b serovar kurstaki | |
| 02/066 | B. thuringiensis | 1 | H52 serovar kim | 98/021 | B. thuringiensis | 4 | H14 serovar israelensis | |
| 02/067 | B. thuringiensis | 1 | H53 serovar asturiensis | 99/028 | B. thuringiensis | 1 | Bt407 | |
| 02/068 | B. thuringiensis | 1 | H54 serovar poloniensis |
Sources of 131 different strains were as follows: 1, Institut Pasteur, Paris, France; 2, INRA, Avignon, France; 3, Hôpital des Armées Bégin, Paris, France; 4, Abbott Laboratories, France. ID, serovar and subspecies; *, eight strains used for the testing of 17 primer pairs.
TABLE 2.
B. cereus group isolates investigated in this studya
| Reference | Species | Additional data/reference | Source | Reference | Species | Additional data/reference | Source |
|---|---|---|---|---|---|---|---|
| 02/216 | B. cereus | 8D1 | 1 | 02/561 | B. cereus | 94.9.16.344 | 2 |
| 02/217 | B. cereus | 4F1 | 1 | 02/562 | B. cereus | 94.9.19.398 | 2 |
| 02/219 | B. cereus | 18G4 | 1 | 02/563 | B. cereus | 95.7.27.304 | 2 |
| 02/220 | B. cereus | Italian lactic ferment | 1 | 02/564 | B. cereus | 95.5.14.39 | 2 |
| 02/221 | B. cereus | Barégine | 1 | 02/565 | B. cereus | 95.4.24.34 | 2 |
| 02/222 | B. cereus | 406 | 1 | 02/566 | B. cereus | 91.3.11.1388 | 2 |
| 02/223 | B. cereus | 6C2 | 1 | 02/567 | B. cereus | None | 2 |
| 02/224 | B. cereus | 7A21 | 1 | 02/569 | B. cereus | 92.6.18.420 | 2 |
| 02/225 | B. cereus | 9A7 | 1 | 02/570 | B. cereus | 92.1.2.183 | 2 |
| 02/226 | B. cereus | 10H5 | 1 | 02/571 | B. cereus | 92.4.19.242 | 2 |
| 02/227 | B. cereus | 2 | 1 | 02/572 | B. cereus | 3.53 | 2 |
| 02/228 | B. cereus | 19A5 round | 1 | 02/573 | B. cereus | 95.3.20.507 | 2 |
| 02/229 | B. cereus | 19A5 filamentous | 1 | 02/574 | B. cereus | 46 | 2 |
| 02/230 | B. cereus | X44 | 1 | 02/575 | B. cereus | 95.1.6.117 | 2 |
| 02/231 | B. cereus | 109 | 1 | 02/576 | B. cereus | 95.7.24.357 | 2 |
| 02/236 | B. cereus | W4II | 1 | 02/577 | B. cereus | None | 2 |
| 02/481 | B. cereus | 7C1 nebulous | 1 | 02/578 | B. cereus | 91.3.19.1367 | 2 |
| 02/482 | B. cereus | 6A14 | 1 | 02/579 | B. cereus | 92.4.12.227 | 2 |
| 02/483 | B. cereus | 7A2 | 1 | 02/580 | B. cereus | 31.3.19.1292 | 2 |
| 02/484 | B. cereus | 7A11 | 1 | 02/582 | B. cereus | 92.7.23.208 | 2 |
| 02/485 | B. cereus | 7A17 | 1 | 02/583 | B. cereus | 95.4.24.253 | 2 |
| 02/486 | B. cereus | 6B11 | 1 | 02/584 | B. cereus | 95.2.23.398 | 2 |
| 02/487 | B. cereus | 7B2 | 1 | 02/585 | B. cereus | 92.04 | 2 |
| 02/488 | B. cereus | 17B4 | 1 | 02/586 | B. cereus | 94.9.9.360 | 2 |
| 02/489 | B. cereus | 7C2 nebulous | 1 | 02/622 | B. cereus | Biotox 18/12/02 | 3 |
| 02/490 | B. cereus | 8C2 nebulous | 1 | 02/623 | B. cereus | Biotox 18/12/02 | 3 |
| 02/491 | B. cereus | 8C4 pearl | 1 | 02/624 | B. cereus | Biotox 18/12/02 | 3 |
| 02/492 | B. cereus | 8C12 | 1 | 04/021 | B. cereus | 1 | 4 |
| 02/493 | B. cereus | 2M4 | 1 | 04/022 | B. cereus | 2 | 4 |
| 02/495 | B. cereus | 15G9 | 1 | 04/023 | B. cereus | 3 | 4 |
| 02/496 | B. cereus | 16K3 | 1 | 04/024 | B. cereus | 4 | 4 |
| 02/498 | B. cereus | Anthracoides horsehair | 1 | 04/0254 | B. cereus | 8 | 4 |
| 02/499 | B. cereus | Anthracoides water | 1 | 04/026 | B. cereus | 10 | 4 |
| 02/500 | B. cereus | Anthracoides g and j | 1 | 04/027 | B. cereus | 26 | 4 |
| 02/501 | B. cereus | Anthracoides s | 1 | 04/028 | B. cereus | 52 | 4 |
| 02/542 | B. cereus | None | 2 | 04/029 | B. cereus | Bt001 | 4 |
| 02/543 | B. cereus | 94.9.16.344 | 2 | 04/030 | B. cereus | Bc1 | 4 |
| 02/544 | B. cereus | 92.9.25.319 | 2 | 97/076 | B. cereus | TZ-4 | 5 |
| 02/545 | B. cereus | 92.9.25.294 | 2 | 97/077 | B. cereus | T6-6 | 5 |
| 02/546 | B. cereus | 95.7.125.165 | 2 | 97/078 | B. cereus | T5-2 | 5 |
