Abstract
Rhizobia, the root-nodule endosymbionts of leguminous plants, also form natural endophytic associations with roots of important cereal plants. Despite its widespread occurrence, much remains unknown about colonization of cereals by rhizobia. We examined the infection, dissemination, and colonization of healthy rice plant tissues by four species of gfp-tagged rhizobia and their influence on the growth physiology of rice. The results indicated a dynamic infection process beginning with surface colonization of the rhizoplane (especially at lateral root emergence), followed by endophytic colonization within roots, and then ascending endophytic migration into the stem base, leaf sheath, and leaves where they developed high populations. In situ CMEIAS image analysis indicated local endophytic population densities reaching as high as 9 × 1010 rhizobia per cm3 of infected host tissues, whereas plating experiments indicated rapid, transient or persistent growth depending on the rhizobial strain and rice tissue examined. Rice plants inoculated with certain test strains of gfp-tagged rhizobia produced significantly higher root and shoot biomass; increased their photosynthetic rate, stomatal conductance, transpiration velocity, water utilization efficiency, and flag leaf area (considered to possess the highest photosynthetic activity); and accumulated higher levels of indoleacetic acid and gibberellin growth-regulating phytohormones. Considered collectively, the results indicate that this endophytic plant-bacterium association is far more inclusive, invasive, and dynamic than previously thought, including dissemination in both below-ground and above-ground tissues and enhancement of growth physiology by several rhizobial species, therefore heightening its interest and potential value as a biofertilizer strategy for sustainable agriculture to produce the world's most important cereal crops.
A large diversity of endophytic bacteria, including nitrogen fixers, can be isolated from healthy plant tissues after they are surface sterilized (16, 24, 30, 32). Among these, Rhizobium is most thoroughly studied because of its ability to induce root nodules on legume plants of agricultural importance and provide these plant hosts with fixed nitrogen, enabling them to grow productively in nitrogen-limited agricultural soils (12, 21, 34, 36). Recent studies indicate that Rhizobium also endophytically colonizes roots of certain cereal crop plants, promoting their growth and grain yield at harvest while reducing their dependence on chemical fertilizer inputs, independent of root nodulation and biological N2 fixation (2, 3, 5, 15, 17, 22, 23, 26, 27, 37, 38). For instance, the beneficial growth responses of rice to rhizobia include increased seed germination, rate of radical elongation, seedling vigor, root architecture (length, branching, biovolume, surface area), shoot growth, photosynthetic activity, stomatal conductance, shoot and grain N content, harvest index, agronomic N fertilizer use efficiency, and grain yield. These cereal plants represent many of the world's most important grain food crops, including rice, maize, wheat, and barley.
The potential agronomic benefits of using selected strains of endophytic rhizobia as biofertilizer inoculants for cereal production is being examined in the Egyptian Nile delta where it was first discovered (37). To date, 22 field inoculation studies (37, 38; Y. G. Yanni and F. B. Dazzo, unpublished data) have demonstrated increased crop yield by up to 29 to 30% for rice and wheat above what is reached using inorganic fertilizers alone, with a reduction of up to one-third of the cost for currently used fertilizer inputs at levels previously assessed and recommended without any biofertilizer. The potential benefit of exploiting this endophytic plant-bacterium association for cereal production also extends to decreased environmental pollution and health risks originating from excessive use of the agrochemical N fertilizers to achieve high grain yield.
Despite a widespread occurrence of this natural endophytic rhizobium-cereal association, much remains unknown about its infection and colonization processes (8, 27, 28, 37, 38). Important issues central to development of this plant-microbe association are the primary portals of bacterial entry into the plant tissues, the extent of their dissemination (especially ascending migration) within the plant host after primary root infection, and their population dynamics in planta. The first clue suggesting an ascending migration of an endophytic strain of R. leguminosarum bv. trifolii within rice came from early microscopic studies that found the bacteria within leaf whirls at the stem base above the roots that were inoculated and grown in gnotobiotic culture (37). Here, we used computer-assisted microscopy and viable plating methods to quantify the colonization, dispersion, and growth dynamics of several gfp-tagged species of rhizobia within below-ground and above-ground tissues of healthy rice plants after inoculation of their rhizosphere with these marked rhizobia. We also assessed the potential benefit to the growth physiology of rice after inoculation of their roots with these same gfp-tagged rhizobium strains
(A portion of this work was presented at the 14th International Congress on Nitrogen Fixation in Beijing, China [7].)
