Abstract
Twenty Mimosa-nodulating bacterial strains from Brazil and Venezuela, together with eight reference Mimosa-nodulating rhizobial strains and two other β-rhizobial strains, were examined by amplified rRNA gene restriction analysis. They fell into 16 patterns and formed a single cluster together with the known β-rhizobia, Burkholderia caribensis, Burkholderia phymatum, and Burkholderia tuberum. The 16S rRNA gene sequences of 15 of the 20 strains were determined, and all were shown to belong to the genus Burkholderia; four distinct clusters could be discerned, with strains isolated from the same host species usually clustering very closely. Five of the strains (MAP3-5, Br3407, Br3454, Br3461, and Br3469) were selected for further studies of the symbiosis-related genes nodA, the NodD-dependent regulatory consensus sequences (nod box), and nifH. The nodA and nifH sequences were very close to each other and to those of B. phymatum STM815, B. caribensis TJ182, and Cupriavidus taiwanensis LMG19424 but were relatively distant from those of B. tuberum STM678. In addition to nodulating their original hosts, all five strains could also nodulate other Mimosa spp., and all produced nodules on Mimosa pudica that had nitrogenase (acetylene reduction) activities and structures typical of effective N2-fixing symbioses. Finally, both wild-type and green fluorescent protein-expressing transconjugant strains of Br3461 and MAP3-5 produced N2-fixing nodules on their original hosts, Mimosa bimucronata (Br3461) and Mimosa pigra (MAP3-5), and hence this confirms strongly that Burkholderia strains can form effective symbioses with legumes.
Although it was generally accepted for many years that legumes (and the nonleguminous plant Parasponia) were nodulated exclusively by members of the Rhizobiaceae in the α-Proteobacteria (including the genera Allorhizobium, Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium) (30, 34), recently there has been an increasing number of reports of members of the β-Proteobacteria being isolated from nodules. So far, these include Burkholderia tuberum strain STM678 and Burkholderia phymatum strain STM815 (originally isolated from Aspalathus carnosa in South Africa and Machaerium lunatum in French Guiana, respectively [26, 39]), Ralstonia taiwanensis strains (isolated from Mimosa pudica in Taiwan and India and Mimosa diplotricha in Taiwan [8, 41] and now renamed Cupriavidus taiwanensis [38]), and several Burkholderia strains isolated from Mimosa casta, Mimosa pigra (synonym, Mimosa pellita), M. pudica, and another mimosoid legume, Abarema macradenia, in Panama (3). Although symbiotic genes (nifH and nodA) have been identified in the Burkholderia strains STM678 and STM815, so far there is very little physiological and structural evidence of their symbiotic nature, and they have been shown to form only ineffective nodules on the promiscuous legume Macroptilium atropurpureum (26). More convincingly, not only have some of the Panamanian Burkholderia strains been shown to possess symbiosis-related genes (nodB and nifD), but initial nodulation studies have suggested that they may indeed be N-fixing symbionts within nodules on M. pigra (3). However, these last observations await confirmation by microscopy. The evidence of effective nodulation by C. taiwanensis is much stronger than that presented so far for Burkholderia, as Chen et al. (9) have demonstrated using a green fluorescent protein (GFP)-tagged strain and detailed light and electron microscopic studies that this bacterium can readily form N-fixing nodules on Mimosa spp.
Chen et al. (10) have shown that the genus Mimosa has a particular affinity for nodulation by β-rhizobia. For example, in their study of 190 isolates from symbiotic nodules on M. pudica and M. diplotricha in Taiwan, the vast majority of the isolates were identified as β-rhizobia, and these consisted mostly of C. taiwanensis (>93%), with the remainder being made up of a small number of “conventional” α-rhizobia (Rhizobium and Sinorhizobium) as well as two strains of Burkholderia caribensis, a previously described species that was not known to nodulate legumes (39).
On the basis of their data from Taiwanese Mimosa isolates, Chen et al. (10) even suggested that C. taiwanensis may actually be the “specific symbiont” of M. pudica and M. diplotricha. However, this may only hold true in Taiwan, as the legume genus Mimosa probably originated in tropical America (2). Mimosa pudica was introduced to Taiwan by Europeans in 1645 as an ornamental, which then escaped and colonized all parts of the island, and M. diplotricha was first reported in Taiwan in 1965 (43). Given that M. pudica and M. diplotricha are nonnative invasive species in Taiwan, there are two likely mechanisms to explain their apparent affinity for C. taiwanensis and other β-rhizobial symbionts: (i) the bacteria were brought with them from tropical America or (ii) they have coopted local Taiwanese bacteria. The fact that C. taiwanensis has also been isolated from M. pudica nodules in India (41) supports the former hypothesis, and therefore it is likely that β-rhizobia are also present in Mimosa nodules in other parts of the tropics, particularly in tropical America, where the genus originated. In the present study, we show, by comparing the sequences of their 16S rRNA genes with those of genes from reference strains, that 20 strains isolated from nodules of various Mimosa spp. in South America are all members of the genus Burkholderia. Moreover, evidence is also presented that five of these Burkholderia strains possess symbiotic genes (nifH, nodA, and nod box) and that all five can form functional, symbiotic nodules on Mimosa spp. In particular, we focus on genetically modified transconjugant variants of two of the strains, Br3461 and MAP3-5, both marked with the gfp marker gene, and hence demonstrate definitively that these two Burkholderia strains are functional symbionts of Mimosa.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The Mimosa strains used in these studies are listed in Table 1. The strains with “Br” prefixes were originally isolated from various Mimosa spp. in regions of Brazil, especially the Atlantic Forest (Mata Atlantica) in the Southeast, but also from Amazonia and the Cerrado (11-13; S. M. de Faria et al., unpublished). The Venezuelan strains, MAP3-1 through MAP3-6, were isolated from nodules of Mimosa pigra growing in the seasonally flooded forests adjacent to the Mapire River (a tributary of the Orinoco) during the study of Barrios and Herrera (4). All strains were revived from glycerol stocks kept at −70°C and then grown on yeast extract-mannitol plates at 28°C. All cultures used for subsequent DNA extraction, amplification, phylogenetic analysis, and inoculation onto various Mimosa spp. were derived from single colonies (8).
TABLE 1.