| 02/547 | B. cereus | 92.1.3.53 | 2 | 97/079 | B. cereus | TH-3 | 5 |
| 02/548 | B. cereus | 94.9.9.360 | 2 | 97/080 | B. cereus | 97/080 | 6 |
| 02/549 | B. cereus | 95.4.28.73 | 2 | 97/081 | B. cereus | 97/081 | 6 |
| 02/550 | B. cereus | 95.4.28.73 | 2 | 97/082 | B. cereus | 97/082 | 6 |
| 02/551 | B. cereus | 91.3.9.1292 | 2 | 97/083 | B. cereus | 97/083 | 6 |
| 02/552 | B. cereus | 94.12.16.385 | 2 | 97/084 | B. cereus | 97/084 | 6 |
| 02/554 | B. cereus | 92.4.19.242 | 2 | 97/085 | B. cereus | 97/085 | |
| 02/557 | B. cereus | 95.5.12.269 | 2 | 97/091 | B. cereus | 97/091 | 7 |
| 02/558 | B. cereus | 92.4.24.7342 | 2 | 99/043 | B. cereus | 99.819.0254 | 2 |
| 02/560 | B. cereus | 92.6.18.420 | 2 |
The sources of 99 isolates were as follows: 1, soil and dairy, INRA, Jouy-en-Josas, France; 2, clinical isolates, Hôpital des Armées Bégin, Paris, France; 3, soil, Biotox analyses; 4, clinical isolates, Centre Hospitalier, Toulouse, France; 5, CNEVA, France; 6, soil, Institut Pasteur, Paris, France; 7, clinical isolate, Hôpital des Armées Percy, Paris, France.
The 131 reference strains (Table 1) originate from well-characterized collections and comprise 90 B. thuringiensis and 41 B. cereus strains. In particular, the B. thuringiensis collection used contains one strain for each B. thuringiensis serovar described in reference 26. Similarly, the B. cereus strains are a representative panel described in reference 13. The 99 newly investigated isolates (Table 2) were initially identified by the laboratory of origin as being B. cereus members of the B. cereus group. This was confirmed on the basis of phenotypic criteria (motility and hemolytic capacity) by biochemical criteria analysis with the Vitek system using Bacillus Biochemical (BAC) cards (bioMérieux, Marcy l'Etoile, France), by the gram-positive plates and the “dangerous pathogens” database of the Biolog system (AES, Combourg, France) and in some instances by direct microscopic observation (detection of the crystal after Coomassie blue staining).
DNA purification.
A total of 5 ml of 2YT broth were inoculated by picking a single colony. The culture was incubated overnight at 37°C, and bacteria were harvested by centrifugation at 4,000 × g for 20 min. The pellet was suspended in 200 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]) with lysozyme (50 mg/μl) and incubated at 37°C for 1 h. The solution was grounded with 0.1-mm glass beads at 2,500 rpm for 90 s in a Mini-Beadbeater (Biospec Products, Bartlesville, Ohio). The supernatant was recovered, and the beads were washed with 250 μl of TE buffer. An RNase solution was added at a final concentration of 500 μg/ml, and the mixture was incubated for 15 min at 50°C. One volume of lysis buffer containing 20 mM Tris (pH 8), 2 mM EDTA, 20 mM NaCl, and 1% sodium dodecyl sulfate was added, along with 100 μg of proteinase K per ml, and the mixture was incubated overnight at 55°C. Chromosomal DNA was extracted by phenol-chloroform by using a Phase Lock gel (Eppendorf, Hamburg, Germany) and precipitated with 2 volumes of ice-cold 95% ethanol. The precipitated DNA was collected by centrifugation for 20 min at 15,000 × g. The dried DNA was then dissolved in TE buffer. The quality (average size) of the DNA was checked by 0.7% agarose gel electrophoresis, and the DNA concentration was measured with the DyNA Quant 200 fluorimeter (Hoefer, San Francisco, Calif.).
VNTR PCR amplification and genotyping.
PCRs were performed in 15 μl containing 2 ng of DNA, 1× PCR buffer, 1 U of Taq DNA polymerase, 200 μM concentrations of each deoxynucleoside triphosphate, and 0.3 μM concentrations of each flanking primer. The Taq DNA polymerase was obtained from Qbiogen (Illkirch, France) and used as recommended by the manufacturer.