MATERIALS AND METHODS
Bacteria, plasmids, and plants.
Rhizobium strains and plasmids used in this study are listed in Table 1. The vector pHC60 (6) encodes for tetracycline resistance and contains the gfp gene that is constitutively expressed at fairly constant levels from a constitutive lacZ promoter without the required expression of lacZ. This vector was similar to the one used earlier by Gage et al. (13), except that it contains a stability region so that expression of its gfp is more stably maintained within bacterial cells in the absence of selective pressure (6). The half-life of the green fluorescent protein (GFP) in some organisms is approximately 1 day (35). For construction of the gfp-tagged strains, the pHC60 vector was transferred to the wild-type rhizobia strains using the triparental mating method (9). Japonica rice (Oryza sativa L.) varieties Zhonghua 8 and Nipponbare were obtained from the Institute of Crop Science, Chinese Academy of Agriculture, China, and National Institute of Agrobiological Sciences of Japan. Medicago sativa L., Sesbania rostrata, Astragalus sinicus L., and Pisum sativum L. were used as the legume hosts.
TABLE 1.
Plasmids and bacterial strains used in this study
| Plasmid or Rhizobium strain | Characteristics (amt of antibiotic used [μg/ml])a | Source |
|---|---|---|
| pHC60 | Broad-host-range plasmid with gfp; Tcr (10) | City University of New York (6) |
| Sinorhizobium meliloti 1021 | Smr (50) | Institute of Plant Physiology, Shanghai, People's Republic of China |
| Azorhizobium caulinodans ORS 571 | Ampr (100), Cbr (100) | Academy of Agriculture, Beijing, People's Republic of China |
| Sinorhizobium meliloti USDA 1002 | Kmr (50) | Chinese University of Agriculture, Beijing, People's Republic of China |
| Rhizobium leguminosarum USDA 2370 | Smr (50) | Chinese University of Agriculture, Beijing, People's Republic of China |
| Mesorhizobium huakui 93 | Smr (10) | Nanjing University of Agriculture, Nanjing, People's Republic of China |
| Sinorhizobium meliloti 1021 (gfp) | Smr (50), Tcr (10) | City University of New York, New York, N.Y. |
| Azorhizobium caulinodans ORS 571 (gfp) | Ampr (100), Tcr (10) | This study |
| Sinorhizobium meliloti USDA 1002 (gfp) | Kmr (50), Tcr (10) | This study |
| Rhizobium leguminosarum USDA 2370 (gfp) | Smr (50), Tcr (10) | This study |
| Mesorhizobium huakui 93 (gfp) | Smr (10), Tcr (10) | This study |
Tcr, tetracycline resistance; Smr, streptomycin resistance; Cbr, carbenicillin resistance; Kmr, kanamycin resistance; Ampr, ampicillin resistance; Tcr, tetracycline resistance.
Growth conditions.
For gnotobiotic culture of rice with rhizobia, seeds were de-hulled, treated with 70% ethanol for 10 min, washed three times with sterile water, surface sterilized with 0.1% HgCl2 for 10 min, and then washed three times again with sterile water. Surface-sterilized seeds were germinated on Luria-Bertani (LB) plates in the dark for 2 days at 28°C to verify that they were axenic. Four to five small axenic seedlings were transferred aseptically into sterilized glass tubes (40 cm in height, 5 cm in diameter) containing 250 cm3 of sterile vermiculite and 125 ml of half-strength Hoagland's no. 2 plant growth medium. Bacteria were cultured for 2 days at 28°C on TY agar (1) containing the appropriate antibiotic(s), and then suspended in phosphate-buffered saline (PBS; pH 7.4) to 108 cells/ml. A total of 5 ml of bacterial inoculum was carefully introduced into the seedling rhizosphere using a pipette tip inserted 1 cm below the vermiculite surface in each tube in order to avoid contamination of the above-ground rice tissues. Each inoculated tube was covered with transparent tissue culture paper (12 cm by 12 cm) and incubated in a growth chamber programmed with a 14-h photoperiod and 28/25°C day/night cycle. Three inoculated seedlings were replicated for each strain.