α- and β-rhizobial strains examined by ARDRA
| Rhizobial strain | Host plant | Geographical origin | Reference(s) |
|---|---|---|---|
| Ralstonia taiwanensis LMG19424a | Mimosa pudica | Taiwan | 8 |
| Ralstonia taiwanensis LMG19425a | Mimosa diplotricha | Taiwan | 8 |
| Sinorhizobium sp. strain TJ170 | Mimosa pudica | Taiwan | 10 |
| Rhizobium sp. strain TJ167 | Mimosa diplotricha | Taiwan | 10 |
| Rhizobium sp. strain TJ171 | Mimosa diplotricha | Taiwan | 10 |
| Rhizobium sp. strain TJ172 | Mimosa diplotricha | Taiwan | 10 |
| Rhizobium sp. strain TJ173 | Mimosa diplotricha | Taiwan | 10 |
| Burkholderia caribensis TJ182 | Mimosa diplotricha | Taiwan | 10 |
| B. phymatum STM815 | Machaerium lunatum | French Guiana | 26, 38 |
| B. tuberum STM678 | Aspalathus carnosa | South Africa | 26, 38 |
| Burkholderia sp. strain Br 3429 | Mimosa acutistipula | Brazil | This study |
| Burkholderia sp. strain Br 3432 | Mimosa acutistipula | Brazil | This study |
| Burkholderia sp. strain Br 3470 | Mimosa bimucronata | Brazil | This study |
| Burkholderia sp. strain Br 3461 | Mimosa bimucronata | Brazil | This study |
| Burkholderia sp. strain Br 3469 | Mimosa camporum | Brazil | This study |
| Burkholderia sp. strain Br 3407 | Mimosa caesalpiniaefolia | Brazil | This study |
| Burkholderia sp. strain Br 3405 | Mimosa caesalpiniaefolia | Brazil | This study |
| Burkholderia sp. strain Br 3462 | Mimosa flocculosa | Brazil | This study |
| Burkholderia sp. strain Br 3464 | Mimosa flocculosa | Brazil | This study |
| Burkholderia sp. strain Br 3446 | Mimosa laticifera | Brazil | This study |
| Burkholderia sp. strain Br 3467 | Mimosa pigra | Brazil | This study |
| Burkholderia sp. strain Br 3437 | Mimosa scabrella | Brazil | This study |
| Burkholderia sp. strain Br 3454 | Mimosa scabrella | Brazil | This study |
| Burkholderia sp. strain Br 3466 | Mimosa tenuiflora | Brazil | This study |
| Burkholderia sp. strain MAP 3-1 | Mimosa pigra | Venezuela | This study |
| Burkholderia sp. strain MAP 3-2 | Mimosa pigra | Venezuela | This study |
| Burkholderia sp. strain MAP 3-3 | Mimosa pigra | Venezuela | This study |
| Burkholderia sp. strain MAP 3-4 | Mimosa pigra | Venezuela | This study |
| Burkholderia sp. strain MAP 3-5 | Mimosa pigra | Venezuela | This study |
| Burkholderia sp. strain MAP 3-6 | Mimosa pigra | Venezuela | This study |
Now renamed Cupriavidus taiwanensis (38).
DNA manipulation.
Amplified rRNA gene restriction analysis (ARDRA) was performed as described previously (8), except that AluI, CfoI, HinfI, and MspI were used. For each strain, the normalized restriction patterns obtained were entered into a combined profile and analyzed using the Dice similarity coefficient (SD) and the unweighted-pair group method using average linkages clustering algorithm by MVSP 3.1 software (Kovach Computing Services). Nearly-full-length 16S rRNA genes were amplified and sequenced as previously described (8). A 658-bp fragment containing part of the nifH gene was amplified and sequenced using the primers 5′-CGCIWTYTACGGIAARGGIGG-3′ and 5′-GGIKCRTAYTSGATIACIGTCAT-3′. A 350-bp fragment containing part of the nodA gene was amplified and sequenced using the primers 5′-TGGARVBTNYSYTGGGAAA-3′ and 5′-CCRAAVSCRAAYGGVAC-3′. A 380- to 395-bp fragment containing the intergenic sequences of the nodD gene and the nodB gene was amplified and sequenced using the primers 5′-CAGATCNAGDCCBTTGAARCGCA-3′ (located at the end of nodD in rhizobia) and 5′-GGRTKNGGNCCRTCRTCRAANGT-3′ (located at the beginning of nodB in rhizobia).
Phylogenetic analyses.
Sequences were imported into BioEdit 4.8.4 (18), where amino acid sequences were deduced from the nifH and nodA sequences. ClustalW 1.4 (36), used from within BioEdit, was used to align the 16S rRNA gene, NifH, and NodA sequences and to construct neighbor-joining phylogenies with 1,000 bootstrap replicates, using Kimura distance corrections and discarding positions with gaps in any sequence. Trees were displayed using TREEVIEW (28). Sequence identities were calculated using BioEdit.
Plant tests and microscopy.
Plant cultivation and nodulation tests were carried out as described previously (9), using the tube method of Gibson (16). Seeds of Mimosa acutistipula, M. diplotricha, M. pigra, and M. pudica were surface sterilized with concentrated sulfuric acid for 10 min followed by 3% sodium hypochlorite for 10 min and then washed with sterile water. Seeds were germinated on nutrient agar plates at 28°C in darkness to make sure that there was no contamination. The tubes contained a modified Jensen's N-free plant nutrient medium (33) and were incubated at 35°C under an irradiance of 400 μmol m−2 s−1 with a photoperiod of 16 h. Seven days after germination, the seedlings were inoculated with 100 μl (approximately 105 cells) of a washed suspension of 1 of the 10 bacterial strains. Plants were harvested 60 days after inoculation, and nitrogenase activity was checked using the “closed” acetylene reduction assay according to the method of James and Crawford (21).
A separate tube experiment was performed with GFP-tagged strains of Br3461 and MAP3-5 that had been constructed according to the method of Chen et al. (9), using Escherichia coli S17-1 λpir with pUTmini-Tn5gfp. Mimosa pudica was inoculated with either Br3461-gfp or MAP3-5-gfp, Mimosa bimucronata was inoculated with Br3461-gfp, and M. pigra was inoculated with MAP3-5-gfp. Plants were harvested 35 days after inoculation and tested for nitrogenase activity as described above. For the detection of GFP, fresh nodules were taken and sectioned (50 to 100 μm) using a Vibratome 1500 (Agar Aids, Stansted, United Kingdom). The sections were then mounted on slides and examined under either a Nikon Eclipse epifluorescence microscope or a Zeiss 510META confocal laser scanning microscope (CLSM) according to the method of Chen et al. (9). Some nodules were also fixed and embedded in resin, and sections of these were labeled with immunogold according to the method of James et al. (23), using antibodies raised against the nitrogenase NifH protein (1:100).
Nucleotide sequence accession numbers.
