Abstract
Loop-mediated isothermal amplification (LAMP) assay was effective in detecting Salmonella enterica in naturally contaminated liquid egg samples. Salmonella was detected in 110 samples taken from four egg-breaking plants. The egg samples were pre-enriched in buffered peptone water (BPW) at 37°C for 20 h. The selective enrichment was done in Rappaport-Vassiliadis or tetrathionate broth and plated onto xylose lysine deoxycholate agar and brilliant green agar, modified. In addition, the PCR assay was used to detect Salmonella after pre-enrichment in BPW at 37°C for 20 h. The culture method and PCR assay were compared to the LAMP assay, which was also performed after pre-enrichment in BPW. PCR failed to detect Salmonella in 10% of 110 samples, whereas the culture method and LAMP assay successfully identified Salmonella in all samples. However, the LAMP assay was found to be much more rapid than the culture method and as sensitive in detecting Salmonella from liquid eggs. In all of the egg-breaking plants studied, Salmonella was isolated on most tested days. The positive samples showed that more than 75% of the Salmonella strains had identical genetic patterns when analyzed by pulsed-field gel electrophoresis. This suggests that the same Salmonella strains having survived long periods of time in the plants were contaminating the production line. The LAMP assay is rapid, specific, and sensitive for Salmonella detection in liquid eggs and is able to monitor Salmonella contamination in egg-handling plants more reliably.
Salmonella enterica is a major food-borne pathogen worldwide. Foods such as meat, eggs, and vegetables are reported to be sources of Salmonella infections. Various serotypes of Salmonella are implicated in food-borne infections. In particular, after the 1980s, the serotype Enteritidis has caused infection via contaminated eggs in many countries (2, 12, 21, 23). Hens infected with serotype Enteritidis produce eggs whose contents are contaminated with the microorganism (1, 20). The consumption of these eggs has caused serotype Enteritidis infection (7, 22). Since the conditions of contamination are different among farms (3, 14, 19), the reported frequency of Salmonella contamination in eggs also has a wide range (5, 17). Serotype Enteritidis frequently contaminates unpasteurized liquid eggs. Reports in the early 1990s showed that the rates of Salmonella contamination were 20% in the United States (4, 6) and 10 to 20% in Japan (17).
Loop-mediated isothermal amplification (LAMP) is a novel nucleic acid amplification technique that relies on an autocycling strand displacement DNA synthesis performed by the Bst DNA polymerase large fragment (13, 15, 16, 18). LAMP is different from PCR in that six primers perform the amplification of the target gene, the amplification uses a single temperature step at 60 to 65°C for about 60 min, and the amplification products have many types of structures in large amounts. Thus, LAMP is more specific, rapid, and simple to perform than PCR. Furthermore, gel electrophoresis is not needed because the LAMP method synthesizes a large amount of DNA where the products can be detected by simple turbidity or fluorescence (1). Thus, expensive equipment is not necessary to give a high level of precision, one equivalent to or greater than those of other PCR techniques. Recently, we have shown the efficiency of LAMP assay for detecting Salmonella (unpublished data).
In the present study, liquid eggs naturally contaminated with low numbers of Salmonella were tested by using the LAMP assay. In addition, salmonellae from liquid eggs were characterized to strain and serotype levels.
MATERIALS AND METHODS
Unpasteurized liquid whole-egg samples.
Raw liquid eggs before pasteurization (300 g/sample) were purchased from an egg-breaking plant (plant A) in the spring of 2003 (24 samples) and from four egg-breaking plants (plants A to D; 110 samples) in the autumn of 2003 in Japan. Each 200 g of sample from an unpasteurized liquid egg tank obtained from each of the plants was transferred to a sterilized stomacher bag with a sterilized container. The stomacher bags containing liquid egg samples were sealed and stored at −20°C until tested.
Culture method for the liquid egg sample.
The 24 samples were collected from the egg plant in the spring of 2003 to measure the contamination level of Salmonella. Liquid egg samples were homogenized with a stomacher, and 1-ml aliquots were plated onto five plates of MLCB agar (Oxoid). After incubation at 37°C for 24 h, some of the suspected colonies were tested for antibody agglutination by using a Salmonella antibody kit (Unipath; Oxoid) and confirmed by using the biochemical characteristics on TSI (Eiken Chemical Co., Ltd., Tokyo, Japan) and LIM (Eiken Chemical). For aerobic plate counts, 1 ml of a 101- to 104-fold dilution of each sample in phosphate-buffered saline was poured onto 19 ml of plate count agar (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan), followed by incubation at 36 to 37°C for 24 h.