PCRs were run on a MJ Research PTC200 thermocycler (Waltham, MA). An initial denaturation at 96°C for 5 min was followed by 30 cycles of denaturation at 96°C for 30 s, annealing at 56°C for 1 min, and elongation at 70°C for 1 min, followed by a final extension step of 5 min at 70°C. Then, 5 μl of the PCR products were run on standard 2% agarose gel (Qbiogen) in 0.5× TBE buffer (10× TBE is 890 mM Tris base, 890 mM boric acid, 20 mM EDTA [pH 8.3]) at a voltage of 10 V/cm. Samples were manipulated and distributed (including gel loading) with multichannel electronic pipettes (Biohit, Bonnelles, France) in order to reduce the risk of errors. Gel length of 20 cm were used. Gels were stained with ethidium bromide, visualized under UV light, and photographed. Alleles size was estimated by using a 100-bp ladder (Bio-Rad, Marnes-la-Coquette, France) as a size marker as previously described (27, 28). One reference strain was included for each set of five DNA samples as a control for size assignments. Gel images were analyzed by using the Bionumerics software package version 4.0 (Applied-Maths, Sint-Martens-Latem, Belgium) as previously described (27). The number of repeats in each allele was deduced from the amplicon size. The resulting data were analyzed with Bionumerics as a character data set. Clustering analysis was done by using the categorical parameter and the UPGMA (unweighted pair-group method with arithmetic averages) coefficient.
Sequencing of PCR amplification products and MLST clustering analysis.
PCR was performed in a 60-μl volume. Amplification products of a size greater than 300 bp were purified by polyethylene glycol precipitation as described previously (11). Amplification products shorter than 300 bp were concentrated by ethanol precipitation and treated by ExoSAP-IT from USB Corp. (Cleveland, Ohio) as described by the manufacturer. Sequencing was done by MWG Biotech (Courtaboeuf, France). A portion (10) of the PlcR gene was amplified for sequencing by using 5′-TGGCCATTTTAAGAAGAGTATTGA-3′ and 5′-CACTCTAGCTTTTCTAGGCATTCA-3′. The primers used for the MLST amplification reactions were as described in reference 15. The MLST sequence data was converted to a character data set by using Bionumerics version 4.0 in order to be able to produce a minimum-spanning tree. The inclusion of hypothetical missing links was allowed in the making of the minimum-spanning tree.
Search for B. anthracis prophages.
The four prophages present in B. anthracis and absent in a number of B. cereus strains as observed by (35) were investigated by using, respectively 5′-CGGTGACGTGTTAACTGAGC-3′ and 5′-CGTACGTGTTACTCGCCAAA-3′ (prophage 1), AAGTCAATCCTTCCGGGTTT and TCACCAATCATGGTCAGGAA (prophage 2), CGTTAACCAAACTGGGCAAT and TTATCGTCCTCACGCAGTTG (prophage 3), and TCAGGCATGGGTTATGTGAA and TCATGATGCTCACGGTTATGA (prophage 4) as primers.
RESULTS
Analysis of monomorphic tandem repeats.
Le Flèche et al. (28) previously tested numerous tandem repeat loci in order to identify an appropriate set of polymorphic markers for B. anthracis strain typing. Seventeen tandem repeats proved to be monomorphic in B. anthracis, whereas preliminary investigations demonstrated that at least some were polymorphic in B. cereus-B. thuringiensis strains (data not shown). We thus hypothesized that strains of the B. cereus group presenting the same allelic combination or at least some common allele sizes with B. anthracis for a number of tandem repeats monomorphic within B. anthracis, would be more closely related to B. anthracis. The flanking sequences of each B. anthracis (strain Ames) monomorphic tandem repeat identified by Le Flèche et al. (28) were compared to their homolog in B. cereus ATCC 10987 and ATCC 14579 (20, 34). Primers were chosen to match the two sequenced genomes and were given a Bcms suffix. The 17 primer pairs were tested on a subset of eight B. thuringiensis strains (indicated by asterisks in Table 1). Only eight primer pairs yielded PCR amplification products in all eight strains, and polymorphism was observed for five tandem repeats (Table 3). All five minisatellites are part of putative open reading frames, Bcms 19 and Bcms 20 being inside the same gene.
TABLE 3.
Primers used for PCR amplification of Bcms VNTRs
| Name | Unit length | Primers
|
Allele size range (bp) | Expected length (bp)a | On B. cereus ATCC 14579 genome
|
No. of alleles | ||
|---|---|---|---|---|---|---|---|---|
| Orientationb | Sequence | Tandem repeat location | Corresponding gene | |||||
| Bcms 08 | 18 | F | GTGCTGG(W)GCAAACACAGAC | 460-838 | 739 | 797035-797773 | Enterotoxin/cell | 23 |
| R | TGGTCGCCTGCTTTATAACC | wall binding protein | ||||||
| Bcms 17 | 16 | F | ATTGGACAAGAAAAACAAGGTACTG | 215-272 | 215 | 4148084-4148298 | Stage III sporu | 4 |
| R | CGCTGATCTTCCATTTGCAT | lation pro- tein AH | ||||||
| Bcms 18 | 15 | F | CCTTGTTTTGCACGCTCAG | 230-283 | 268 | 1273504-1273771 | Hypothetical | 7 |
| R | CTGGTCAACAACCTACTGAAAATGT | protein | ||||||
| Bcms 19 | 12 | F | GGAATAGAAGATGAAGAAGAAGTTACG | 291-411 | 363 | 4361476-4361838 | SpoVID- | 18 |
| R | TTTCG(S)TTTTATTGGTGGTTG | dependent spore coat assembly factor SafA | ||||||
| Bcms 20 | 36 | F | CGCCAAATGTATCGAAAGAA | 412-844 | 451 | 4360634-4361084 | SpoVID- | 20 |
| R | TGCTGATATGGCATTTGATATGG | dependent spore coat assembly factor SafA | ||||||
In B. cereus ATCC 14579.