The GenBank accession numbers for 16S rRNA gene sequences obtained as part of this study are as follows: AY773185 (BR3405), AY773186 (BR3407), AY773187 (BR3429), AY773188 (BR3432), AY773189 (BR3437), AY773190 (BR3446), AY773191 (BR3454), AY773192 (BR3461), AY773193 (BR3462), AY773194 (BR3464), AY773195 (BR3466), AY773196 (BR3467), AY773197 (BR3469), AY773198 (BR3470), and AY533859 (MAP3-5). Accession numbers for nodA sequences are as follows: AY533869 (MAP3-5), AY533870 (Br3454), AY533871 (Br3461), AY533872 (Br3407), and AY533873 (Br3469). Accession numbers for nifH sequences are as follows: AY533864 (MAP3-5), AY533865 (Br3454), AY533866 (Br3461), AY533867 (Br3407), and AY533868 (Br3469). Accession numbers for intergenic sequences between nodD and nodB are as follows: AY533874 (MAP3-5), AY533875 (Br3454), AY533876 (Br3461), AY533877 (Br3407), and AY533878 (Br3469).
RESULTS
Mimosa-nodulating strains from South America belong to the genus Burkholderia.
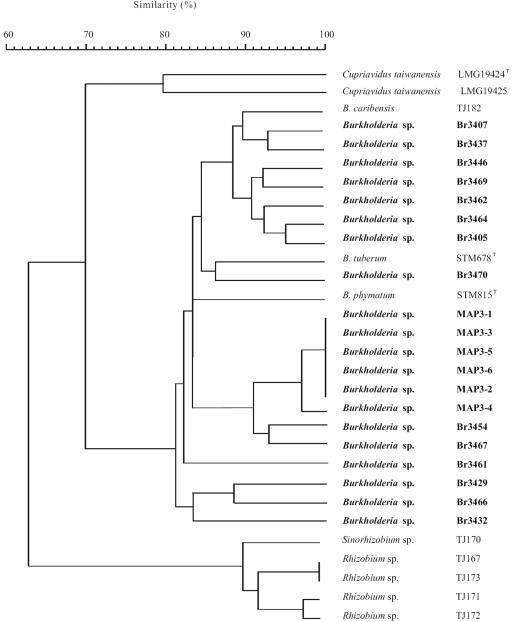
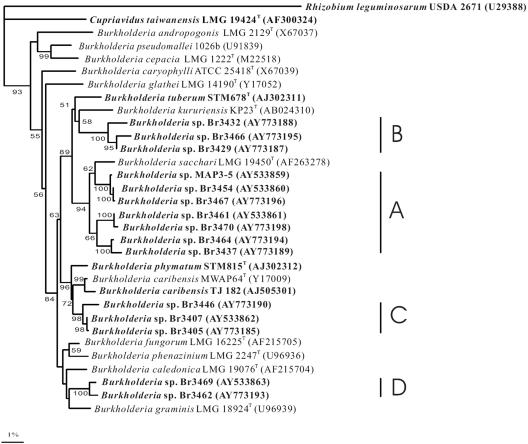
Twenty South American Mimosa-nodulating strains, together with eight reference Mimosa strains and two other β-rhizobial strains, were examined by ARDRA analysis (Table 1). In the numerical analysis of the combined ARDRA patterns (Fig. 1), the South American isolates fell into 16 patterns and formed a single cluster together with B. caribensis, B. phymatum, and B. tuberum (with similarities of >83%). The other reference strains, belonging to Rhizobium, Sinorhizobium, and C. taiwanensis, occupied separate and distinct positions. The 16S rRNA gene sequences of 15 of the 20 strains were determined, and all strains were shown to belong to the genus Burkholderia (Fig. 2). Four clusters could be discerned, as follows. Cluster A consisted of seven strains, including MAP3-5, isolated from M. pigra nodules in Venezuela, as well as the Brazilian strains Br3437, Br3454 (isolated from Mimosa scabrella), Br3461 (isolated from M. bimucronata), Br3464 (isolated from Mimosa flocculosa), Br3467 (isolated from M. pigra), and Br3470 (isolated from M. bimucronata). Cluster B consisted of Br3429, Br3432 (isolated from M. acutistipula), and Br3466 (isolated from Mimosa tenuiflora), cluster C consisted of Br3405, Br3407 (isolated from Mimosa caesalpiniaefolia), and Br3446 (isolated from Mimosa laticifera), and cluster D consisted of Br3462 (isolated from M. flocculosa) and Br3469 (isolated from Mimosa camporum). Published sequences show that B. tuberum STM678 (isolated from Aspalathus carnosa) is related to clusters A and B and that B. caribensis TJ182 and B. phymatum STM815 (isolated from Machaerium lunatum) are related to cluster C.
FIG. 1.
Dendrogram derived from the unweighted-pair group method average linkages of Dice similarity coefficients (SD) between the combined ARDRA patterns of all strains studied. The coefficient is expressed as the percentage of similarity for convenience.
FIG. 2.
Neighbor-joining tree showing phylogenetic positions of South American Mimosa-nodulating strains and Burkholderia species within the β-proteobacteria based on 16S rRNA gene sequence comparisons. Rhizobium leguminosarum USDA 2671 was used as an outgroup. Legume symbionts are shown in bold. Bootstrap values are indicated on branches. Only bootstrap values of >50% are shown. Scale bar, 1% sequence divergence (one substitution per 100 nucleotides). Representative sequences in the dendrogram were obtained from GenBank (accession numbers are given in parentheses).
Nodulation and nitrogen fixation genes.
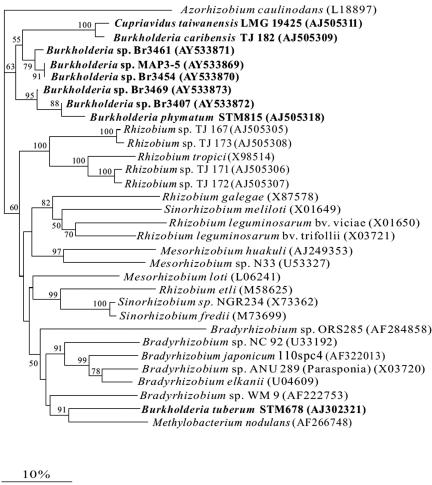
The partial nodA gene sequences (350 bp) of five representative Mimosa-nodulating strains were determined and compared with those of other α- and β-rhizobia (Fig. 3). The NodA protein (116 amino acids) sequence similarities between the five South American Mimosa-nodulating strains showed that they were 89.4 to 100% similar. The nodA sequence similarities to genes encoding other β-rhizobial NodA proteins ranged from 86.5 to 98.0% for B. phymatum, 84.6 to 87.5% for B. caribensis TJ 182, 83.6 to 86.5% for C. taiwanensis, and 60 to 75.0% for other rhizobia. As shown previously (9), the NodA sequence of B. tuberum was quite distant from that of the other β-rhizobia (Fig. 3).