The sensitivity of Salmonella detection by the culture method was compared to the reaction yield by the lamp and PCR assay. A total of 25 g of all samples of liquid egg was homogenized with 225 ml of buffered peptone water (BPW) by using a stomacher (model 400; A. J. Seward, London, United Kingdom), followed by incubation at 37°C for 20 h. After incubation, 0.5 ml of each culture was added to 10 ml of TT broth (Oxoid) and RV broth (Oxoid). The TT and RV broths were incubated at 42°C for 20 h and from this 0.01 ml of each culture was streaked onto xylase lysine deoxycholate agar and BGM agar (Oxoid), followed by incubation at 37°C for 24 h. A portion of the suspected colonies form XLD and BGM agars were confirmed by using antibody agglutination and biochemical characteristics described above. Confirmed isolates were further serotyped for agglutination with Salmonella O and H antigens (Denkaseiken, Tokyo, Japan).
The enrichment culture in BPW was used as samples for the LAMP and PCR assays.
LAMP assay.
The LAMP reaction was performed with a Loopamp DNA amplification kit (Eiken Chemical). A portion (50 μl) of the samples pre-enriched in BPW was taken for the LAMP assay. It was added to 50 μl of the extraction solution (pH 12.5) to extract DNA and heated at 95°C for 5 min. After flash heating, the samples were centrifuged (Tomy, Tokyo, Japan). The supernatant was transferred to a new microtube and used as the template DNA solution for the LAMP assay. The LAMP reaction mixture contained the primers (20 μl) for Salmonella detection, Bst polymerase (1 μl), YO-PRO-1 iodide (0.125 μl. intercalating; Molecular Probes, Eugene, OR), and template DNA solution (5 μl). The reaction mixture was incubated at 65°C for 60 min with a thermal cycler (ABI7700; Applied Biosystems, Foster City, CA) and then heated to 80°C for 2 min to terminate the reaction. The LAMP amplicon was detected as a value of fluorescence (delta Rn) in real-time when there was an increase in fluorescence intensity caused by the intercalating dye. In addition, turbidity produced by magnesium pyrophosphate as a by-product during the reaction was visually monitored. Salmonella DNA extracted from a suspension of serovar Enteritidis cells by heating at 95°C for 5 min was used a as positive control in the LAMP assay.
The pre-enrichment culture with BPW of all samples from egg-breaking plants was tested by using the methods described above.
PCR assay.
The pre-enrichment culture in BPW of the samples was treated to extract DNA in a similar way as that for LAMP assay described above. The template DNA solution was neutralized by adding 1/10 volume of Tris-HCl (0.1 M, pH 7.0). PCR-targeted invA was performed as follows. Primer set SIN-1 and -2 (each 0.5 μl; Takara, Shiga, Japan), deoxynucleoside triphosphate mixture (4 μl), X10 Taq buffer (5 μl), Takara Taq (0.25 μl), template DNA solution (2.5 μl), and distilled water (37.75 μl) were mixed in a reaction tube. The reaction was performed at 94°C for 1 min for denaturing, 55°C for 1 min for annealing, and 72°C for 1 min for extension using a thermal cycler (ABI7000; Applied Biosystems). After 35 cycles and finally heating to 72°C for 10 min, PCR products were separated by agarose gel electrophoresis (3%). After staining with ethidium bromide, the size of the PCR product (378 bp) was compared to that of the serotype Enteritidis strain used as a positive control.
PFGE for Salmonella isolates.
Pulsed-field gel electrophoresis (PFGE) typing of S. Enteritids was performed by the method of Izumiya et al. (8, 9). All isolates were analyzed by using the restriction endonucleases BlnI (Takara) and XbaI (Takara). PFGE was performed with a CHEF-DR II apparatus (Bio-Rad Laboratories, Richmond, CA) at 6 V/cm and 14°C. A linearly ramped switching time from 5 to 50 s was applied for 21 h. A Lambda phage ladder (Bio-Rad Laboratories) was used as a molecular size marker.
A dendrogram was constructed from the PFGE profiles according to the Dice coefficient and an unweighted pair-group method with arithmetic averaging cluster analysis was performed with Finger-Printing II software (Bio-Rad Laboratories). Strains showing more than 80% similarity were defined as the same subtype.
