Abstract
Soil is a highly heterogeneous matrix, which can contain thousands of different bacterial species per gram. Only a small component of this diversity (maybe <1%) is commonly captured using standard isolation techniques, although indications are that a larger proportion of the soil community is in fact culturable. Better isolation techniques yielding greater bacterial diversity would be of benefit for understanding the metabolic activity and capability of many soil microorganisms. We studied the response of soil bacterial communities to carbon source enrichment in small matrices by means of terminal restriction fragment length polymorphism (TRFLP) analysis. The community composition of replicate enrichments from soil displayed high variability, likely attributable to soil heterogeneity. An analysis of TRFLP data indicated that enrichment on structurally similar carbon sources selected for similar bacterial communities. The same analysis indicated that communities first enriched on glucose or benzoate and subsequently transferred into medium containing an alternate carbon source retained a distinct community signature induced by the carbon source used in the primary enrichment. Enrichment on leucine presented a selective challenge that was able to override the imprint left by primary enrichment on acetate. In a time series experiment community change was most rapid 18 hours after inoculation, corresponding to exponential growth. Community composition did not stabilize even 4 days after secondary enrichment. Four different soil types were enriched on four different carbon sources. TRFLP analysis indicated that in three out of four cases communities enriched on the same carbon source were more similar regardless of which soil type was used. Conversely, the garden soil samples yielded similar enrichment communities regardless of the enrichment carbon source. Our results indicate that in order to maximize the diversity of bacteria recovered from the environment, multiple enrichments should be performed using a chemically diverse set of carbon sources.
Enrichment, maintenance, and isolation of bacteria constitute an important component of a large number of industrial, agricultural, and social applications, including wastewater management, composting, biofertilization, bioremediation, and discovery of natural products (5). The composition, function, and dynamics of microbial communities determine their suitability for industrial, agricultural, and environmental applications, yet these parameters are notoriously difficult to measure. Due to a lack of adequate methodologies, microbiologists usually have difficulty determining exactly how many different bacterial types there are in a sample or which population of cells within the sample is responsible for a relevant chemical transformation.
Culturable bacteria account for only a small fraction of the total bacterial count in environmental samples (10, 13, 21), and there is now a plethora of studies demonstrating that the environment holds a great many more bacterial species than can be isolated using standard techniques. Uncultured bacteria are also potentially the largest source of genetic information on the planet. DNA extracted from soil has been shown to be highly heterogeneous by using reassociation kinetics, which indicate the presence of as many as 4,000 different bacterial genomes in 180 g of soil (27, 29).
In recent years emphasis has been placed on the use of novel culture methods in order to cultivate the large proportion of unknown bacteria in the environment. Extinction dilution and low-nutrient methods have, for example, been used to culture numerically abundant but difficult-to-culture oceanic bacteria (3, 4). Other methods that have been applied to this problem include optical tweezers (8) and gel-microdroplet encapsulation (31), but such techniques are probably too specialized to be adopted by the wider scientific community (14). Contrary to mainstream belief, Joseph et al. have argued that simple cultivation techniques are still adequate for culturing many novel/uncultured bacteria from soil (14). In that study, the authors obtained 350 different isolates by using simple solid medium techniques. Of these, 27% belonged to yet-unnamed and poorly studied bacterial groups. These data indicate that proper experimental design can enhance our ability to isolate novel and unknown bacterial strains from the environment by using standard microbiological methodologies.
The purpose of this study was to examine the community composition and change of soil bacterial communities during sequential enrichments on several different carbon sources. Results from these experiments should prove useful in the design of experiments that aim to maximize bacterial diversity recovered from the environment by using traditional enrichment and plating techniques. Specifically, we tested the hypothesis that structurally similar carbon sources will enrich for similar bacterial enrichment communities. Second, we investigated the effect of a sequence of enrichment steps on bacterial community composition by using more than one carbon source. Finally, we investigated whether the soil source material used in an enrichment has a greater effect on the composition of an enrichment community than the carbon source.
MATERIALS AND METHODS
Medium.
All enrichments were performed in minimal salt medium (MMO) (26). The medium was autoclaved and cooled, and a sterile carbon source was added using aseptic technique. The following concentrated stock solutions of carbon sources were prepared and sterilized by autoclaving: 100 mM sodium acetate, 100 mM sodium propionate, 100 mM sodium benzoate, 100 mM p-hydroxybenzoate, 100 mM glucose, 100 mM cellobiose, 2.5 g yeast extract per 100 ml, and 2.5 g tryptone per 100 ml. The amino acid stocks for l-leucine and l-arginine (100 mM) were not autoclaved but were sterilized by filtration through 0.2-μm Acrodisc filters (Pall Life Sciences, Ann Arbor, MI). Benzoate, p-hydroxybenzoate, leucine, arginine, glucose, and cellobiose were added to the medium at a final concentration of 2 mM. Propionate was added at a final concentration of 4 mM and acetate to a final concentration of 6 mM. MMO containing yeast extract or tryptone was prepared by adding 1 ml of the concentrated stock solution to 50 ml of MMO.