FIG. 3.
NodA phylogenetic tree of α- and β-rhizobia. β-Proteobacterial strains are shown in bold. The tree was reconstructed by using a neighbor-joining approach based on a 116-amino-acid sequence alignment. Values along branches indicate bootstrap percentages of >50%, based on 1,000 replicates. nodA sequences for representative sequences are available from GenBank (accession numbers are given in parentheses).
The common nod genes of the five representative Mimosa-nodulating strains were successfully amplified by PCR and sequenced with conserved primers which were located at the end of nodD and the beginning of nodB. The fragments ranged from 380 to 395 bp, and these contained the intergenic region of the nodD and nodB genes, within which there was a NodD-dependent regulatory consensus sequence (nod box) in all five strains. This showed that these five strains were similar to C. taiwanensis and the consensus sequence from other rhizobia (31) (Fig. 4).
FIG. 4.
Comparison of nod box sequences in promoter regions of nodulation genes from five South American Burkholderia strains and C. taiwanensis strain LMG19424. Footnotes: a, Cupriavidus taivanensis LMG19424; b, the consensus nod box sequence (30) is located in the promoter regions of nodulation genes of various rhizobia, including Rhizobium, Sinorhizobium, Mesorhizobium, Bradyrhizobium, and Azorhizobium spp.
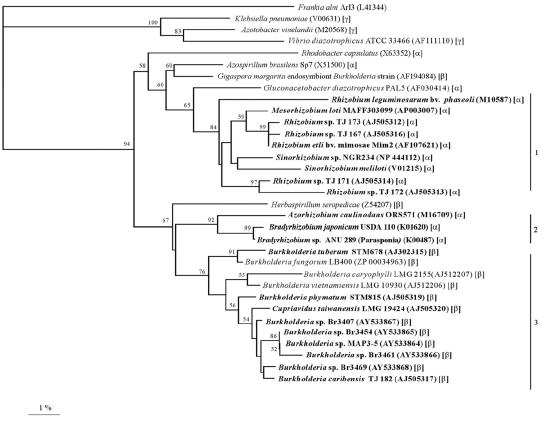
Finally, for each of the five strains, a 658-bp fragment of the nifH gene, which encodes dinitrogenase reductase, a key enzyme in nitrogen fixation, was amplified and sequenced. The NifH sequences of the five strains (Fig. 5) were closest to each other (84.1 to 98.9% identity) and to that of B. caribensis TJ 182 (80.5 to 95.8% identity), with the next closest sequence being that of C. taiwanensis (76.1 to 80.6% identity). Although all the known β-rhizobia clustered together, the NifH sequence of B. tuberum again appeared to be slightly more distant and was more closely aligned with that of Burkholderia fungorum (Fig. 5).
FIG. 5.
NifH phylogenetic tree. The tree was reconstructed by neighbor joining based on a 218-amino-acid alignment. Values along branches indicate bootstrap percentages of >50%, based on 1,000 replicates. The tree was rooted using sequences from Frankia alni, Vibrio diazotrophicus, Klebsiella pneumoniae, and Azotobacter vinelandii. Rhizobia are shown in bold, and the α-, β-, or γ-proteobacterial classification is indicated in parentheses. Clusters 1 and 2 contain α-rhizobia only, while cluster 3 includes both symbiotic and nonsymbiotic diazotrophic β-proteobacteria. nifH sequences for published bacteria are available from GenBank (accession numbers are given in parentheses).
Nodulation of Mimosa spp.
The four Mimosa species tested were readily nodulated by the five representative strains (Table 2). The nodules produced in each case were pink, which indicated the presence of leghemoglobin (Lb) and thus suggested that the bacteria were effectively fixing nitrogen. For M. pudica, this was confirmed using the acetylene reduction assay (Table 2). Pink nodules were also formed on M. diplotricha and M. acutistipula by the five strains, but the number of nodules was generally smaller than that on M. pudica, except for plants inoculated with Br3469 (Table 2). With M. pigra, there were larger numbers of nodules (two- to threefold more) with MAP3-5 than with the other strains (Table 2).
TABLE 2.
Nodulation of Mimosa spp. by South American Burkholderia strains
| Strain | No. of nodules per planta
|
|||
|---|---|---|---|---|
| M. pudicab | M. diplotricha | M. pigra | M. acutistipula | |
| Br3407 | 12.0 ± 1.0 (388) | 10.3 ± 0.6 | 10.0 ± 1.4 | 4.3 ± 0.9 |
| Br3454 | 19.0 ± 4.6 (2,906) | 12.0 ± 5.6 | 9.5 ± 0.7 | 12 ± 3.1 |
| Br3461 | 22.7 ± 6.0 (1,197) | 8.3 ± 2.3 | 17.2 ± 5.6 | 9.6 ± 3.1 |
| Br3469 | 13.7 ± 2.5 (279) | 19.3 ± 4.0 | 9.0 ± 1.7 | 3.6 ± 0.6 |
| MAP3-5 | 23.7 ± 3.1 (511) | 11.7 ± 2.7 | 33.5 ± 6.6 | 7.8 ± 2.8 |
Results are numbers of nodules per plant 21 days after inoculation (mean ± standard deviations; n = 4). All nodules were pink, indicating the presence of Lb. Control plants (uninoculated) had no nodules.
Numbers in parentheses show the nitrogenase (acetylene reduction) activity in nmol C2H4 plant−1 h−1 for 1 to 4 plants.
Structure of nodules on Mimosa spp.