RESULTS
Salmonella was detected by the culture method, LAMP, and PCR assay in all samples from egg-breaking plant A obtained over 12 days in the spring of 2003 (Table 1). The number of contaminated salmonellae in samples ranged from <1 CFU/g to 2.4 × 102 CFU/g. Total bacterial numbers in the samples were from 3.7 × 104 to 1.6 × 107 CFU/g.
TABLE 1.
Detection of salmonellae in liquid egg from plant A in the spring of 2003
| Sample no. | Sampling
|
Initial no. of contaminated organisms (CFU/g)
|
Detection methoda
|
||||
|---|---|---|---|---|---|---|---|
| Day | Time | Salmonella | Bacteria | Culture | LAMP | PCR | |
| 1 | 1 | a.m. | 1.2 × 102 | 3.5 × 105 | + | + | + |
| 2 | p.m. | 1.3 × 102 | 9.2 × 104 | + | + | + | |
| 3 | 2 | a.m. | 1.0 × 101 | 1.8 × 106 | + | + | + |
| 4 | p.m. | <1 | 4.6 × 104 | + | + | + | |
| 5 | 3 | a.m. | 3.0 × 101 | 6.4 × 106 | + | + | + |
| 6 | p.m. | 3.0 × 101 | 7.9 × 104 | + | + | + | |
| 7 | 4 | a.m. | 4.0 × 101 | 2.0 × 106 | + | + | + |
| 8 | p.m. | <1 | 3.7 × 104 | + | + | + | |
| 9 | 5 | a.m. | 7.0 × 101 | 1.5 × 106 | + | + | + |
| 10 | p.m. | 3.0 × 101 | 9.1 × 104 | + | + | + | |
| 11 | 6 | a.m. | 1 | 1.4 × 106 | + | + | + |
| 12 | p.m. | 2 | 6.5 × 104 | + | + | + | |
| 13 | 7 | a.m. | <1 | 6.1 × 105 | + | + | + |
| 14 | p.m. | 3.0 × 101 | 1.1 × 105 | + | + | + | |
| 15 | 8 | a.m. | 7.0 × 101 | 4.5 × 106 | + | + | + |
| 16 | p.m. | 2.4 × 102 | 1.7 × 105 | + | + | + | |
| 17 | 9 | a.m. | 1 | 2.6 × 106 | + | + | + |
| 18 | p.m. | 3 | 1.7 × 105 | + | + | + | |
| 19 | 10 | a.m. | 2 | 4.3 × 106 | + | + | + |
| 20 | p.m. | 2.0 × 101 | 6.8 × 105 | + | + | + | |
| 21 | 11 | a.m. | <1 | 8.4 × 105 | + | + | + |
| 22 | p.m. | 8.0 × 101 | 2.2 × 105 | + | + | + | |
| 23 | 12 | a.m. | 2.1 × 102 | 1.6 × 107 | + | + | + |
| 24 | p.m. | 4 | 6.7 × 104 | + | + | + | |
Culture consisted of enrichment culture in BPW with a 25-g sample, followed by culture with TT and RV, plated onto xylose lysine deoxycholate. For LAMP and PCR, enrichment culture in BPW with a 25-g sample was used for the assays.
Salmonella was also detected by the culture method, LAMP, and PCR assays in the unpasteurized liquid eggs from four plants (A to D) tested in the autumn of 2003 (Table 2). More than 92% of the samples at plants A, B, and D were positive for Salmonella. Although Salmonella detection was lowest at plant C, 72% of the samples had the microorganism. At plant A, Salmonella was detected in 1 of 27 (3.7%) samples by the LAMP and PCR assays but not by the culture method. At plants B, C, and D, 4 of 24 (16.7%), 6 of 22 (27.2%), and 1 of 27 (3.7%) samples, respectively, were positive for Salmonella by the culture method and the LAMP assay but not by the PCR method. When we combined the results of 110 samples from plants A to D, all samples that were negative in the LAMP assay were negative in the culture method and PCR assay. Of 110 samples, 1 (0.9%) was positive in the LAMP and PCR assay but not with the culture method. Of 110 samples, 11 (10%) were positive in the culture method and LAMP assay but not in the PCR assay. Of 110 samples, 10 (9.1%) were determined to be negative by all of the methods: culture, LAMP, and PCR assays. Although we tested the same culture in BPW at plants B and C by both LAMP and PCR assays, the detection with the PCR assay was lower than that of the LAMP assay. There was no difference between the results detected by fluorescence or turbidity.
