Abstract
On-probe sample pretreatment using trifluoroacetic acid as an additional reagent enabled the direct detection of phospholipids in whole bacteria by means of matrix-assisted laser desorption ionization mass spectrometry for not only gram-negative organisms but also gram-positive ones with a thicker peptidoglycan layer.
Matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) has become a powerful method to characterize lipid components in bacterial cells (5). However, ordinary MALDI-MS requires a tedious sample pretreatment, such as solvent extraction of the lipids from the sample matrix or preparation of the sample suspension. Recently, a new analytical protocol for MALDI-MS called “on-probe sample pretreatment” was developed to achieve the direct detection of proteins in bacterial cells (1, 3, 7). In this technique, growing bacterial colonies are transferred directly from agar surfaces to the MALDI sample plate and then overlaid with a microvolume of matrix solution prior to irradiation with an N2 laser. We applied a similar sampling protocol for the direct MALDI-MS measurement of phospholipids in bacterial cells (6) and successfully observed a series of ion peaks derived from phospholipids in gram-negative bacteria in the mass spectra. However, spectra of gram-positive bacteria did not show any lipid-related peaks, presumably due to their thicker peptidoglycan layer. In this work, direct detection of phospholipids in gram-positive bacteria was tried by means of MALDI-MS combined with the on-probe sample pretreatment using trifluoroacetic acid (TFA) as an additional reagent.
Bacillus subtilis subsp. subtilis (NBRC 3007), a gram-positive bacterial sample purchased from National Institute of Technology and Evaluation, Japan, was used for stock cultures. Lyophilized cells of the bacterial sample were rehydrated with distilled water containing 1% NaCl, 0.5% yeast extract, and 1% polypeptone and then used to streak culture plates containing solid growth medium. The bacterial sample was cultured on LB agar and incubated at 37°C for 24 h.
The procedure for on-probe sample pretreatment is basically similar to that described in our previous paper (6). Individual bacterial colonies weighing about 100 μg were carefully removed from the agar surface with a tungsten wire loop and immediately spotted onto the MALDI plate. According to our previous paper (6), 2,5-dihydroxybenzoic acid (Aldrich) and sodium iodide (NaI; Wako) were used as a matrix and cationization reagent, respectively. A mixed solution of the matrix and cationization reagent was prepared by dissolving 2,5-dihydroxybenzoic acid and NaI in the solvent (2:1 mixture of chloroform:methanol) in concentrations of 500 mM and 100 mM, respectively. Here, TFA (Wako) was added to the solution in a concentration of 5% (vol/vol) to enhance the ion formation of phospholipids in the bacterial cell wall. Then, a ∼1-μl droplet of the solution was deposited onto each bacterial sample spot after the sample spot was additionally covered with 1 μl of a TFA solution (5%). Finally, each spot was dried at room temperature and subjected to the MALDI-MS measurement using a Voyager DE-RP time-of-flight mass spectrometer (Applied Biosystems) under the same instrumental conditions described in our previous paper (6).
In order to obtain additional data on fatty acid moieties, the bacterial sample was also subjected to thermally assisted hydrolysis and methylation-gas chromatography (THM-GC) in the presence of an organic alkali (2, 4, 8). About 100 μg of bacterial colony was weighed into a small platinum cup, and then 3 μl of a methanol solution (3.8 M) of tetramethylammonium hydroxide (Aldrich) was added to the same sample cup as a hydrolysis and derivatization reagent. The sample cup was then introduced into the heated center of a microfurnace pyrolyzer (Frontier Lab PY-2020D) maintained at 400°C, which was directly attached to a gas chromatograph (Agilent 6890) equipped with a flame ionization detector.