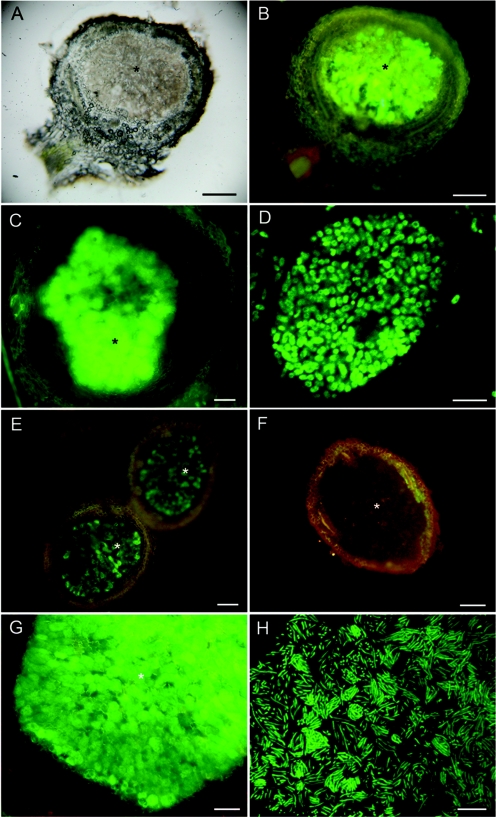
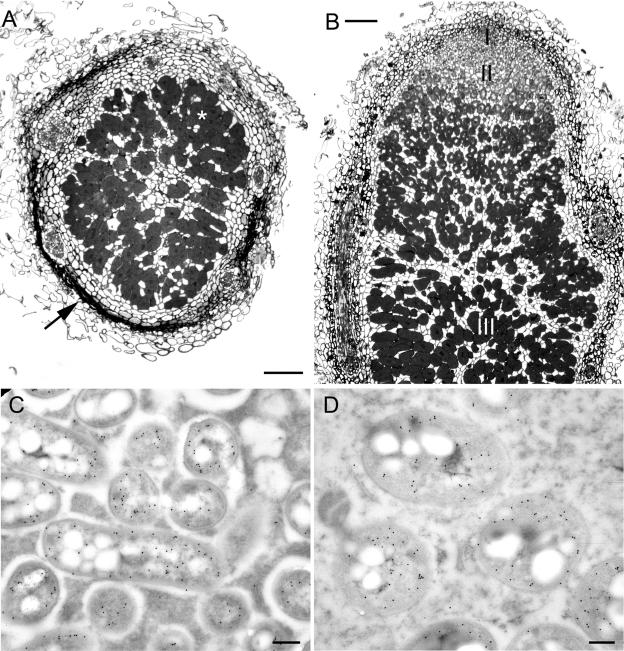
Sections of nodules formed by wild-type Br3407, Br3454, Br3461, Br3469, and MAP3-5 on M. pudica and M. diplotricha showed them to have structures typical of effective N-fixing Mimosa nodules (data not shown, but see the work of Chen et al. [9]). Further microscopy was performed using two gfp transconjugant strains, Br3461-gfp (Fig. 6A to D), and MAP3-5-gfp (Fig. 6E to H), and both strains formed effective, fluorescent nodules on M. pudica (Fig. 6A, B, E, and F) as well as on their original hosts, M. bimucronata (Fig. 6C) and M. pigra (Fig. 6G), respectively, with fluorescent bacteroids being clearly discerned in both plants by CLSM (Fig. 6D and H). At the time of harvest (30 days after inoculation), M. bimucronata plus Br3461-gfp and M. pigra plus MAP3-5-gfp had nitrogenase (acetylene reduction) activities of (mean ± standard error) 14.2 ± 4.6 and 46.3 ± 8.2 nmol C2H4 plant−1 h−1, respectively. More details of the structure of the nodules formed by Br3461 and MAP3-5 on their original hosts, M. bimucronata and M. pigra, respectively, are shown in Fig. 7A and B. They are essentially similar to the nodules formed on M. pudica (9) in that they are indeterminate, with a meristem at the tip, an invasion zone containing cells being invaded by infection threads, and then an infected, N-fixing zone (40). However, as is usual for nodules on woody species, their nodules (particularly those on M. bimucronata) differ from those on M. pudica in having a very pronounced and heavily lignified cortex (Fig. 7A). The expression of nitrogenase in symbiotic bacteroids was confirmed for Br3461 and MAP3-5 by use of an antibody raised against the Fe (NifH) protein (Fig. 7C and D). Bacteria reisolated from the GFP-expressing nodules had identical ARDRA patterns to those of the original inoculated strains.
FIG. 6.
Light microscopy of fresh nodules of Mimosa spp. inoculated with gfp-tagged or wild-type (WT) Burkholderia strains Br3461 (A to D) and MAP3-5 (E to H). A Mimosa pudica nodule infected with Br3461-gfp was viewed with transmitted light (A) or epifluorescence (B). Panels C and D show a Mimosa bimucronata nodule infected with Br3461-gfp viewed under epifluorescence (C) and an infected cell from the same nodule containing fluorescent bacteroids viewed using CLSM (D). (E and F) Mimosa pudica nodules infected with MAP3-5-gfp (E) or MAP3-5 WT (F) viewed under epifluorescence. Note that the nodule containing WT MAP3-5 does not fluoresce (F). (G) Mimosa pigra nodule infected with MAP3-5-gfp viewed with epifluorescence. Note that the infected zone fluoresces intensely green (*), but also that the meristematic region has red fluorescence, possibly due to the presence of polyphenolic compounds. (H) CLSM of fluorescent bacteroids within an infected cell from an M. pigra nodule infected with MAP3-5-gfp. Bars, 100 μm (A, B, C, E, and F), 5 μm (D), 50 μm (G), and 10 μm (H).
FIG. 7.
Transverse (A) and longitudinal (B) sections of fixed and embedded Mimosa bimucronata (A) and M. pigra (B) nodules formed after inoculation with Burkholderia strains Br3461 and MAP3-5, respectively. Both nodules are indeterminate (see panel B), with a meristem (zone I), an invasion zone (zone II), and a N2-fixing zone (zone III and * in panel A). Note the heavily lignified cortex in panel A (arrow). (C and D) Transmission electron micrographs of bacteroids from nodules of M. pudica plus Br3461 (C) and M. pigra plus MAP3-5 (D). Both panels C and D were immunogold labeled with an antibody against the NifH protein of nitrogenase. Bars, 100 μm (A and B) and 300 nm (C and D).
DISCUSSION
Phylogeny.