TABLE 2.
Detection of salmonellae in liquid egg from four plants and serotypes of the isolates in the autumn of 2003
| Parameter | Egg-breaking plant
|
|||
|---|---|---|---|---|
| A | B | C | D | |
| No. of tested samples | 28 | 26 | 28 | 28 |
| No. of samples in which salmonellae were detected (%) | ||||
| Culture method | 26 (92.9) | 24 (92.3) | 22 (78.6) | 27 (96.4) |
| LAMP assay | 27 (96.4) | 24 (92.3) | 22 (78.6) | 27 (96.4) |
| PCR assay | 27 (96.4) | 20 (76.9) | 16 (57.1) | 26 (92.9) |
| Total | 27 (96.4)a | 24 (92.3)b | 22 (78.6)c | 27 (96.4)d |
| No. of samples in which salmonellae were undetected (%) | 1 (3.6) | 2 (7.7) | 6 (21.4) | 1 (3.6) |
| No. of sampling days | 14 | 13 | 14 | 14 |
| No. of days salmonellae were detectede | 14 | 12 | 13 | 14 |
| No. of days serovar Enteritidis was isolated | 12 | 5 | 10 | 12 |
| Mean bacterial no. ± SD (log CFU/g) | 4.20 ± 0.30 | 3.92 ± 0.41 | 5.03 ± 0.59 | 4.29 ± 0.52 |
| Serotype of isolate (no. of strains)f | Enteritidis (17) | Braenderup (11) | Enteritidis (13) | Enteritidis (16) |
| Cerro (6) | Enteritidis (8) | Cerro (10) | Mbandaka (7) | |
| Emek (4) | Typhimurium (3) | Infantis (8) | Thompson (5) | |
| Infantis (3) | Virchow (3) | Thompson (2) | Infantis (4) | |
| O35:Z4,Z23:− (3) | Cerro (2) | Haifa (1) | Typhimurium (3) | |
| Corvallis (2) | Infantis (2) | Mbandaka (1) | Cerro (2) | |
| Derby (1) | Lockleaze (2) | Montevideo (1) | Corvallis (2) | |
| Havana (1) | Agona (1) | Hadar (1) | ||
| Livingstone (1) | Bareilly (1) | Heidelberg (1) | ||
| Mbandaka (1) | Yoruba (1) | Schwarzengrund (1) | ||
| Saintpaul (1) | Virchow (1) | |||
| Thompson (1) | O8:Z4,Z24:− (1) | |||
| Virchow (1) | ||||
Contained one sample which was positive by LAMP and PCR assays but not by culture method.
Contained four samples which were positive by culture method and LAMP assay but not by PCR assay.
Contained six samples which were positive by culture method and LAMP assay but not by PCR assay.
Contained one sample which was positive by culture method and LAMP assay but not by PCR assay.
Detected by some of the three methods.
Isolates of the same PFGE pattern in same samples was regard as one strain. A total of 25 serotypes were isolated.
Salmonella was detected during most sampling days, and serotype Enteritidis was detected in more than 10 of 14 days at plants A, C, and D. On the other hand, serotype Enteritidis was detected only on 5 days at plant B.
Serotypes of the 156 isolates were determined (Table 2). A total of 25 serotypes were detected in the samples from four plants. Further analyses of the isolates were performed by using PFGE. The strains that showed the same pattern in PFGE were regarded as a single strain. At plants A, C, and D serotype Enteritidis was the major serotype, and more than 11 serovar Braenderup strains were detected at plant B specifically and predominantly. A few serotypes were predominant in all plants where 7 to 13 serotypes were isolated.