F, forward; R, reverse.
MLVA with these five markers was performed using DNA from 230 strains belonging to the B. cereus group including B. thuringiensis CEB97/27 strain (17, 31). The allele sizes were converted to repeat unit numbers by using conventions defined earlier (27). A dendrogram was produced that also includes the theoretical alleles of five sequenced genomes: B. anthracis Ames, B. cereus ATCC 14579, B. cereus ATCC 10987, B. cereus G9241, and B. cereus ZK (GenBank accession numbers AE017334, AE016877, AE017194, AAEK00000000, and NC_006274, respectively). The 234 strains were distributed into 199 genotypes, essentially owing to the large number of alleles observed at the Bcms 08 (23 alleles), Bcms 19 (18 alleles), and Bcms 20 (20 alleles) loci (see online data at http://minisatellites.u-psud.fr/).
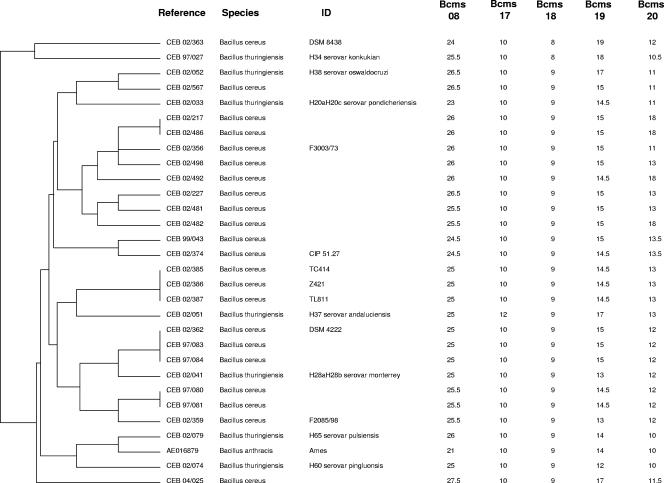
Three major clusters—A, B, and C—were differentiated by MLVA clustering analysis. Interestingly, the clustering assignment fits very well with the Bcms 17 alleles. Clusters A, B, and C have 10, 8, and 9 motifs, respectively, with rare exceptions. Cluster A, containing B. anthracis and 29 other strains, is shown in Fig. 1. Two B. thuringiensis strains CEB02/079 (serovar pulsiensis) and CEB02/074 (serovar pingluonsis) have, respectively four and three alleles of five in common with B. anthracis (hence, MLVA scores of 4/5 and 3/5, respectively). Strain CEB97/27 is in cluster A, although only the Bcms 17 allele has the B. anthracis size. Cluster B contains 125 strains, including the sequenced strain B. cereus ATCC 14579 and the two B. thuringiensis strains used as a biopesticide (BtH14 [israelensis], listed as CEB98/020 and BtH3a3b [kurstaki] listed as CEB98/021); cluster C contains 79 strains, including strains B. cereus ATCC 10987, ZK, and G9241 (online data at http://minisatellites.u-psud.fr). B. cereus and B. thuringiensis strains could not be distinguished by MLVA, a finding in agreement with previous reports using other molecular methods. Apart from the strains closely related to B. anthracis cited before, no other strains have a score of 3/5 or more. Thus, the strains belonging to cluster A are B. anthracis close-neighbor candidates. The strains derived from patients (Table 2) appear to be equally distributed among all groups.
FIG. 1.
Cluster A of the dendrogram deduced from VNTR typing of 234 B. cereus group strains. The first column indicates the reference of the strains (see Tables 1 and 2). The second column indicates the species. The third column indicates the known identity number. The columns Bcms show the number of motifs obtained for each minisatellite.
Comparison with MLST analysis.