The ARDRA patterns and 16S rRNA gene sequences of the 20 South American Mimosa-nodulating strains showed conclusively that they belong to the genus Burkholderia and also that they are closely related to other nodulating Burkholderia species, such as B. caribensis TJ 182, B. phymatum, and B. tuberum. The ARDRA patterns of the six Venezuelan M. pigra-nodulating strains (MAP3-1 to MAP3-6) clustered together along with those of Br3454 (isolated from M. scabrella) and a Brazilian strain isolated from M. pigra, Br3467. Moreover, the 16S rRNA gene sequence of the “representative” MAP strain, MAP3-5, was very close to those of Br3454 and Br3467 (group A), and therefore it is possible that the MAP strains, Br3454, and Br3467 are all in the same species. However, this awaits confirmation via a polyphasic analysis which is currently being undertaken by P. Vandamme et al. (unpublished). The other four strains in group A, i.e., Br3461, Br3470 (isolated from M. bimucronata), Br3464 (isolated from M. flocculosa), and Br3437 (isolated from M. scabrella), were slightly more distant from the three aforementioned strains and might even represent different species. The two M. bimucronata-nodulating strains in group A, Br3461 and Br3470, were particularly close to each other. The three Mimosa-nodulating strains in group B, i.e., Br3429, Br3432 (isolated from M. acutistipula), and Br3466 (isolated from M. tenuiflora), were close to each other, and all were quite distant from the closest nodule-forming isolate described previously, the Aspalathus carnosa strain B. tuberum STM678 from South Africa (39), and were also related to the free-living diazotroph Burkholderia kururiensis (14). Group C consisted of three Brazilian Mimosa-nodulating strains, namely, Br3446 (isolated from M. laticifera), Br3405, and Br3407 (both isolated from M. caesalpiniaefolia), with the last two being particularly close to each other. Finally, group D consisted of only two nodule-forming strains, Br3462 (isolated from M. flocculosa) and Br3469 (isolated from M. camporum), which were close to each other but separate from non-nodulating Burkholderia strains, such as Burkholderia graminis. Overall, the 15 South American Mimosa-nodulating strains did not cluster closely with non-nodulating Burkholderia strains, and also they were not particularly close to previously discovered β-rhizobia, such as B. caribensis TJ182, B. phymatum, and B. tuberum. Instead, they formed clusters of their own, with strains isolated from the same host species usually clustering very closely, and these clusters may represent new species.
The phylogenetic relationships of NodA sequences suggest that β-rhizobia have acquired their symbiosis genes from the “conventional” α-rhizobia and that there has been more than one independent transfer between the α- and β-proteobacteria because the β sequences fall into two separate clades (10), with C. taiwanensis, B. caribensis TJ182, and B. phymatum in one clade and B. tuberum in the other. The nodulation genes (nodA and nod box) of all the South American strains were very similar to each other and to those of C. taiwanensis, B. caribensis TJ182, and B. phymatum, but they were quite distant from those of B. tuberum, suggesting that the strains had most likely acquired these genes from the same transfer event as the C. taiwanensis-B. caribensis TJ182-B. phymatum clade. However, the considerable divergence of NodA sequences within these β-rhizobia indicates that this transfer event was a long time ago, implying that β-rhizobia have been established for many millions of years. The NodA phylogeny provides strong evidence of a transfer of nodulation genes between the α- and β-proteobacteria, but the widely held assumption that the direction of transfer was from the α to the β group has not yet been rigorously proven. Further studies are under way to establish whether long-range horizontal gene transfer has occurred repeatedly and whether the symbiosis genes in each β-rhizobium can be traced to a single donor source in the α-rhizobia.
Within the α-rhizobia, the nif genes seem to have a different evolutionary history from that of the nod genes, as their phylogenies are not congruent (19). Indeed, there is some evidence that the nif genes of Bradyrhizobium japonicum may have originated in the β-Proteobacteria (46). It is therefore interesting to consider that the nif genes of the β-rhizobia might not, in fact, have the same α-proteobacterial origin as the nod genes. This should be evident from phylogenetic analysis, and the indication for strains examined so far is that NifH is indeed the “local” β-proteobacterial form (10). In the present study, the NifH sequences of all five strains examined were very close to each other and to those of C. taiwanensis, B. caribensis, and B. phymatum but were not so close to that of B. tuberum. Therefore, as with the NodA sequences, this suggests that the former group of bacteria obtained their nif genes from a different source than that of B. tuberum.
Origin of strains.
All 14 Brazilian strains were originally isolated over 20 years ago by de Faria and coworkers (unpublished) and have long been known to have the ability to effectively nodulate their original hosts (all Brazilian natives) as well as other Mimosa spp. Indeed, they have all been extensively tested for high N fixation in an evaluation of tropical woody legumes (including Mimosa spp.) for use in the reclamation of degraded and deforested areas in Brazil (15). With specific regard to the four representative Brazilian strains, Br3454 is of particular interest, as details of its ability to infect and nodulate M. scabrella were reported by de Faria et al. (12). This study showed that “Rhizobium” strain Br3454 had an unusual ability to infect the roots of M. scabrella by direct epidermal infection, and so far M. scabrella is the only legume reported to be infected in this manner. The hosts of the other three strains, M. caesalpiniaefolia (Br3407), M. bimucronata (Br3461), and M. camporum (Br3469), are all common in forested areas of Brazil (2, 13, 32) and have long been known to be nodulated (1, 5, 7, 20, 29, 34). Mimosa caesalpiniaefolia, a fast-growing tree, has been well studied owing to its ability to grow and nodulate in acidic soils (35) and for its potential for use in the reclamation of degraded areas (15, 17), while the smaller, invasive shrubs M. bimucronata and M. camporum are being evaluated for their potential to assist in the recovery of drastically disturbed lands (29) and to withstand prolonged shading (20), respectively.
Mimosa pigra (syn. M. pellita) is likely to have originated in South America (2), where it has long been known to undergo nodulation, particularly in wetland regions (5, 24). However, in contrast to the hosts of the four Brazilian strains, M. pigra is highly invasive and is now becoming a major pest in some countries, such as in wetland regions in tropical Australia (6), Thailand (45), and southern Taiwan (44). Several Burkholderia strains have recently been isolated from M. pigra nodules in Taiwan (9a) and Panama (3), but strain MAP3-5 and the other MAP strains were isolated from nodules of M. pigra growing alongside the Mapire river, a tributary of the Orinoco in Venezuela, by Barrios (unpublished). Strain MAP3-5 was shown to effectively nodulate M. pigra in a preliminary study by James et al. (22). This last study also suggested that M. pigra, at least when grown under flooded conditions, was infected by MAP3-5 via enlarged epidermal/aerenchyma cells and not via root hairs. This is a particularly interesting observation given the very close relationship between MAP3-5 and Burkholderia strain Br3454, which infects M. scabrella via direct epidermal penetration (12), and hence further infection/microscopy studies are being undertaken with the gfp transconjugant strain of MAP3-5 to determine more exactly how it infects different Mimosa spp.
Geographical distribution of Mimosa-nodulating β-rhizobia.