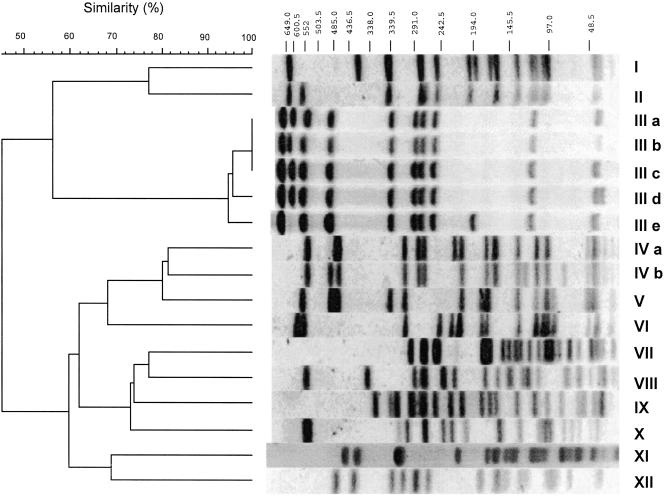
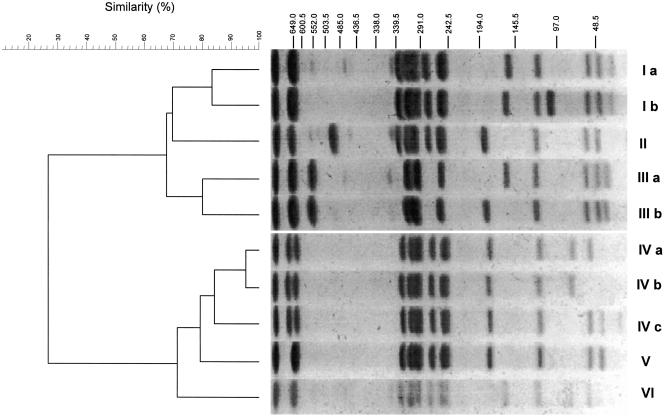
Fifty-four strains of serotype Enteritidis from liquid egg samples from all of the plants in the autumn of 2003 were analyzed. We found 12 types (I to XII) in the patterns by using PFGE with BlnI (Fig. 1). The patterns with BlnI were distributed in two main clusters with a similarity percentage of <45%. Further, using the result with similar percentages of >80%, types III and IV were resolved to five (a to e) and two (a and b) subtypes, respectively. On the other hand, the patterns with XbaI were distributed into two main clusters with a similar percentage of <27%, which were constructed with six patterns (I to VI) (Fig. 2). Types I and III and type IV were resolved to two (a and b) and three (a to c) subtypes, respectively.
FIG. 1.
PFGE typing with BlnI in Salmonella sp. serotype Enteritidis strains isolated from unpasteurized liquid egg. PFGE patterns of S. Enteritidis strains were distributed among 12 major types (I to XII). Types III and IV consisted of five and two subtypes, respectively.
FIG. 2.
PFGE typing with XbaI in Salmonella sp. serotype Enteritidis strains isolated from unpasteurized liquid egg. PFGE patterns of serotype Enteritidis strains were distributed among six major types (I to VI). Types I, III, and IV consisted of two, two, and three subtypes, respectively.
With BlnI, type III was the majority in the samples from plants A, B, and D (Table 3). Type IV was predominantly found in the samples from plant C, but the patterns detected at this plant were not detected in any other plants. With XbaI, types I, IV, and V were predominant in the samples from plants C, D, and A, respectively (Table 3). At plant B, types IV and V were the major patterns seen with PFGE.
TABLE 3.