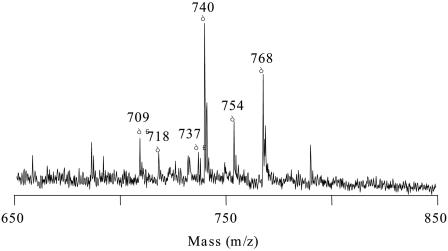
Figure 1 shows the typical MALDI mass spectrum in the phospholipids region of the B. subtilis colony sample obtained by the on-probe sample pretreatment using TFA. In this mass spectrum, a series of ion peaks presumed to be phospholipids were clearly observed, whereas no characteristic peak was observed on the spectra obtained without adding TFA. This drastic change in the spectral pattern might be derived from the promoted decomposition of bacterial cell wall by the addition of TFA, which in turn resulted in enhanced ion formation of phospholipids. Table 1 summarizes the assignments of the ion peaks observed on the mass spectrum together with their relative peak intensities. As shown in Table 1, the observed ions were assigned from the observed m/z values to the disodiated adducts (M + 2Na+ − H) of phosphatidylglycerols (PGs; major components [84%]) and phosphatidylethanolamines (PEs; minor components [16%]), consisting of a total of 30 to 32 carbons in their two acyl groups, which had been known as the main phospholipids in B. subtilis (9).
FIG. 1.
Typical MALDI mass spectrum of B. subtilis colony sample obtained by on-probe sample pretreatment using TFA. Peak assignments are listed in Table 1.
TABLE 1.
Assignments of representative peaks on the spectrum of B. subtilis colony sample
| m/z | Relative intensity (%)a | Assignment |
|---|---|---|
| 709 | 9.7 | [PE (C30:0) + 2Na − H]+ |
| 737 | 6.3 | [PE (C32:0) + 2Na − H]+ |
| Subtotal | 16.0 | |
| 718 | 6.2 | [PG (C30:0) + Na]+ |
| 740 | 37.8 | [PG (C30:0) + 2Na − H]+ |
| 754 | 14.2 | [PG (C31:0) + 2Na − H]+ |
| 768 | 25.7 | [PG (C32:0) + 2Na − H]+ |
| Subtotal | 84.0 | |
| Total | 100.0 |
Relative intensities were obtained on the basis of the peak intensities observed on the mass spectra.
Although the additional use of TFA in the on-probe sample pretreatment allowed the direct detection of phospholipids for gram-positive bacteria, the same preparation applied to E. coli (a gram-negative bacterium) resulted in a much poorer spectral signal-to-noise ratio than that obtained without using TFA (6). This fact suggests that the additional TFA could cause hydrolysis of phospholipids in gram-negative bacteria, which have a thinner peptidoglycan layer.
Finally, the observed peaks in the MALDI mass spectra of B. subtilis shown in Fig. 1 were then interpreted in terms of their constituting fatty acid moieties by taking into consideration the fatty acid compositions obtained by the THM-GC measurement for the same B. subtilis colony. Table 2 lists the relative abundances of phospholipids, sorted by the total number of carbons and double bonds or rings in the constituting acyl moieties, obtained by on-probe MALDI-MS and THM-GC along with the probable combinations of fatty acid components based on the THM-GC measurements. As shown in Table 2, the relative abundances obtained by on-probe MALDI-MS were similar to those obtained by THM-GC, although the individual values calculated were to some extent different between the two methods. This dissimilarity could be explained by the fact that diphosphatidylglycerols (DPGs), which had been reported to be abundant in Bacillus species (9), were not observed in the MALDI mass spectrum of B. subtilis, mainly due to the difference in the optimum MALDI conditions for PGs and DPGs. Further studies to detect DPGs in gram-positive bacterial colonies are currently in progress.
TABLE 2.
Relative abundances of phospholipids for B. subtilis estimated by MALDI-MS and THM-GC
| Technique or combination | Relative abundance (%) of phospholipids or probable fatty acid combinationsa
|
||||
|---|---|---|---|---|---|
| C30:0 | C31:0 | C32:0 | Othersb | Total | |
| On-probe MALDIc | 53.7 | 14.2 | 32.0 | 100 | |
| THM-GCd | 32.2 | 23.4 | 32.3 | 12.1 | 100 |
| Combinatione | aC15:0, aC15:0; aC15:0, iC15:0 | aC15:0, C16:0; iC15:0, C16:0 | C16:0, C16:0; aC15:0, iC17:0 | ||
Abundance is shown based on the number of carbons and double bonds or rings in constituting acyl groups.