Enrichments.
Dark, organic-rich soil was sampled freshly prior to each enrichment experiment. Soil originated from the area surrounding Passion Puddle at Cook College of Rutgers University, New Brunswick, N.J. With the exception of soil used for the experiments shown in Fig. 6, all soils used in a particular experiment were sampled at the same time from the same location. Four different soils were sampled for the experiment shown in Fig. 6: Passion Puddle lake sediment, garden soil from a managed flower bed, low-organic-matter sand (yellow/brown), and low-organic-matter clay (red). The last three of these samples were taken from within 1 km of Passion Puddle. Primary enrichments (EI) were obtained by adding 50 mg of soil to 5 ml of MMO containing different carbon sources. All samples were incubated in 25- by 150-mm culture tubes at 26°C in a shaking incubator at 200 rpm and were sampled after 24 h. With the exception of the leucine enrichments, 24-h incubations produced turbid cultures. Samples containing leucine as a carbon source took longer to grow to turbidity and were incubated for 48 h. Secondary (EII) and tertiary (EIII) enrichments were obtained by transferring 50 μl of the enrichment culture to 5 ml of fresh medium. The experiment shown in Fig. 5 was performed by enriching 0.5 g of soil in 100 ml of MMO containing acetate in a 500-ml Erlenmeyer flask. The enrichment was incubated at 26°C in a shaking incubator at 200 rpm. The EII enrichment was obtained by transferring 1 ml of the EI enrichment to a new flask containing fresh MMO.
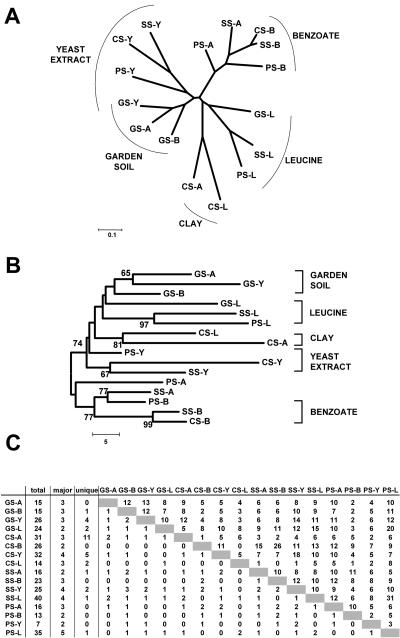
FIG. 6.
Effect of soil type on carbon source enrichment. Carbon sources: A, acetate; B, benzoate; L, leucine; Y, yeast extract. Soils: GS, garden soil; CS, clay soil; SS, sandy soil; PS, pond sediment. For example, GS-B indicates an enrichment of garden soil in MMO containing benzoate. All TRFLPs originated from secondary enrichments (EII) on the respective carbon sources. (A) Dendrogram generated by neighbor-joining analysis of similarity values (Ps) calculated from TRFLP data by applying a search window of ±0.5 bp to match peaks. A distance of 0.1 indicates 10% dissimilarity between two TRFLP patterns. (B) Consensus tree of 10,000 bootstrap replicates of binned and catenated (two restriction enzymes) TRFLP data. The scale bar indicates the number of differences. Only bootstrap values over 50 are shown. (C) TRFLP data matrix for one of the enzymes used in the analysis. Shown are the total number of peaks, the major peaks (peaks that account for more then 50% of the total TRFLP area), and the number of peaks unique to each TRFLP trace. The upper right portion of the matrix indicates the number of shared peaks between the two TRFLP patterns, while the lower left indicates the number of shared major peaks.
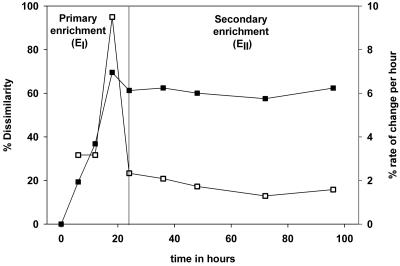
FIG. 5.
Community change over time in an enrichment culture. One gram of soil was inoculated into 100 ml of MMO medium containing acetate as a carbon source. Samples were taken from the primary enrichment (EI) every 6 hours. One milliliter of primary enrichment was transferred to a new flask containing 100 ml of MMO medium for the secondary enrichment (EII) after 24 h. Samples were then taken at 12, 24, 48, and 72 h. The x axis indicates the time elapsed since the initial soil inoculation in EI. Shown are the percent dissimilarity of TRFLP patterns of samples and the original soil TRFLP patterns (▪) and the percent rate of change in TRFLP patters between successive samples normalized to time (per hour) (□). The vertical line indicates the time that EII was inoculated.
DNA extraction and PCR.