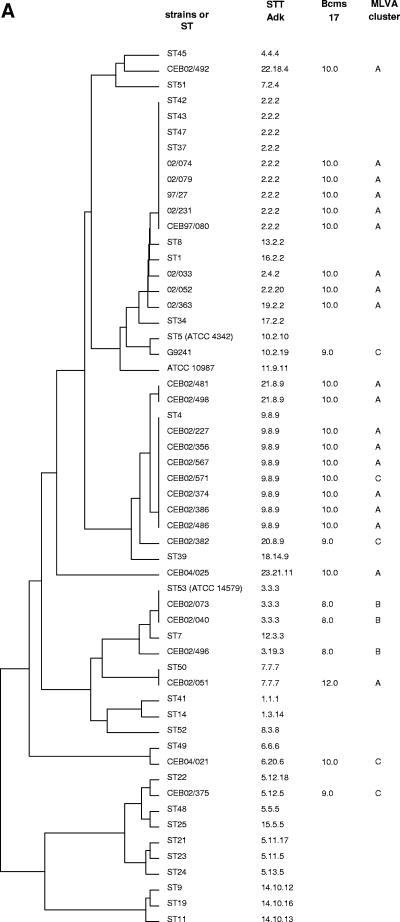
To estimate the validity of the classification observed by VNTR typing, selected strains were analyzed by MLST as developed by Helgason et al. (15). The dendrograms derived from the adk sequences alone, or from all seven loci, were previously shown to be in very good agreement, at least in the vicinity of B. anthracis (15). For this reason, we first analyzed the adk sequence of all strains in cluster A and of some strains from clusters B and C. A dendrogram was produced from the resulting adk sequences together with some sequences from Helgason et al. (Fig. 2A) and with the fully sequenced strains. The cluster A strains are separated into two groups, one being very close to B. anthracis. Strains CEB02/079 (H65), CEB02/074 (H60), CEB97/27, and CEB97/080 have the same adk sequence as the strains of sequence types (STs) 37, 42, 43 and 47, respectively. Strains CEB02/231 and CEB02/571, which were assigned, respectively, to MLVA clusters B and C, in spite of their allele 10 at Bcms 17, have adk alleles falling within the cluster A strains. CEB02/051, which was assigned to cluster A, is distant in the adk tree. This strain has a Bcms 17 allele 12. Strain CEB04/021, with a Bcms 17 allele 10 and belonging to cluster C, appears distant in the adk tree as well.
FIG. 2.
(A) Genetic relationship among some strains presented here and elsewhere (15) obtained with the adk sequence. The dendrogram was constructed by using UPGMA parameters. The first column indicates the reference of the strains (see Tables 1 and 2) or the ST according to (15). The second column indicates the split ST (STT) of adk gene sequences as presented previously (15). The two last columns indicate the Bcms 17 allele assignment and the cluster assignment of the strain in the MLVA analysis. (B) Minimum-spanning tree deduced from MLST data among B. anthracis closely related strains described here and previously (15). Open circles are hypothetical missing links, which reduce the overall tree length. ST numbers are as presented previously (15). ST1 (B. anthracis) is grayed. Strain 02/079 with MLVA score 4/5 is marked with an arrow.
A complete MLST scheme was applied to the typing of the strains with adk alleles closest to B. anthracis (Fig. 2B). It confirms that the strains identified by MLVA as being close to B. anthracis have a very similar MLST type. Among them, strain CEB02/079 (H65) seems to be the most closely related to ST1 (B. anthracis) (in agreement with the MLVA analysis and the 4/5 score) and to the strains with ST37 in the analysis of Helgason et al. Strains 97/080 and CEB97/27, on the other hand, seem to be closely related, respectively, to ST43 and ST8.
Sequencing of the Bcms 17 alleles.
Since Bcms 17 allele size alone seemed to efficiently identify a subset of strains closely related to B. anthracis (strains with allele 10), we sought to verify whether the sequence of allele 10 was identical in strains of cluster A and different from that of strains with a similarly sized allele in cluster C. We thus further analyzed this locus by sequencing. This minisatellite has a very poor internal motif conservation. Figure 3A shows the manual alignment of part of the locus bearing Bcms 17 in the sequenced B. anthracis genome, the three sequenced B. cereus (ATCC 14579, ATCC 10987, and G9241), B. thuringiensis CEB97/27, and some strains from the present study. In strain CEB04/021, the motif arrangement was clearly different from that of the other alleles of identical size, and many mutations were observed as in B. cereus strains of clusters B and C. Similarly, allele 12 of Bcms 17 in strain CEB02/051 showed a very different organization and many mutations compared to alleles of strains closely related to B. anthracis. Except for CEB04/021, all of the strains bearing allele 10 had the same motif organization as B. anthracis. In the minisatellite region, they were highly similar to B. anthracis. CEB97/27, CEB02/079, and CEB02/033 share the identical 10_1 alleles. Allele 10_2 (CEB97/080, CEB97/081, CEB02/363, and CEB02/052) differs by one point mutation. Alleles 10_3 (CEB04/025) and 10_4 (CEB02/074 and CEB02/231) differ by, respectively, one or two distinct additional point mutations (Fig. 3B).
FIG. 3.