Although a few species are native to Asia (e.g., Mimosa himalayana) and Africa (particularly Madagascar), most of the 480 or so species of Mimosa are native to Central and South America (2), with the Cerrado region of Central Brazil being the major center of diversification for the genus (2, 32). It has long been known that Mimosa plants may be nodulated by a variety of rhizobia (1, 37), but few have been typified, and prior to 2000, all had been ascribed to known α-rhizobial genera (3, 25, 27, 42, 47). Since the initial report of Moulin et al. (26) on Burkholderia strains STM815 and STM678, many other strains of “β-rhizobia” have been isolated, but the majority of them have come from Mimosa spp. in Asia (8, 9a, 10, 41), and relatively few have been found in the Americas (3). Even though they came from geographically very distant parts of South America, i.e., the Brazilian Atlantic Forest (most of the “Br” strains) and the flooded forests of the Orinoco basin (MAP3-1 to MAP3-6), and from 10 different Mimosa spp., all 20 strains examined in the present study were β-rhizobia. Obviously, considerably more South American Mimosa nodule isolates need to be examined before any firm conclusions can be made, but our study not only gives confirmation that they have a particular preference for Mimosa spp. (10 different species) but also suggests that they may be the “dominant” rhizobial type involved with the genus Mimosa across large parts of South America. Indeed, this is supported by the recent report of 27 Burkholderia strains isolated from mimosoid legumes in Central America (3). Furthermore, it is interesting that none of the 20 strains were in the genus Cupriavidus, which appears strange considering that C. taiwanensis is so dominant in Taiwan (10) and possibly India (41). Could it be that C. taiwanensis is an Asian bacterium that has acquired its symbiosis genes from Burkholderia strains resident within Mimosa nodules that were brought from tropical America and the Caribbean by European colonists from 1645 onwards (43)? Clearly, further studies are urgently needed to compare South American β-rhizobia with those from other continents.
Acknowledgments
We thank Lionel Moulin and Marcelo F. Simon for discussions, Paul Ludden for the NifH antibody, and Margaret Gruber for technical assistance.
W.-M. Chen was supported by grants from the National Science Council, Taipei, Taiwan, Republic of China (NSC 93-2320-B-022-001 and −003), and A. Prescott, G. N. Elliott, J. I. Sprent, J. P. W. Young, and E. K. James were supported by the Natural Environment Research Council (NERC).
REFERENCES
- 1.Allen, O. N., and E. K. Allen. 1981. The Leguminosae: a source book of characteristics, uses, and nodulation. University of Wisconsin Press, Madison, Wis.
- 2.Barneby, R. C. 1991. Sensitivae censitae: a description of the genus Mimosa Linnaeus (Mimosaceae) in the New World. Mem. N. Y. Botanic. Garden 65:1-835. [Google Scholar]
- 3.Barrett, C. F., and M. A. Parker. 2005. Prevalence of Burkholderia sp. nodule symbionts on four mimosoid legumes from Barro Colorado Island, Panama. Syst. Appl. Microbiol. 28:57-65. [DOI] [PubMed] [Google Scholar]
- 4.Barrios, E., and R. Herrera. 1994. Nitrogen cycling in a Venezuelan tropical seasonally flooded forest: soil nitrogen mineralization and nitrification. J. Trop. Ecol. 10:399-416. [Google Scholar]
- 5.Barrios, S., and V. Gonzalez. 1971. Rhizobial symbioses on Venezuelan savannas. Plant Soil 34:707-719. [Google Scholar]
- 6.Braithwaite, R. W., W. M. Lonsdale, and J. A. Estbergs. 1989. Alien vegetation and native biota in tropical Australia: the spread and impact of Mimosa pigra. Biol. Conserv. 48:189-210. [Google Scholar]
- 7.Campêlo, A. B., and C. R. Campêlo. 1970. Eficiência da inoculacao cruzada entre espêcies da subfamília Mimosoideae. Pesq. Agropec. Bras. 5:333-337. [Google Scholar]
- 8.Chen, W. M., S. Laevens, T. M. Lee, T. Coenye, P. de Vos, M. Mergeay, and P. Vandamme. 2001. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. Evol. Microbiol. 51:1729-1735. [DOI] [PubMed] [Google Scholar]
- 9.Chen, W. M., E. K. James, A. R. Prescott, M. Kierans, and J. I. Sprent. 2003. Nodulation of Mimosa spp. by the β-proteobacterium Ralstonia taiwanensis. Mol. Plant-Microbe Interact. 16:1051-1061. [DOI] [PubMed] [Google Scholar]
- 9a.Chen, W. M., E. K. James, J. H. Chou, S. Y. Sheu, S. Z. Yang, and J. I. Sprent. August2005, posting date. β-Rhizobia from Mimosa pigra, a newly discovered invasive plant in Taiwan. New Phytol. [Online.] doi: 10.1111/j.1469-8137.2005.01533.x. [DOI] [PubMed]
- 10.Chen, W. M., L. Moulin, C. Bontemps, P. Vandamme, G. Béna, and C. Boivin-Masson. 2003. Legume symbiotic nitrogen fixation by β-Proteobacteria is widespread in nature. J. Bacteriol. 185:7266-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Faria, S. M., H. C. de Lima, A. A. Franco, E. S. F. Mucci, and J. I. Sprent. 1987. Nodulation of legume trees from South East Brazil. Plant Soil 99:347-356. [Google Scholar]
- 12.de Faria, S. M., G. T. Hay, and J. I. Sprent. 1988. Entry of rhizobia into roots of Mimosa scabrella Bentham occurs between epidermal cells. J. Gen. Microbiol. 134:2291-2296. [Google Scholar]
- 13.de Faria, S. M., and H. C. de Lima. 1998. Additional studies of the nodulation status of legume species in Brazil. Plant Soil 200:185-192. [Google Scholar]
- 14.Estrada de Los Santos, P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco, A. A., and S. M. de Faria. 1997. The contribution of N2-fixing tree legumes to land reclamation and sustainability in the tropics. Soil Biol. Biochem. 29:897-903. [Google Scholar]
- 16.Gibson, A. H. 1963. Physical environment and symbiotic nitrogen fixation. I. The effect of root temperature on recently nodulated Trifolium subterraneum L. plants. Aust. J. Biol. Sci. 16:28-42. [Google Scholar]
- 17.Goi, S. R., J. I. Sprent, and J. Jacob-Neto. 1997. Effect of different sources of N2 on the structure of Mimosa caesalpiniaefolia root nodules. Soil Biol. Biochem. 29:983-987. [Google Scholar]