The Ultraclean soil DNA kit (MO BIO Laboratories, Carlsbad, CA) was used to extract DNA from soil. DNA was extracted from 0.2 g of soil according to the manufacturer's instructions. Enrichments from soil were extracted by first centrifuging 2 ml of enrichment culture for 10 min and removing the supernatant. The pellets were frozen at −20°C and later extracted by first resuspending cells in 200 μl IRS solution, provided in the MO BIO kit. DNA was then extracted as per the manufacturer's instructions. Five nanograms of DNA from each extraction was then added to 50-μl PCR mixtures. The total bacterial community was analyzed using the 16S rRNA gene PCR primers 27F (5′-GAGTTTGATCMTGGCTCAG-3′) and 1525R (5′-AAGGAGGTGATCCAGCC-3′) (19). The primer 27F was carboxyfluorescein labeled for terminal restriction fragment length polymorphism (TRFLP) analysis. PCR conditions were as follows: 200 nM of each PCR primer, 2.5 mM MgCl, 5 U Taq DNA polymerase (Promega, Madison, WI) and 0.4 μl of 10-mg ml−1 acetylated bovine serum albumin (BSA) (Promega). Cycling times were 5 min at 95°C followed by 30 cycles of 1 min at 95°C, 1 min at 55°C, and 1.5 min at 72°C. A 15-min extension step at 72°C was applied after PCR amplification.
TRFLP and data analysis.
Thirty nanograms of carboxyfluorescein-labeled PCR products were independently digested using 2 units of the 4-base-pair-cutting restriction enzyme MnlI or AluI (NEB, Beverly, MA). Digests were precipitated with glycogen as a carrier and resuspended in 20 μl deionized formamide. Samples were then denatured at 95°C for 5 min, and the lengths of the individual restriction fragments were determined using an ABI 310 automated sequencer (Applied Biosystems, Foster City, CA) to determine their TRFLP patterns. TRFLPs generated in this manner contained maximum peak heights in the range of 4 × 103 to 6 × 103 fluorescence units (FU) and cumulative peak areas of between 2 × 105 and 4 × 105 FU2. Data were exported into Matlab (Natick, MA) and analyzed for the presence and absence of peaks only. A search window of ±0.5 bp was applied in order to match peaks between individual TRFLP runs. Two different methods for generating similarity matrices were used to confirm that the results were not an artifact of the analysis method. First, pairwise similarities were calculated as previously described (25) using the Sorenson index: Cs = 2Nab/(Na + Nb), where Nab is the number of shared peaks and Na and Nb are the numbers of peaks in each respective sample (20, 22). Alternatively, the percent similarity was calculated as Ps = Sa/Na, where Sa is the number of peaks in sample A that are also found in sample B and Na is the total number of peaks in sample A. The top left and bottom right of these matrices were then averaged and exported. Distances for both methods and both enzymes were then calculated as 1 − Cs or 1 − Ps and exported into MEGA (Molecular Evolutionary Genetics Analysis software, version 2.1) (17) to generate neighbor-joining dendrograms. Overall, the same general topologies were obtained for all dendrograms, whether similarities were calculated using the Sorenson index (Cs) or by calculating the percentage of shared peaks (Ps) (data not shown). During the analysis we observed that the Sorenson index can occasionally produce percentages greater than 100%, particularly in low-complexity samples. This is likely due to the fact that we chose not to bin our data. Two peaks which are only a fraction of a base pair apart can sometimes be binned differently (mathematical rounding artifact) even though visual inspection of the TRFLP patterns indicates that they likely correspond. This can affect the calculation of similarity profiles and skew or ruin pair-wise TRFLP analysis (16). In addition, Ps displayed smaller variability than Cs if the same DNA sample was analyzed in triplicate (data not shown). Dendrograms were thus generated by using a pairwise matrix of values of 1 − Ps.
Bootstrap analysis.
Bootstrap analysis was performed to further confirm that the dendrogram topologies observed in this study were not an artifact of our analysis method. To do so, data from each TRFLP run were first binned by rounding each peak size to the nearest integer. TRFLP data generated with two different restriction enzymes from each sample were catenated. The data from each experiment were then exported as an alignment in which each position corresponds to an integer nucleotide length. This alignment was then imported into MEGA, and 10,000 bootstrap replicates were performed. Distance matrices were calculated by determining the number of differences between samples, and dendrograms were generated by neighbor-joining analysis.
Minimum peak height analysis.
An important problem during TRFLP data analysis arises because a minimum peak height cutoff needs to be chosen. Loading of differential amounts of DNA can result in the detection of a different number of peaks (16), and it has been noted that small peaks which either are artifacts or are present/absent because of differential DNA loading can skew similarity profiles (6, 15). To address this problem a best minimum peak height was determined for each experimental data set by maximizing the average similarity among TRFLP patterns instead of using an arbitrary value of 50 or 100 FU. Noise peaks are likely to be small. Noise will also have a tendency to make two related TRFLP patterns (in the present case two different treatments of the same soil sample) more dissimilar unless a very large amount of noise is present. Raising the minimum peak height should thus increase the average similarity of TRFLPs of a data set and increase the signal-to-noise ratio.