Alignment of Bcms 17 nucleotide sequence from Bacillus strains. (A) Alignment of Bcms 17 nucleotide sequence from other Bacillus strains. The reference strains are as follows: ATCC 14579, B. cereus ATCC 14579; ATCC 10987, B. cereus ATCC 10987; G9241, B. cereus G9241; Bant, B. anthracis strain Ames (allele 10_1). The other strains are identified by their CEB number (see Tables 1 and 2). The flanking sequences are italicized. The residues are grouped according to the Bcms 17 motifs identified in the tandem repeats database (8) for B. anthracis strain Ames (35). A dot indicates identity with B. anthracis sequence, a dash indicates a gap, and a nucleotide letter indicates positions showing a polymorphism. The numbers in parentheses indicate the theoretical number of motifs according to the MLVA analysis: 08, 215 bp; 09, 227 bp; 10, 242 bp; 12, 262 bp. *, Strain CEB 04/021 was assigned a Bcms 17 allele 10, but the actual PCR product size is 245 bp, and this rare allele is indeed quite distinct from the ordinary allele 10. (B) Alignment of different sequences obtained for the 242-bp Bcms 17 allele 10, which had the same motif organization as B. anthracis. Flanking sequences are italicized. A dot indicates identity with B. anthracis sequence; nucleotide letters indicate positions showing polymorphisms. Allele 10_1 was obtained for B. anthracis Ames, CEB97/27, CEB02/079, and CEB02/033; allele 10_2 was obtained for CEB97/80, CEB97/81, CEB02/363, and CEB02/052; allele 10_3 was obtained for CEB04/025; and allele 10_4 was obtained for CEB02/074 and CEB02/231.
Prophages and PlcR status.
All strains analyzed in detail and presented in Fig. 2B were hemolytic, as tested by plating on blood agar plates. Sequencing of part of the PlcR gene containing the nonsense mutation responsible for the absence of hemolytic activity in B. anthracis shows that strains 02/079, 02/033, 02/052, 02/231, and 02/074 (Fig. 2B) are identical to B. anthracis except for the nonsense point mutation. These strains have “allele 3” as described in reference 10. In addition, all strains in Fig. 3B are missing the four prophages identified by Read et al. (35; data not shown).
DISCUSSION
In this study, MLVA was used to develop an assay for the identification of strains from the B. cereus group most closely related to B. anthracis. For this purpose, MLVA markers were selected among loci that were previously shown (28) to be monomorphic in B. anthracis but that are polymorphic in the B. cereus group. It was then speculated that strains (if they exist) which would show B. anthracis-like alleles would be candidate B. anthracis closest neighbors. If such an assay could be produced, then it would be easy to run on a large scale because it would require only PCR amplifications followed by agarose gel electrophoresis and allele size measurement. Consequently, it could be run in any laboratory with very basic and standard molecular genetics capacities at a low cost. The approach was tested on a collection of 234 B. cereus group strains using five selected markers and proved to be highly successful. On the basis of MLVA polymorphism, strains were distributed in three main clusters A, B, and C. B. anthracis strains are included in cluster A (Fig. 1). Incidentally, this MLVA investigation further confirms that the distinction between B. cereus and B. thuringiensis is not supported by chromosomal polymorphism analysis and only reflects a plasmid content. No strain showed an MLVA profile identical to B. anthracis, and only one strain (CEB02/079, cluster A) of 234 was identical to B. anthracis strains at four out of five markers. One strain from cluster A has alleles in common with B. anthracis for three VNTRs, twenty-five strains from cluster A have alleles in common with B. anthracis for two VNTRs, and two strains from cluster A have alleles in common with B. anthracis for one VNTR only. CEB97/27 is among these two, although it was shown by MLST to be very close to B. anthracis (Fig. 2B). This illustrates the discriminatory power of the MLVA assay in spite of the fact that loci shown to be monomorphic in B. anthracis were purposefully used here.
Cluster A strains were further investigated by partial (Fig. 2A) or complete (Fig. 2B) MLST analysis, which demonstrates that indeed CEB02/079 is most closely related to B. anthracis. Only six point mutations in the MLST analysis (covering close to 3 kb) separate CEB02/079 (B. thuringiensis, H65 serovar pulsiensis) from B. anthracis, but this is still a significant gap: multiple genetic events separate CEB02/079 from B. anthracis, CEB02/079 is hemolytic, and CEB02/079 lacks the four prophages observed in the B. anthracis genome (35). One of these prophages contains a ThyX (Thy1) gene (29). Owing to this prophage acquisition, B. anthracis happens to be equipped with two alternate pathways for thymidylate synthesis. Interestingly, among the sequenced B. cereus strains, the so-called Zebra killer strain is the only one to share this property (acquired by a seemingly different evolutionary pathway), and ThyX proteins are found in many pathogenic bacteria (29). This suggests that at least some of these prophages may have played a role in the evolution of B. cereus-B. thuringiensis toward the highly pathogenic B. anthracis and that many additional strains from different geographic origins will need to be analyzed in order to precisely decipher the chronological order of these different events.