- 18.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 19.Haukka, K., K. Lindström, and J. P. W. Young. 1998. Three phylogenetic groups of nodA and nifH genes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America. Appl. Environ. Microbiol. 64:419-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izaguirre-Mayoral, M. L., A. I. Vivas, and T. Oropeza. 1995. New insights into the symbiotic performance of native tropical legumes. 1. Analysis of the response of thirty-seven native legume species to artificial shade in a neotropical savanna. Symbiosis 19:111-129. [Google Scholar]
- 21.James, E. K., and R. M. M. Crawford. 1998. Effect of oxygen availability on nitrogen fixation by two Lotus species under flooded conditions. J. Exp. Bot. 49:599-610. [Google Scholar]
- 22.James, E. K., E. Barrios, and R. M. M. Crawford. 1995. Effect of flooding on N2-fixation by the Orinoco flooded forest species Mimosa pellita, p. 695. In I. A. Tikhonovich, N. A. Provorov, V. I. Romanov, and W. E. Newton (ed.), Nitrogen fixation: fundamentals and applications. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 23.James, E. K., P. P. M. Iannetta, P. J. Nixon, A. J. Whiston, L. Peat, R. M. M. Crawford, J. I. Sprent, and N. J. Brewin. 1996. Photosystem II and oxygen regulation in Sesbania rostrata stem nodules. Plant Cell Environ. 19:895-910. [Google Scholar]
- 24.James, E. K., M. de Fatima Loureiro, A. Pott, V. J. Pott, C. M. Martins, A. A. Franco, and J. I. Sprent. 2001. Flooding-tolerant legume symbioses from the Brazilian Pantanal. New Phytol. 150:723-738. [Google Scholar]
- 25.Moreira, F. M. S., M. Gillis, B. Pot, K. Kersters, and A. A. Franco. 1993. Characterization of rhizobia isolated from different divergence groups of tropical Leguminosae by comparative polyacrylamide gel electrophoresis of their total proteins. Syst. Appl. Microbiol. 16:135-146. [Google Scholar]
- 26.Moulin, L., A. Munive, B. Dreyfus, and C. Boivin-Masson. 2001. Nodulation of legumes by members of the β-subclass of proteobacteria. Nature 411:948-950. [DOI] [PubMed] [Google Scholar]
- 27.Oyaizu, H., S. Matsumoto, K. Minamisawa, and T. Gamou. 1993. Distribution of rhizobia in leguminous plants surveyed by phylogenetic identification. J. Gen. Appl. Microbiol. 39:339-354. [Google Scholar]
- 28.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:358. [DOI] [PubMed] [Google Scholar]
- 29.Patreze, C. M., and L. Cordeiro. 2004. Nitrogen-fixing and vesicular-arbuscular mycorrhizal symbioses in some tropical legume trees of tribe Mimosae. Forest Ecol. Manage. 196:275-285. [Google Scholar]
- 30.Sawada, H., L. D. Kuykendall, and J. M. Young. 2003. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J. Gen. Appl. Microbiol. 49:155-179. [DOI] [PubMed] [Google Scholar]
- 31.Schlaman, H.-R. M., D.-A. Phillips, and E. Kondorosi. 1998. Genetic organization and transcriptional regulation of rhizobial nodulation genes, p. 361-386. In H. Spaink, A. Kondorosi, and P. J. J. Hooykaas (ed.), The Rhizobiaceae. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 32.Simon, M. F., and C. Proença. 2000. Phytogeographic patterns of Mimosa (Mimosoideae, Leguminosae) in the Cerrado biome of Brazil: an indicator genus of high altitude centers of endemism? Biol. Conserv. 96:279-296. [Google Scholar]
- 33.Somasegaran, P., and H. J. Hoben. 1994. Handbook for rhizobia: methods in legume-rhizobium technology. Springer-Verlag, New York, N.Y.
- 34.Sprent, J. I. 2001. Nodulation in legumes. Royal Botanic Gardens, Kew, London.
- 35.Stamford, N. P., A. D. Ortega, F. Temprano, and D. R. Santos. 1997. Effects of phosphorus fertilization and inoculation of Bradyrhizobium and mycorrhizal fungi on growth of Mimosa caesalpiniaefolia in an acid soil. Soil Biol. Biochem. 29:959-964. [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trinick, M. J. 1980. Relationships amongst the fast-growing rhizobia of Lablab purpureus, Leucaena leucocephala, Mimosa spp., Acacia farnesiana and Sesbania grandiflora and their affinities with other rhizobial groups. J. Appl. Bacteriol. 49:39-53. [Google Scholar]
- 38.Vandamme, P., and T. Coenye. 2004. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int. J. Syst. Evol. Microbiol. 54:2285-2289. [DOI] [PubMed] [Google Scholar]
- 39.Vandamme, P., J. Goris, W. M. Chen, P. de Vos, and A. Willems. 2002. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov. nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25:507-512. [DOI] [PubMed] [Google Scholar]
- 40.Vasse, J., F. de Billy, S. Camut, and G. Truchet. 1990. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma, S. C., S. P. Chowdhury, and A. K. Tripathi. 2004. Phylogeny based on 16S rDNA and nifH sequences of Ralstonia taiwanensis strains isolated from nitrogen-fixing nodules of Mimosa pudica, in India. Can. J. Microbiol. 50:313-322. [DOI] [PubMed] [Google Scholar]
- 42.Wang, E. T., M. A. Rogel, A. García-de los Santos, J. Martínez-Romero, M. A. Cevallos, and E. Martínez-Romero. 1999. Rhizobium etli bv. Mimosae, a novel biovar isolated from Mimosa affinis. Int. J. Syst. Bacteriol. 49:1479-1491. [DOI] [PubMed] [Google Scholar]
- 43.Wu, S.-H., S.-M. Chaw, and M. Rejmánek. 2003. Naturalized Fabaceae (Leguminosae) species in Taiwan: the first approximation. Bot. Bull. Acad. Sin. 44:59-66. [Google Scholar]
- 44.Yang, S.-Z., and C.-I. Peng. 2001. An invading plant in Taiwan—Mimosa pigra L. Q. J. Forest Res. Taiwan 23:1-6. [Google Scholar]
- 45.Yoneyama, T., T. Murakami, N. Boonkerd, P. Wadisirisuk, S. Siripin, and K. Kouno. 1990. Natural 15N abundance in shrub and tree legumes, Casuarina, and non N2 fixing plants in Thailand. Plant Soil 128:287-292. [Google Scholar]
- 46.Young, J. P. W. 2005. The phylogeny and evolution of nitrogenases. p. 221-241. In R. Palacios and W. E. Newton (ed.), Genomes and genomics of nitrogen-fixing organisms. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 47.Zurdo-Piñeiro, J. L., E. Velázquez, M. F. Lorite, G. Brelles-Mariño, E. C. Schröder, E. J. Bedmar, P. F. Mateos, and E. Martinez-Molina. 2004. Identification of fast-growing rhizobia nodulating tropical legumes from Puerto Rico as Rhizobium gallicum and Rhizobium tropici. Syst. Appl. Microbiol. 27:469-477. [DOI] [PubMed] [Google Scholar]