RESULTS
Analysis method.
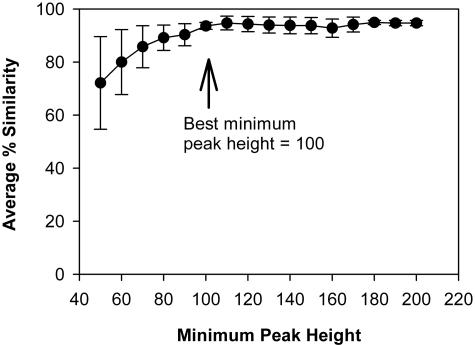
Average similarities among triplicate TRFLP patterns were low (in the range of 70%) if a cutoff of 50 FU was used (Fig. 1 [not all data shown]). This is mainly due to variability in peak heights of small peaks found in the TRFLP traces. However, if the minimum peak height was increased from 50 to 100 FU, a large and significant increase in average similarity among triplicate TRFLPs was observed, approaching a plateau at between 90 and 95% similarity. An example of this is shown in Fig. 1 (acetate enrichment, AluI-digested TRFLP pattern). Analysis of data generated from subsequent enrichment experiments in this study (see Fig. 2 to 6) produced plots analogous to that shown in Fig. 1 (data not shown). These data indicate that blindly choosing a minimum peak height of 50 FU can lead to difficulties in data analysis, as previously suggested (16), and that it can be useful to optimize this parameter. The data also indicate that for the data in this study, about 10% of the variability observed was inherent in the technique and did not represent true differences in the microbial community composition. Minimum peak height was therefore subsequently optimized for each experimental data set by choosing a value as low as possible that fell within the plateau region but was smaller than 5% of the highest peak in the TRFLP pattern or 200 FU (whichever was smaller).
FIG. 1.
Example of best-minimum-peak-height analysis. Soil samples were enriched for 24 h in MMO containing acetate, and 1 ml of culture was sampled in triplicate. DNA was extracted independently from subsamples and then analyzed for their TRFLP patterns. Shown are the average similarities of three TRFLP patterns generated from the same enrichment sample as a function of minimum peak height. One milliliter of culture was collected from the same enrichment in triplicate, and the samples were analyzed by TRFLP independently. The data were analyzed for presence/absence of peaks only. All peaks smaller than minimum peak height were not used for analysis. Error bars indicate one standard deviation.
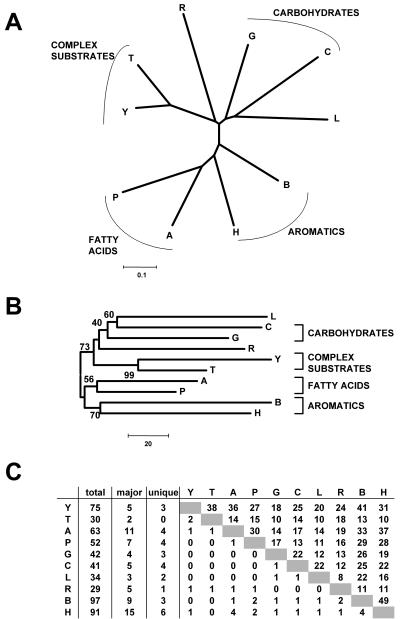
FIG. 2.
Comparison of TRFLP data for communities enriched from soil on 10 structurally similar and different carbon sources. Carbon sources: A, acetate; P, propionate; B, benzoate; H, p-hydroxybenzoate G, glucose; C, cellobiose; R, arginine; L, leucine; Y, yeast extract; T, tryptone. All TRFLPs originated from secondary enrichments (EII) on the respective carbon sources. (A) Dendrogram generated by neighbor-joining analysis of similarity values (Ps) calculated by applying a search window of ±0.5 bp to match peaks. A distance of 0.1 indicates 10% dissimilarity between two TRFLP patterns. (B) Consensus tree of 10,000 bootstrap replicates generated from binned and catenated (two restriction enzymes) TRFLP data. The scale bar indicates the number of differences. Only bootstrap values over 50 are shown. (C) TRFLP data matrix for one of the enzymes used in the analysis. Shown are the total number of peaks, the major peaks (peaks that account for more then 50% of the total TRFLP area), and the number of peaks unique to each TRFLP trace. The upper right portion of the matrix indicates the number of peaks shared between the two TRFLP patterns, while the lower left indicates the number of shared major peaks.
Effect of carbon sources.