Examples of others are C28:0, C29:0, C33:0, etc.
Abundances were estimated from the peak intensities observed by MALDI-MS using on-probe sample pretreatment.
Abundances were calculated based on the fatty acid compositions for the same B. subtilis colony obtained by THM-GC.
Combinations were estimated based on the fatty acid distribution for the same B. subtilis colony sample by THM-GC.
Acknowledgments
We are grateful to S. Iijima and K. Nishijima (Graduate School of Engineering, Nagoya University) and to K. Miyake (EcoTopia Science Institute, Nagoya University) for helpful comments during the course of this investigation.
This work was supported in part by Grants-in-Aid for Scientific Research (B) (16350081) and Young Scientists (B) (16750132) of the Japan Society for the Promotion of Science and the 21st Century COE Program “Nature-Guided Materials Processing” of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Claydon, M. A., S. N. Davey, V. Edwards-Jones, and D. B. Gordon. 1996. The rapid identification of intact microorganisms using mass spectrometry. Nat. Biotechnol. 14:1584-1586. [DOI] [PubMed] [Google Scholar]
- 2.Dworzanski, J. P., L. Berwald, W. H. McClennen, and H. L. C. Meuzelaar. 1991. Mechanistic aspects of the pyrolytic methylation and transesterification of bacterial cell wall lipids. J. Anal. Appl. Pyrolysis 21:221-232. [Google Scholar]
- 3.Evason, D. J., M. A. Claydon, and D. B. Gordon. 2000. Effects of ion mode and matrix additives in the identification of bacteria by intact cell mass spectrometry. Rapid Commun. Mass Spectrom. 14:669-672. [DOI] [PubMed] [Google Scholar]
- 4.Hendricker, A. D., C. Abbas-Hawks, F. Basile, K. J. Voorhees, and T. L. Hadfield. 1999. Rapid chemotaxonomy of pathogenic bacteria using in situ thermal hydrolysis and methylation as a sample preparation step coupled with a field-portable membrane-inlet quadrupole ion trap mass spectrometer. Int. J. Mass Spectrom. 190/191:331-342. [Google Scholar]
- 5.Ho, Y., and C. Fenselau. 1998. Applications of 1.06-μm IR laser desorption on a Fourier transform mass spectrometer. Anal. Chem. 70:4890-4895. [DOI] [PubMed] [Google Scholar]
- 6.Ishida, Y., A. J. Madonna, J. C. Rees, M. A. Meetani, and K. J. Voorhees. 2002. Rapid analysis of intact phospholipids from whole bacterial cells by matrix-assisted laser desorption/ionization mass spectrometry combined with on-probe sample pretreatment. Rapid Commun. Mass Spectrom. 16:1877-1882. [DOI] [PubMed] [Google Scholar]
- 7.Madonna, A. J., F. Basile, I. Ferrer, M. A. Meetani, J. C. Rees, and K. J. Voorhees. 2000. On-probe sample pretreatment for the detection of proteins above 15 KDa from whole bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 14:2220-2229. [DOI] [PubMed] [Google Scholar]
- 8.Nakanishi, O., Y. Ishida, S. Hirao, S. Tsuge, H. Ohtani, J. Urabe, T. Sekino, M. Nakanishi, and T. Kimoto. 2003. Highly sensitive detection of lipid components including polyunsaturated fatty acids in individual zooplankters by one-step thermally assisted hydrolysis and methylation-gas chromatography in the presence of trimethylammonium hydroxide. J. Anal. Appl. Pyrolysis 68-69:187-195. [Google Scholar]
- 9.O'Leary, W. M., and S. G. Wilkinson. 1988. Gram-positive bacteria, E. The family Bacillaceae, 1. The genus Bacillus, p. 155-164. In C. Ratledge and S. G. Wilkinson (ed.), Microbial lipids, vol. 1. Academic Press, San Diego, Calif. [Google Scholar]