Interestingly, it appears that differentiation using only the Bcms 17 allele size is sufficient to identify strains closely related to B. anthracis (allele 10). There are four agarose gel size classes of this minisatellite in the complete collection of strains, with allele 12 being present in only one strain. The high degree of internal divergence inside the minisatellite suggests that this is a very old tandem repeat and that no insertion or deletion events occurred in the recent past. This high internal heterogeneity is likely to further prevent the production of new alleles by recombination. A group of five B. thuringiensis isolates (CEB02/074 [B. thuringiensis subsp. pingluonsis], 02/079 [B. thuringiensis subsp. pulsiensis], 02/041 [B. thuringiensis subsp. monterrey], 02/033 [B. thuringiensis subsp. pondicheriensis], and 02/052 [B. thuringiensis subsp. oswaldocruzi]) with Bcms 17 allele 10 were also grouped by 16S RNA restriction fragment length polymorphism showing that this characteristic is relevant (21). In Fig. 4, we show how this observation can be used to propose a simple typing scheme. In the first step, strains with a Bcms 17 allele 10 are identified by PCR and agarose gel electrophoresis. Only this subset of strains (13% of the population investigated here) needs to be screened for the other four markers (step 2). Strains with at least four loci showing the B. anthracis allele are then investigated by MLST analysis (step 3). In the present investigation, only one strain of 234 fulfilled this criteria, and the whole screening process would have required in the present case not more than 350 PCR amplifications and size analyses and the sequencing of seven PCR products. Alternatively (Fig. 4, right side), the Bcms 17 allele of strains showing an “allele 10” could be directly sequenced (step 2). Strains with an allele identical to B. anthracis Bcms 17 will be investigated by MLST (three strains in the present collection). This alternate pathway would have required 230 PCR amplification and sizing and sequencing of approximately 50 PCR products. The proportions and figures given are expected to vary according to the population of strains. For instance, a higher proportion of close neighbors is expected to be observed in the geographic area from which B. anthracis emerged. The “Bcms 17” assay is very easy to run since the allele assignment can be derived from PCR amplifications, followed by agarose gel electrophoresis. Kim et al. (23) have used the vrrA VNTR marker to determine the genetic relatedness between B. anthracis and closely related species. However, vrrA is a polymorphic marker for B. anthracis strains, showing a divergence between B. anthracis strains and a greater one in the B. cereus group (1). Moreover, in some strains described here, two amplicons were produced or PCR was weak. In comparison, Bcms 17 was efficiently amplified for the 230 strains, showed only four alleles, and was previously shown to be monomorphic for all B. anthracis strains tested.
FIG. 4.
Typing scheme to screen B. cereus-B. thuringiensis for B. anthracis close relatives. (First step) PCR amplification at the Bcms 17 locus and identification of “allele 10” strains by agarose gel electrophoresis. (Second step, left) Typing of the four additional markers and identification of strains matching the B. anthracis pattern at three or more of these markers. (Second step, right, [alternatively]) Sequencing of the Bcms 17 PCR product. (Third step) MLST analysis of the very rare selected strains, and additional more detailed investigations.
Acknowledgments
We thank Alexandra Gruss (INRA, Jouy-en-Josas, France), Hélène Guinebretière (INRA, Avignon, France), and Maryse Archambaud (CHU, Toulouse, France) for the provision of strains.
This study is supported by Délégation Générale pour l'Armement PEA 02-36-01.
REFERENCES
- 1.Andersen, G. L., J. M. Simchock, and K. H. Wilson. 1996. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J. Bacteriol. 178:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ash, C., J. A. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 3.Carlson, C. R., and A. B. Kolsto. 1994. A small (2.4 Mb) Bacillus cereus chromosome corresponds to a conserved region of a larger (5.3 Mb) Bacillus cereus chromosome. Mol. Microbiol. 13:161-169. [DOI] [PubMed] [Google Scholar]
- 4.Cherif, A., S. Borin, A. Rizzi, H. Ouzari, A. Boudabous, and D. Daffonchio. 2003. Bacillus anthracis diverges from related clades of the Bacillus cereus group in 16S-23S ribosomal DNA intergenic transcribed spacers containing tRNA genes. Appl. Environ. Microbiol. 69:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daffonchio, D., S. Borin, A. Consolandi, and C. Sorlini. 1999. Restriction site insertion-PCR (RSI-PCR) for rapid discrimination and typing of closely related microbial strains. FEMS Microbiol. Lett. 180:77-83. [DOI] [PubMed] [Google Scholar]
- 6.Daffonchio, D., S. Borin, G. Frova, R. Gallo, E. Mori, R. Fani, and C. Sorlini. 1999. A randomly amplified polymorphic DNA marker specific for the Bacillus cereus group is diagnostic for Bacillus anthracis. Appl. Environ. Microbiol. 65:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damgaard, P. H. 1995. Diarrhoeal enterotoxin production by strains of Bacillus thuringiensis isolated from commercial Bacillus thuringiensis-based insecticides. FEMS Immunol. Med. Microbiol. 12:245-250. [DOI] [PubMed] [Google Scholar]