It is commonly assumed that enriching on dissimilar carbon sources will select for dissimilar bacterial communities. It is also assumed that structurally similar carbon sources, which are metabolized by related biochemical pathways, select for similar microbial communities. To test these hypotheses, we enriched freshly sampled soil in minimal medium on two carbohydrates, two amino acids, two fatty acids, two aromatic molecules, and two complex carbon sources. Figure 2 shows the TRFLP analysis of the EII enrichments. All 10 enrichments contained unique bacterial communities after enrichment (average similarity among all samples, 34%), and all but the enrichment on tryptone contained TRFLP peaks unique to the carbon source used (Fig. 2C), supporting the idea that dissimilar carbon sources will select for dissimilar communities. The TRFLP data support the hypothesis that structurally similar carbon sources select for similar microbial communities. TRFLP patterns of communities enriched on the two fatty acids, two aromatics, and two complex substrates were notably more similar to each other than to the remaining enrichments (58, 52, and 82% similar to each other, respectively), forming attracting branches in cluster analysis, which was supported by high bootstrap values (56, 70, and 99%, respectively). TRFLP patterns of glucose- and cellobiose-enriched samples also clustered in the dendrogram (Fig. 2), but the similarity between these two samples was within the range of values observed for structurally unrelated carbon sources (considering the variability of up to 10% observed in our experiments), and bootstrap support for this node was comparatively low (40%). The samples enriched on the two amino acids leucine and arginine were no more similar to each other than to the remaining enrichment samples (36% similar).
Effect of enrichment sequence.
Two enrichment experiments were performed to examine if changing the carbon sources in successive enrichment steps has an effect that is reflected by a shift in the enrichment community composition. In the first experiment (Fig. 3A, B, and C), soil was enriched in medium containing either glucose or benzoate (EI enrichment) and subsequently transferred to new medium (EII) alternately containing these two carbon sources. Analysis of TRFLP data generated from these enrichments indicated that the carbon source in the primary enrichment affects the position of the EII enrichments in dendrogram. All samples that were grown on benzoate in the EI enrichment formed a cluster (Fig. 3A), irrespective of the carbon source used for the secondary (EII) enrichment (bootstrap support was 81%). There was also a clustering of EIII samples which had received the same carbon source during the EII enrichment (bootstrap support was 55, 65, 100, and 100%).
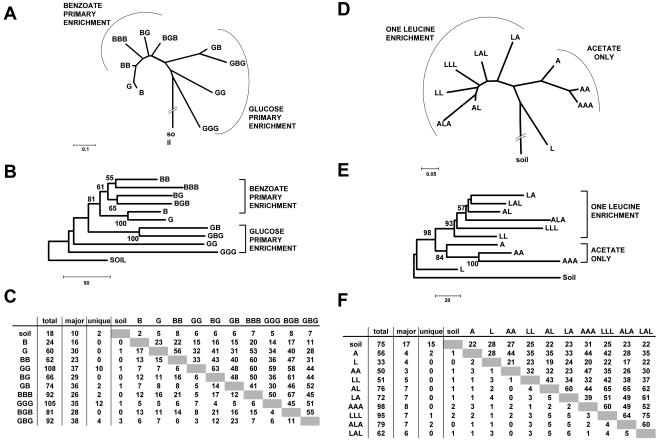
FIG. 3.
Effect of swapping carbon sources during enrichments. The dendrogram was generated by neighbor-joining analysis of similarity values calculated from TRFLP data for communities enriched from soil. Carbon sources: B, benzoate; G, glucose; L, leucine; A, acetate. Multiple letters indicate a series of enrichment steps. For example, BGB indicates an enrichment sequences with a primary enrichment (EI) on benzoate, a secondary enrichment (EII) on glucose, and a tertiary enrichment (EIII) on benzoate, where the sample originates from the EIII enrichment. (A to C) benzoate and glucose; (D to F) leucine and acetate. The dendrograms in (A) and (D) were generated by neighbor-joining analysis of similarity values (Ps) calculated from TRFLP data by applying a search window of ±0.5 bp to match peaks. A distance of 0.1 indicates 10% dissimilarity between two TRFLP patterns. The branch connecting the soil community was shortened to condense the figure. (B) and (E) are consensus trees of 10,000 bootstrap replicates of binned and catenated (two restriction enzymes) TRFLP data. The scale bars indicate the number of differences. Only bootstrap values over 50 are shown. (C) and (F) show TRFLP data matrices for one of the enzymes used in the analysis. Shown are the total number of peaks, the major peaks (peaks that combined account for more then 50% of the total TRFLP area), and the number of peaks unique to each TRFLP trace. The upper right portion of the matrix indicates the number of shared peaks between the two TRFLP patterns, while the lower left indicates the number of shared major peaks.
An analogous experiment was performed using acetate and leucine, and a different situation was observed (Fig. 3D, E, and F). Samples from enrichments with a history of at least one enrichment step on leucine clustered to the exclusion of enrichment sequences that included acetate only. Bootstrap support for this branching was 84, 98, and 93% (Fig. 3E). An enrichment step on leucine thus appears to have an overriding influence on the microbial community, at least in part canceling the effect of previous enrichment steps.