GFP-tagged rhizobia recovered from rice were tested for nodulation ability on their respective legume host (M. sativa L. for S. meliloti, S. rostrata for A. caulinodans, A. sinicus L. for M. huakui, and P. sativum L. for R. leguminosarum bv. viciae) in a gnotobiotic tube culture. Conditions were the same as described above for rice except that the gnotobiotic tube cultures contained 100 cm3 of vermiculite and 60 ml of Fahraeus nitrogen-free nutrient medium, and the rhizobial inoculum density was 106 cells/ml. At 30 days postinoculation (dpi) in the growth chamber, the roots were checked for nodules that exhibited green fluorescence.
For potted soil culture of rice with rhizobia, seeds (without surface sterilization) were imbibed in tap water for 2 days, and then transferred to a small pot containing watered soil. After 20 days of growth under outdoor ambient conditions, the seedlings were transplanted to large pots, each containing 13 liters of watered, nonsterilized porous media (soil-vermiculite mix [1:1]). After 7 and 45 days of growth, the seedlings were inoculated with 100 ml of the same rhizobial cell suspension (optical density at 600 nm = 0.8) into each pot (three replicates per strain), being careful not to contaminate above-ground plant tissues. Potted plants were grown in the open outdoors for 160 dpi before harvesting.
Microscopy and image analysis.
The GFP accumulated within the gfp-tagged bacteria produces a sufficiently bright fluorescent signal allowing for their single-cell detection and quantification en masse by computer-assisted fluorescence microscopy. Since all the rhizobial cells carry the plasmid and express the gfp gene at approximately the same level, their local population sizes can be analyzed by measuring the integrated density of emitted fluorescent light from within a defined area or volume. Tissues of rice roots, leaf sheaths at the leaf base, and leaves (Fig. 1) were excised from plants removed from tubes at regular intervals and rolled over TY plates to check whether bacteria could be cultured from their external surfaces. The knife was washed between cuttings with sterile water and 70% alcohol and wiped with sterile absorbent paper to avoid cross-contamination of tissues during excision. Free-hand sections of the excised tissues were rinsed clean with sterile water, mounted on slides and examined using a Bio-Rad MRC 1024 laser confocal microscope with 488- and 568-nm band-pass filters to capture the green fluorescence from gfp-tagged bacteria and the red autofluorescence from host tissue, respectively. The images were acquired as confocal Z-section series using a Nikon E800 scanner and digital camera and then merged into loss-less montage images using Confocal Assistant Software (version 4.02; Todd Clerke Brelje [www.Imf.ludwig.ucl.ac.uk/CAScontent.htm]). Only regions of infected tissues and plant cells immediately surrounding them located at least 20 μm beneath the cut surfaces were included in these extended focus images prepared from Z-series of optisections used to measure the local cell density of endophytic bacteria. This sampling design excluded any fluorescent bacteria lying directly on the cut surface that may have been redistributed while sectioning the tissues and extraneous tissue inaccessible to the bacteria in order to achieve as high of a signal-to-noise ratio of input data as possible for calculating the local population density.
FIG. 1.
Various rice tissues used in these studies.
Images were spatially calibrated, color segmented, and analyzed using CMEIAS Image Analysis software (8, 20, 28) to measure the local population density and spatial distribution of fluorescent gfp-tagged bacteria in situ within the plant tissues. The voxel dimensions were computed from the two-dimensional projected area of fluorescent bacteria within the Z-series of image optisections containing infected tissue, the cumulative Z thickness of each image stack, and 0.5 μm3 as the average biovolume of each vegetative rhizobial cell (computed from an image analysis of 10 individual, well-isolated bacteria in planta).
Geostatistical analysis was performed on confocal images to examine the connectivity between local population density and spatial distribution of the endophytic bacteria (8). For these analyses, each digital image was processed by CMEIAS Color Segmentation software (28) to create a new image containing all of the foreground pixels that represent the fluorescent green microbes of interest on a noise-free background and then digitally cut into 144 square quadrats created by a 12-by-12 grid overlay using CMEIAS Quadrat Maker software. Each image quadrat was then analyzed using CMEIAS Image Analysis software to extract its object count as the Z variate, and the Cartesian x and y coordinates of its representative sample posting within the original image, weighted by the x and y coordinate positions of the objects' local density within it (8). These plot-based, georeferenced data of local bacterial density were tested for spatial autocorrelation, and the derived geostatistical model that best fits the computed spatial variance in local density was then used to compute the spatial scale in which bacteria influence each other's distribution in situ and the corresponding two-dimensional kriging interpolation map of this regional variable over the same spatial domain (8).