- 8.Denoeud, F., and G. Vergnaud. 2004. Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: a Web-based resource. BMC Bioinformatics 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Easterday, W. R., M. N. Van Ert, T. S. Simonson, D. M. Wagner, L. J. Kenefic, C. J. Allender, and P. Keim. 2005. Use of single nucleotide polymorphisms in the plcR gene for specific identification of Bacillus anthracis. J. Clin. Microbiol. 43:1995-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Embley, T. M. 1991. The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett. Appl. Microbiol. 13:171-174. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, J. M., Jr., B. J. Brown, and B. C. Carlton. 1982. Transfer of Bacillus thuringiensis plasmids coding for delta-endotoxin among strains of B. thuringiensis and B. cereus. Proc. Natl. Acad. Sci. USA 79:6951-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guinebretiere, M. H., V. Broussolle, and C. Nguyen-The. 2002. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 40:3053-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helgason, E., N. J. Tourasse, R. Meisal, D. A. Caugant, and A. B. Kolsto. 2004. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez, E., F. Ramisse, J. P. Ducoureau, T. Cruel, and J. D. Cavallo. 1998. Bacillus thuringiensis subsp. konkukian (serotype H34) superinfection: case report and experimental evidence of pathogenicity in immunosuppressed mice. J. Clin. Microbiol. 36:2138-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill, K. K., L. O. Ticknor, R. T. Okinaka, M. Asay, H. Blair, K. A. Bliss, M. Laker, P. E. Pardington, A. P. Richardson, M. Tonks, D. J. Beecher, J. D. Kemp, A. B. Kolsto, A. C. Wong, P. Keim, and P. J. Jackson. 2004. Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 70:1068-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmaster, A. R., J. Ravel, D. A. Rasko, G. D. Chapman, M. D. Chute, C. K. Marston, B. K. De, C. T. Sacchi, C. Fitzgerald, L. W. Mayer, M. C. Maiden, F. G. Priest, M. Barker, L. Jiang, R. Z. Cer, J. Rilstone, S. N. Peterson, R. S. Weyant, D. R. Galloway, T. D. Read, T. Popovic, and C. M. Fraser. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. USA 101: 8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inglesby, T. V., D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. M. Friedlander, J. Hauer, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, K. Tonat, et al. 1999. Anthrax as a biological weapon: medical and public health management. JAMA 281:1735-1745. [DOI] [PubMed] [Google Scholar]
- 20.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 21.Joung, K. B., and J. C. Cote. 2001. A phylogenetic analysis of Bacillus thuringiensis serovars by RFLP-based ribotyping. J. Appl. Microbiol. 91:279-289. [DOI] [PubMed] [Google Scholar]
- 22.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, W., Y. P. Hong, J. H. Yoo, W. B. Lee, C. S. Choi, and S. I. Chung. 2002. Genetic relationships of Bacillus anthracis and closely related species based on variable-number tandem repeat analysis and BOX-PCR genomic fingerprinting. FEMS Microbiol. Lett. 207:21-27. [DOI] [PubMed] [Google Scholar]
- 24.Ko, K. S., J. M. Kim, J. W. Kim, B. Y. Jung, W. Kim, I. J. Kim, and Y. H. Kook. 2003. Identification of Bacillus anthracis by rpoB sequence analysis and multiplex PCR. J. Clin. Microbiol. 41:2908-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Duc, M. T., M. Satomi, N. Agata, and K. Venkateswaran. 2004. gyrB as a phylogenetic discriminator for members of the Bacillus anthracis-cereus-thuringiensis group. J. Microbiol. Methods 56:383-394. [DOI] [PubMed] [Google Scholar]
- 26.Lecadet, M. M., E. Frachon, V. C. Dumanoir, H. Ripouteau, S. Hamon, P. Laurent, and I. Thiery. 1999. Updating the H-antigen classification of Bacillus thuringiensis. J. Appl. Microbiol. 86:660-672. [DOI] [PubMed] [Google Scholar]
- 27.Le Flèche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Flèche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myllykallio, H., G. Lipowski, D. Leduc, J. Filee, P. Forterre, and U. Liebl. 2002. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 297:105-107. [DOI] [PubMed] [Google Scholar]
- 30.Qi, Y., G. Patra, X. Liang, L. E. Williams, S. Rose, R. J. Redkar, and V. G. DelVecchio. 2001. Utilization of the rpoB gene as a specific chromosomal marker for real-time PCR detection of Bacillus anthracis. Appl. Environ. Microbiol. 67:3720-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radnedge, L., P. G. Agron, K. K. Hill, P. J. Jackson, L. O. Ticknor, P. Keim, and G. L. Andersen. 2003. Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 69:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramisse, V., G. Patra, H. Garrigue, J. L. Guesdon, and M. Mock. 1996. Identification and characterization of Bacillus anthracis by multiplex PCR analysis of sequences on plasmids pXO1 and pXO2 and chromosomal DNA. FEMS Microbiol. Lett. 145:9-16. [DOI] [PubMed] [Google Scholar]
- 33.Ramisse, V., G. Patra, J. Vaissaire, and M. Mock. 1999. The Ba813 chromosomal DNA sequence effectively traces the whole Bacillus anthracis community. J. Appl. Microbiol. 87:224-228. [DOI] [PubMed] [Google Scholar]
- 34.Rasko, D. A., J. Ravel, O. A. Okstad, E. Helgason, R. Z. Cer, L. Jiang, K. A. Shores, D. E. Fouts, N. J. Tourasse, S. V. Angiuoli, J. Kolonay, W. C. Nelson, A. B. Kolsto, C. M. Fraser, and T. D. Read. 2004. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L.Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 36.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slamti, L., S. Perchat, M. Gominet, G. Vilas-Boas, A. Fouet, M. Mock, V. Sanchis, J. Chaufaux, M. Gohar, and D. Lereclus. 2004. Distinct mutations in PlcR explain why some strains of the Bacillus cereus group are nonhemolytic. J. Bacteriol. 186:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]