Reproducibility of soil enrichments.
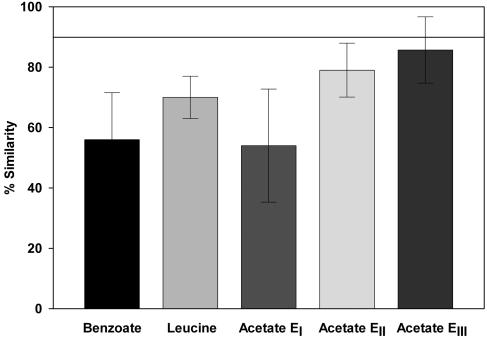
A soil sample was enriched in triplicate on acetate, benzoate, and leucine (n = 9). Analysis of TRFLPs generated from the EI enrichments indicated that the community composition of soil enrichments can be highly variable. Triplicate TRFLP patterns shared only 54, 56, and 70% of peaks for acetate, benzoate, and leucine EI enrichments, respectively (Fig. 4). One of the acetate EI enrichments was subsampled in triplicate into secondary acetate enrichment tubes (EII), and triplicate tertiary enrichments were generated by subsampling of just one of the acetate EII tubes. TRFLP analysis of replicate EII and EIII enrichments indicated that successively lower variability was associated with downstream enrichment steps (Fig. 4). TRFLP patterns of EII enrichments were on average only 79% similar. Triplicate EIII enrichments were on average 86% similar as determined by TRFLP, approaching the 90 to 95% similarity observed for replicate sampling (Fig. 1).
FIG. 4.
Average similarity calculated from TRFLP patterns originating from replicate soil enrichment experiments. Shown are average similarities of triplicate independent primary enrichments (EI) on acetate, benzoate, and leucine. Also shown are the average similarities of triplicate subsamples of acetate secondary (EII) and tertiary (EIII) enrichments (all three tubes in the EII and EIII enrichments received inocula from the from the same EI and EII tubes). Error bars indicate one standard deviation calculated from similarity percentage values. A line at 90% indicates the average similarity of samples taken from the same enrichment tube. Similarity values greater than this line indicate that the communities of two separate samples are not different based on their TRFLP pattern.
Community change over time.
The behavior of a community during the enrichment process was investigated by sampling a time series from an enrichment grown on acetate (Fig. 5). TRFLP data indicate that the community change in the primary enrichment was most rapid between 12 and 18 hours after inoculation of the soil, coinciding with exponential growth of the enrichment culture. The enrichment community was no more than 30% similar to the original soil sample within 18 hours of inoculation. No similar spike in the rate of community change was observed in the EII enrichment. Instead, gradual yet appreciable change in the community composition was observed during 3 days after inoculation, ranging between 31 and 50% per 24 h.
Effect of soil type on carbon source enrichments.
Four different soil types were collected from within 1 km of one another, and each was enriched on four different carbon sources (EII). The experiment was performed to test if the soil source material or the growth substrate had a greater effect on the final composition of an enrichment community. Cluster analysis of TRFLP patterns generated from the EII enrichments (Fig. 6) indicated that the carbon sources used in the experiment had a greater effect on the enrichment community composition than the differences in soil types. All samples enriched on leucine or yeast extract and three out of four samples enriched on benzoate clustered together. Only the samples enriched on acetate did not form a cluster. Interestingly, we observed that three out of four garden soil enrichments formed a cluster (with the exception of the leucine-enriched sample).
DISCUSSION
A wealth of evidence has now been accumulated to indicate that bacterial diversity in the environment is immensely greater than traditional plating techniques would suggest. Soil in particular has been shown to contain an enormously diverse bacterial population (1, 11, 18, 28, 32). A large diversity of bacteria has been isolated by many different techniques, but no single technique appears to capture all bacteria present in a typical sample (12). We investigated the response of soil communities to enrichment in small matrices of a variety of carbon sources. Knowledge of community dynamics under these conditions will aid in the design of better isolation techniques, which will help to maximize the diversity of organisms recovered from the environment.
It is usually assumed that differences in the biochemical capabilities and tolerances of bacteria in the environment are the basis for our ability to enrich, select, and ultimately isolate a single organism. Alternatively, isolation would be a game of chance, where stochastic principles would favor the isolation of more abundant community members and sample heterogeneity would determine reproducibility. Our data provide evidence for both viewpoints. Most importantly, we observed excellent support for the biochemical hypothesis. Clustering of TRFLP patterns generated from enrichment cultures containing structurally similar carbon sources (Fig. 2) would not be expected if the enrichment community structure was solely determined by physical or stochastic principles and limitations. Studies aiming to isolate the largest possible diversity from soil would thus benefit from the use of the greatest possible growth substrate diversity.