PFGE patterns of Salmonella sp. serovar Enteritidis isolates during sampling for 16 days at each egg-breaking plant
| Sampling day | Time | PFGE pattern at planta:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| A
|
B
|
C
|
D
|
||||||
| BlnI | XbaI | BlnI | XbaI | BlnI | XbaI | BlnI | XbaI | ||
| 1 | a.m. | IIIa | V | IIIb | Vb | ND | ND | IIIb | IVa |
| p.m. | IIIa | V | IIIc | V | IVa | Ia | ND | ND | |
| 2 | a.m. | ND | ND | ND | ND | ND | ND | IIIb | IVa |
| p.m. | IIIb | V | ND | ND | VI | Ia | VII | Ia | |
| 3 | a.m. | ND | ND | NC | NC | IVa | Ia | ND | ND |
| p.m. | XII | Ia | NC | NC | IVa | Ia | ND | ND | |
| 4 | a.m. | ND | ND | ND | ND | ND | ND | ND | ND |
| p.m. | IIIc | V | IIIc | V | ND | ND | VII | Ia | |
| 5 | a.m. | NC | NC | NC | NC | NC | NC | NC | NC |
| p.m. | NC | NC | NC | NC | NC | NC | NC | NC | |
| 6 | a.m. | ND | ND | ND | ND | ND | ND | ND | ND |
| p.m. | ND | ND | ND | ND | IVa | Ia | IIIb | IVa | |
| 7 | a.m. | IIIa | V | ND | ND | IVa | Ia | ND | ND |
| p.m. | XI | VI | IIIb | IVb | ND | ND | IIIc | IVc | |
| 8 | a.m. | ND | ND | ND | ND | I | Ia | ND | ND |
| p.m. | IIIa | V | ND | ND | IVb | Ia | IIIb | IVa | |
| 9 | a.m. | ND | ND | ND | ND | ND | ND | IIIb | IVa |
| p.m. | ND | ND | IIIc | V | ND | ND | ND | ND | |
| 10 | a.m. | X | Ia | ND | ND | ND | ND | ND | ND |
| p.m. | IIIa | V | IIId | IIIb | ND | ND | ND | ND | |
| 11 | a.m. | VIII | IIIa | ND | ND | ND | ND | ND | ND |
| p.m. | IIIc | V | ND | ND | ND | ND | IIIb | IVa | |
| 12 | a.m. | NC | NC | NC | NC | NC | NC | NC | NC |
| p.m. | NC | NC | NC | NC | NC | NC | NC | NC | |
| 13 | a.m. | ND | ND | ND | ND | ND | ND | IIIc | IVc |
| p.m. | IIIc | V | ND | ND | IX | Ib | IIIb | IVa | |
| 14 | a.m. | IIIe | II | ND | ND | ND | ND | IIIc | IVc |
| p.m. | ND | ND | ND | ND | V | Ia | IIIb | IVa | |
| 15 | a.m. | ND | ND | IIIb | IVa | VI | Ia | IIIb | IVa |
| p.m. | IIIa | V | IIIb | IVa | VI | Ia | IIIb | IVa | |
| 16 | a.m. | IIIa | V | ND | ND | ND | ND | ND | ND |
| p.m. | IIIa | V | ND | ND | II | Ia | IIIb | IVa | |
NC, sample not collected; ND, Salmonella serotype Enteritidis not detected.
DISCUSSION
The specificity of the LAMP assay was high. LAMP assay was not inhibited by the constituents of liquid egg, and the sensitivity was very high. Salmonella control is important for safety in foods containing liquid egg, and rapid and sensitive methods are therefore needed for its detection in liquid-egg plants.
Based on the results of Salmonella detection in 110 samples of unpasteurized liquid eggs, the LAMP assay was the most effective of the three methods used. In one (0.9%) sample, Salmonella was detected by LAMP assay but not by the culture method. Although there was a possibility of false positives, the positive reaction was considered to be true because of the high specificity and sensitivity of LAMP assay due to the mechanics of the LAMP. The PCR method failed to detect Salmonella in 10% of samples. At plants B, C, and D, the PCR assay but not the LAMP assay failed to detect Salmonella, although the culture enrichment in BPW was performed for both the PCR and the LAMP assays. Therefore, the LAMP assay seems to be more efficient and sensitive than PCR.
In unpasteurized liquid eggs naturally contaminated with Salmonella at low levels (1 to 25 cells/25 g of liquid egg), all methods—LAMP assay, PCR assay, or the culture method—were successful in detecting the pathogen (Table 1). The initial number of Salmonella might be increased to more than 1,000 cells/ml after enrichment.
The detection levels in plants A and D by the culture method, LAMP, and PCR assays were >92.9%. Although the detection levels at plants A, B, and D of the culture method and LAMP assay were >92.3%, the detection levels at plant B by the PCR assay were 76.9% and by the culture method and the LAMP assay were 92.3%. Thus, the PCR assay was not as effective for samples from plant B. In three of four samples in which Salmonella was detected by the LAMP assay but not by the PCR assay, serotype Braenderup was isolated but not serotype Enteritidis. However, there were no differences between the PCR sensitivities of serotypes Braenderup and Enteritidis (data not shown). At plant C, the detection level was lower than at the other plants. It may be reasoned that here that the total bacterial count was larger than those of the other plants. These results indicate that the LAMP assay should be used because it is rapid and simple and because the possibility of false-positive reactions is similar to that which can be expected with other methods. To confirm the specific amplicons by LAMP, reactions showing many size patterns and ladder patterns on electrophoresis are useful. In the present study, one sample from plant A, which was detected by LAMP assay but not PCR assay and the culture method, was tested and showed ladder patterns on electrophoresis. In addition, analysis of the sequence of a part of the amplicon might be more useful to confirm the results of LAMP assay.