In addition to similarities between enrichment communities induced by the choice of carbon sources, there was substantial variability in the data sets that could not be accounted for by differences in growth conditions. TRFLP patterns generated from replicate enrichment tubes that had received 50-mg subsamples of the same soil and 5-ml subsamples of the same medium stock (Fig. 4) were up to 50% dissimilar (in the case of acetate and benzoate; 30% for leucine). A large proportion of these observed differences are potentially explained by the fact that soil is a highly heterogeneous matrix, both physically and chemically. This heterogeneity would be reflected by the 50-mg subsamples used for inoculating the enrichments. Soil heterogeneity both results from and is responsible for high biological heterogeneity (23), and community composition is likely to be controlled by conditions in local microenvironments at the submillimeter scale (23), resulting in a distribution of bacterial cells that is highly patchy. Actinomycetes, for example, have been shown to be primarily associated with larger soil aggregates, while pseudomonads are more numerous in the smaller size fraction (reference 30 as cited in reference 23). As a result, soil heterogeneity and microbe-soil interaction have been recognized as important limitations to quantitative and representative analysis of soil communities (2, 24). Various techniques have been devised to help in homogenization of soil samples, including buffered dilution, chelation, homogenization, and ultrasonication (see reference 2 and references therein). Such methods are clearly necessary if a representative sample is desired. Our data, however, also indicate that soil heterogeneity was not the only factor contributing to the observed differences. Secondary enrichments (EII) were generated by triplicate sampling of the same primary enrichment (EI). The resulting EII communities were on average less than 79% similar based on their TRFLP pattern (Fig. 4). Differences among replicate EII tubes should be no greater than the error associated with the method, because the EII tubes received negligible soil carryover (<0.01%, wt/vol) and cells were suspended and well mixed by shaking at 200 rpm. These data suggest that the outcome of enrichment experiments may not be entirely predictable by mathematical competition models such as the traditional Lotka-Volterra equations. Less variability was associated with subsequent transfer steps, as differences between EIII enrichments (subsampled from a single EII tube [Fig. 4]) approached the variability associated with replicate subsamples taken from the same enrichment tube (Fig. 1).
If a soil community is enriched sequentially on more than one carbon source, one might expect that changes in the composition of the growth medium will be reflected by changes in the composition of the enrichment community. Our data (Fig. 3) support this notion and illustrate two important concepts. In the glucose/benzoate enrichment (Fig. 3A), we observed that all secondary and tertiary enrichment communities clustered based on the source of carbon in the primary enrichment tube; i.e., the enrichment communities had a type of memory of their initial treatment. Liquid batch culture experiments typically select for fast-growing organisms (7, 9). As a result, one might expect that the EI enrichment imposed a selective bottleneck on the microbial population by favoring fast-growing cells that are able to use the substrate more directly and efficiently. Populations in subsequent transfers are then biased toward the initial enrichment population. Alternatively, one would expect a switching of communities in response to and correlating with the sequence of carbon sources used. For example, the sequence glucose-benzoate should have produced a community of cells more similar to those with the sequence benzoate-benzoate than to those with the sequence glucose-benzoate-glucose, a situation that clearly did not arise. For studies aiming to isolate the greatest possible diversity from a soil sample, these data imply that there is a hierarchy with respect to carbon source influence on selection (i.e., leucine > benzoate > acetate) and that it would be useful to use multiple and different sequences of carbon sources during preenrichments. The glucose-benzoate, glucose-glucose, benzoate-glucose, and benzoate-benzoate enrichments were only between 39% and 76% similar based on TRFLP analysis, indicating that each captured unique parts of the total bacterial community in soil. A second interesting observation is illustrated by the experiment shown in Fig. 3B. In this experiment leucine was able to override the bottleneck effect imposed by acetate in the primary enrichment, and all samples that had experienced at least one enrichment step on leucine clustered. Such a situation can arise if the ability to metabolize a particular substrate is limited to only a small portion of the community. The bottleneck effect imposed by leucine is much stronger than that imposed by acetate, and the small proportion of leucine-metabolizing bacteria that survive enrichment on acetate can then become dominant once leucine is present.
An often-invoked principle in microbial ecology is the notion that “everything is everywhere, and the environment selects.” Under this paradigm, the expectation is that different soils will yield similar bacterial communities if enriched using similar conditions. Enrichments of different soil samples with the same carbon source would be expected to cluster in community analysis. The alternative hypothesis is that soils contain distinctive bacterial communities and that enrichments will select for only a subset of this unique population. Under this scenario, samples should cluster by soil type in enrichment community comparative analysis. Data from enrichments on leucine, yeast extract, and benzoate mostly support the “everything is everywhere scenario” (Fig. 6), as samples clearly clustered by the carbon source used during enrichment. However, samples taken from garden soil, sand, and clay also displayed some tendency to cluster together, indicating a component of the microbial community that is not shared between these soil samples.
The ability to enrich and isolate bacteria from the environment thus appears to be determined by an interplay between substrate utilization and species distribution. Our data indicate that a greater diversity of bacteria may be recovered from the environment by using a structurally diverse selection of growth substrates as well as multiple and diverse soil samples.
Acknowledgments
This work was supported by the Fogarty International Center of the NIH under grant U01 TW006674 for the International Cooperative Biodiversity Groups (ICBG).
REFERENCES
- 1.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bull, A. T., A. C. Ward, and M. Goodfellow. 2000. Search and discovery strategies for biotechnology: the paradigm shift. Microbiol. Mol. Biol. Rev. 64:573-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Button, D. K., B. R. Robertson, P. W. Lepp, and T. M. Schmidt. 1998. A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl. Environ. Microbiol. 64:4467-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Button, D. K., F. Schut, P. Quang, R. Martin, and B. R. Robinson. 1993. Viability and isolation of marine bacteria by dillution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell, D. E., G. M. Wolfaardt, D. R. Korber, and J. R. Lawrence. 1997. Cultivation of microbial consoritia and communities, p. 79-90. In C. J. Hurst, G. R. Knudsen, M. J. Mcinerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 6.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunbar, J., S. White, and L. Forney. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol. 63:1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frohlich, J., and H. Konig. 2000. New techniques for isolation of single prokaryotic cells. FEMS Microbiol. Rev. 24:567-572. [DOI] [PubMed] [Google Scholar]
- 9.Harder, W., and L. Dijkhuizen. 1982. Strategies of mixed substrate utilization in microorganisms. Philos. Trans. R. Soc. London Ser. B 297:459-480. [DOI] [PubMed] [Google Scholar]
- 10.Hoppe, H. G. 1976. Determination and properties of actively metabolizing heterotrophic bacteria in the sea investigated by means of micro-auto-radiography. Mar. Biol. 36:291-302. [Google Scholar]
- 11.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. (Erratum, 180: 6793.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter-Cevera, J. C., and A. Belt. 2002. Isolation of cultures, p. 3-20. In A. L. Demain and J. E. Davies (ed.), Manual of industrial microbiology and biotechnology. ASM Press, Washington, D.C.
- 13.Jannasch, H. W., and G. E. Jones. 1959. Bacterial populations in seawater as determined by different methods of enumeration. Limnol. Oceanogr. 4:128-139. [Google Scholar]
- 14.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerkhof, L., M. Santoro, and J. Garland. 2000. Response of soybean rhizosphere communities to human hygiene water addition as determined by community level physiological profiling (CLPP) and terminal restriction fragment length polymorphism (TRFLP) analysis. FEMS Microbiol. Lett. 184:95-101. [DOI] [PubMed] [Google Scholar]
- 16.Kitts, C. L. 2001. Terminal restriction fragment patterns: a tool for comparing microbial communities and assessing community dynamics. Curr. Issues Intest. Microbiol. 2:17-25. [PubMed] [Google Scholar]
- 17.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 18.Kuske, C. R., L. O. Ticknor, M. E. Miller, J. M. Dunbar, J. A. Davis, S. M. Barns, and J. Belnap. 2002. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane, D. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 20.Magurran, A. E. 1998. Ecological diversity and its measurement. Princeton Univerity Press, Princeton, N.J.
- 21.Meyer-Reil, L. A. 1978. Autoradiography and epifluorescence microscopy combined for the determination of number and spectrum of actively metabolizing bacteria in natural water. Appl. Environ. Microbiol. 36:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray, A. E., C. M. Preston, R. Massana, L. T. Taylor, A. Blakis, K. Wu, and E. F. DeLong. 1998. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl. Environ. Microbiol. 64:2585-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicol, G. W., L. A. Glover, and J. I. Prosser. 2003. Spatial analysis of archaeal community structure in grassland soil. Appl. Environ. Microbiol. 69:7420-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Donnell, A. G., M. Goodfellow, and D. L. Hawksworth. 1994. Theoretical and practical aspects of the quantification of biodiverity among microorganisms. Philos. Trans. R. Soc. London Ser. B 345:65-73. [DOI] [PubMed] [Google Scholar]
- 25.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 27.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torsvik, V., J. Goksoyr, F. L. Daae, R. Sorheim, J. Michalsem, and K. Salte. 1996. Total bacterial diveristy in sediment and soil: a review. J. Ind. Microbiol. Biotechnol. 17:170-178. [Google Scholar]
- 29.Torsvik, V., K. Salte, R. Sorheim, and J. Goksoyr. 1990. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl. Environ. Microbiol. 56:776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watts, J. E. M. 1999. Analysis of microbial diversity in polluted an nonpolluted soil: a comparison of genetic, funtional, and culture based techniques. University of Warwick, Warwick, United Kingdom.
- 31.Zengler, K., G. Toledo, M. Rappe, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15684-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, J., B. Xia, H. Huang, A. V. Palumbo, and J. M. Tiedje. 2004. Microbial diversity and heterogeneity in sandy subsurface soils. Appl. Environ. Microbiol. 70:1723-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]