The liquid egg at the egg-breaking plants tested in the present study was finally pasteurized before shipping. However, the contamination of Salmonella should be monitored prior to pasteurization and minimized to produce a safer liquid egg product. The LAMP method is effective for monitoring unpasteurized and pasteurized liquid products.
Although many serotypes of Salmonella were detected in each plant, each plant was highly infected with Salmonella. Furthermore, in the serotype Enteritidis strains that were analyzed by PFGE totals of 12 and 6 distinct patterns were shown with BlnI and XbaI, respectively. In each plant, the PFGE patterns detected were limited. Using BlnI, only one type, including three subtypes, and two types, including two subtypes, were detected at plants B and D, respectively. Using XbaI, the major PFGE type was individually different among three plants (A, C, and D). Although the patterns by BlnI were more distributed than those by XbaI, the result of PFGE with XbaI seems to adapt in epidemiological analysis of serotype Enteritidis isolates.
Because egg is thick and rich in lipid and protein, it is difficult to perfectly remove liquid egg from the equipment in a product line. Salmonella, known for its survival in dried conditions (10, 11), can live in liquid egg remaining in a product line. We thought the high frequency of Salmonella detection in the liquid egg samples was due to contamination of the product line. Brushing and washing with abundant water, followed by treatment with disinfectants, is effective for removing the rest of the liquid. To thoroughly wash the angles of the equipment, dismantling the equipment at regular intervals is a way to reduce Salmonella contamination.
Because the eggs were supplied from various farms, strains showing the same pattern seem to come from the unsanitary production line in the plant rather than from the eggs themselves. These results were similar at plants A, C, and D when evaluated using XbaI. However, type III, including five subtypes (a to e) by BlnI was the predominant pattern at plants A, B, and D. Because shelled eggs were supplied from different farms to each plant, the strains shown to be type III may spread in Japan. Of 91 serotype Enteritidis strains isolated from patients in hospitals in the central area of Japan during 2003, 14 were type III, although the strains have not been analyzed by PFGE with BlnI (unpublished data).
The present study demonstrated that the LAMP assay was effective for detecting Salmonella in egg samples rapidly and at a high sensitivity. Since Salmonella control is important for safety in foods containing egg, the effective strategy could be improved hygiene in places related with egg such as egg breaking plant.
Acknowledgments
This study was supported by a grant from the Ministry of Health, Labour, and Welfare in Japan.
REFERENCES
- 1.Barrow, P. A., and M. A. Lovell. 1991. Experimental infection of egg-laying hens with Salmonella enteritidis phage type 4. Avian Pathol. 20:335-348. [DOI] [PubMed] [Google Scholar]
- 2.Duguid, J. P., and R. A. North. 1991. Eggs and Salmonella food poisoning: an evaluation. J. Med. Microbiol. 34:65-72. [DOI] [PubMed] [Google Scholar]
- 3.Ebel, E. D., M. J. David, and J. Mason. 1992. Occurrence of Salmonella enteritidis in the U.S. commercial egg industry: report on a national spent hen survey. Avian Dis. 36:646-654. [PubMed] [Google Scholar]
- 4.Ebel, E. D., J. Mason, L. A. Thomas, K. E. Ferris, M. G. Beckman, D. R. Cummins, L. Scroeder-Tucker, W. D. Sutherlin, R. L. Glasshoff, and N. M. Smithhisler. 1993. Occurrence of Salmonella enteritidis in unpasteurized liquid egg in the United States. Avian Dis. 37:135-142. [PubMed] [Google Scholar]
- 5.Ebel, E., and W. Schlosser. 2000. Estimating the annual fraction of eggs contaminated with Salmonella Enteritidis in the United States. Int. J. Food Microbiol. 61:51-62. [DOI] [PubMed] [Google Scholar]
- 6.Hogue, A. T., E. D. Ebel, L. A. Thomas, W. Schlosser, N. Bufano, and K. E. Ferris. 1997. Surveys of Salmonella enteritidis in unpasteurized liquid egg and spent hens at slaughter. J. Food Prot. 60:1194-1200. [DOI] [PubMed] [Google Scholar]
- 7.Humphrey, T. J., A. Whitehead, A. H. Gawler, A. Henley, and B. Rowe. 1991. Numbers of Salmonella enteritidis in the contents of naturally contaminated hens' eggs. Epidemiol. Infect. 106:489-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izumiya, H., J. Terajima, A. Wada, Y. Inagaki, K. Itoh, K. Tamura, and H. Watanabe. 1997. Molecular typing of enterohemorrhagic Escherichia coli O157:H7 isolates in Japan by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 35:1675-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izumiya, H., J. Terajima, S. Matsushita, K. Tamura, and H. Watanabe. 2001. Characterization of multidrug-resistant Salmonella enterica serovar Typhimurium isolated in Japan. J. Clin. Microbiol. 39:2700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janning, B., P. H. in't Veld, S. Notermans, and J. Kramer. 1994. Resistance of bacterial strains to dry conditions: use of anhydrous silica gel in a desiccation model system. J. Appl. Bacteriol. 77:319-324. [DOI] [PubMed] [Google Scholar]
- 11.Kusumaningrum, H. D., G. Riboldi, W. C. Hazeleger, and R. R. Beumer. 2003. Survival of food-borne pathogens on stainless steel surfaces and cross-contamination to foods. Int. J. Food Microbiol. 85:227-236. [DOI] [PubMed] [Google Scholar]
- 12.Mishu, B., J. Koehler, L. A. Lee, D. C. Rodrigue, F. H. Brenner, P. Blake, and R. V. Tauxe. 1994. Outbreak of Salmonella enteritidis infection in the United States, 1985-1991. J. Infect. Dis. 169:547-552. [DOI] [PubMed] [Google Scholar]
- 13.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 14.Murase, T., K. Senjyu, T. Maeda, M. Tanaka, H. Sakae, Y. Matsumoto, Y. Kaneda, T. Ito, and K. Otsuki. 2001. Monitoring of chicken houses and an attached egg-processing facility in a laying farm for Salmonella contamination between 1994-1998. J. Food Prot. 64:1912-1916. [DOI] [PubMed] [Google Scholar]
- 15.Nagamine, K., K. Watanabe, K. Ohtsuka, T. Hase, and T. Notomi. 2001. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 47:1742-1743. [PubMed] [Google Scholar]
- 16.Nagamine, K., Y. Kuzuhara, and T. Notomi. 2002. Isolation of single-stranded DNA from loop-mediated isothermal amplification products. Biochem. Biophys. Res. Commun. 290:1195-1198. [DOI] [PubMed] [Google Scholar]
- 17.Nakanishi, H. 1993. Present status of gastroenteritis due to Salmonella Enteritidis and its preventive measures. J. Food Hyg. Soc. Jpn. 34:318-322. (In Japanese.) [Google Scholar]
- 18.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riemann, H., S. Himathongkham, D. Willoughby, R. Tarbell, and R. Breitmeyer. 1998. A survey for Salmonella by drag swabbing manure piles in California egg ranches. Avian Dis. 42:67-71. [PubMed] [Google Scholar]
- 20.Shivaprasad, H. L., J. F. Timoney, S. Morales, B. Lucio, and R. C. Baker. 1990. Pathogenesis of Salmonella enteritidis infection in laying chickens. I. Studies on egg transmission, clinical signs, fecal shedding, and serological responses. Avian Dis. 343:548-557. [PubMed] [Google Scholar]
- 21.St. Luis, M. E., D. L. Morse, M. E. Portter, T. M. DeMeflfi, J. J. Guzewich, R. V. Tauxe, and P. A. Blake. 1988. The emergence of grade A eggs as a major source of Salmonella enteritidis infections. New implications for the control of salmonellosis. JAMA 259:2103-2107. [PubMed] [Google Scholar]
- 22.Telzak, E. E., L. D. Budnick, M. S. Greenberg, S. Blum, M. Shayegani, C. E. Benson, and S. Schultz. 1990. A nosocomial outbreak of Salmonella enteritidis infection due to the consumption of raw eggs. N. Engl. J. Med. 323:394-397. [DOI] [PubMed] [Google Scholar]
- 23.U.S. Department of Health and Human Services. 1992. Outbreak of Salmonella enteritidis infection associated with consumption of raw shell eggs, 1991. Morb. Mortal. Wkly. Rep. 41:369-372. [PubMed] [Google Scholar]