Viable plating experiments.
The rice roots from gnotobiotic seedlings were carefully removed from each tube, excised, washed with sterile water, blotted dry, divided into two parts, and weighed. One portion (1:20 wt/vol) was surface sterilized by vortexing for 1 min in a solution of 1% bleach, 0.1% sodium dodecyl sulfate, and 0.2% Tween 20 in PBS (10). These samples were then rinsed four times with sterile water, placed on agar plates of LB medium for 1 h, and then removed. These plates developed no colonies when incubated for 2 days at 28°C, verifying that the excised roots were surface sterilized. To enumerate the endophytic rhizobia, excised surface-sterilized roots were macerated with a sterile mortar and pestle, diluted in PBS solution containing 20% glycerol, and spread on TY plates supplemented with tetracycline (10 μg/ml) and the other appropriate antibiotic(s) for each test strain. The other excised roots were macerated without surface sterilization and plated as described above to enumerate the combined, viable populations of the rhizobial test strain on the root surface and internal tissue.
To verify that the test strains had not contaminated aerial plant surfaces in these gnotobiotic cultures, fragments of leaf sheaths at the leaf base and 2-cm fragments of leaves were transferred directly without prior surface sterilization onto LB plates for 1 h, and then the plates were incubated at 28°C. After removal from the LB plates, the excised plant tissues were macerated, diluted in PBS, and plated out as described above to enumerate the endophytic population of viable bacterial cells.
To enumerate the viable endophytic rhizobia within tissues of rice grown for 125 days in open potted soil, 15 cm of the leaf sheaths and 15 cm of the leaf fragments were excised 5 and 20 cm, respectively, above the taproots (Fig. 1) from three replicate plants per treatment. These excised tissues were surface sterilized separately in 15% NaOCl for 10 min, washed four times with sterile water, and rolled over the surface of LB solid medium as a test to check whether their surfaces were free of contamination. The tissues were then homogenized in sterile PBS-glycerol and centrifuged briefly at slow speed (785 g) to remove the bulk of macerated plant tissue without affecting the colony counts of suspended bacteria. The supernatant suspensions were diluted in sterile PBS and plated on TY plates containing tetracycline plus the appropriate antibiotic for each rhizobial test strain (Table 1). After 2 days of incubation at 28°C, the colonies were counted from each of the three replicate tissue samples, and 10 colonies from each sample set were checked for green autofluorescence using fluorescence microscopy and nodulation ability on the homologous legume host in gnotobiotic culture as described above.
PCR analysis of rice endophytes after colony isolation.
To further verify Koch's postulates, the same colonies of endophytic bacteria recovered from the potting experiments described above were also analyzed for the gfp and 16S rRNA genes of the respective inoculant strains. The PCR conditions were as follows: 30 cycles of 94°C for 5 min, 94°C for 30 s, 57°C for 30 s, 72°C for 1 min, and 72°C for 10 min. The GFP 5′ primer was 5′-ATGGCTAGCAAAGGA GAAGAACTC-3′, and the 3′ primer was 5′-TAGGTACCCCTTTCAGCAAAA AACCC-3′. The 16S rRNA 5′ primer was 5′-GGTAGTCCACGCCGTAAA CG-3′, and the 3′ primer was 5′-GCGGGACTTAACCCAACATCT-3′.
Analysis of rice growth responses after inoculation with rhizobia.
Axenic seedlings derived from surface-sterilized seeds were transferred to pots containing 13 liters of nonsterilized and watered porous media (soil-vermiculite [1:1]) and grown outdoors. Each pot was planted with 18 seedlings, replicated in three pots, and inoculated with a single strain. The inoculum for each pot consisted of rhizobial cells cultured in TY liquid medium, suspended in 100 ml of PBS buffer to a density of optical density at 600 nm of 0.8, and poured into each planted pot. After a short period of growth, seedlings were tillered to 2 to 3 plants that ultimately produced around 45 to 54 plants per pot at maturity. After 130 days of growth, five top flag leaves of the rice plants selected from each replicate of three pots were analyzed nondestructively for net photosynthetic rates using a portable photosynthesis system LI-COR6400 (LICOR Biosciences) according to the manufacturer's instructions. Simultaneously, the stomatal conductance, transpiration velocity, and CO2 concentration within the flag leaves were recorded using the same instrument. Plants were then harvested after 160 days of rice growth. The mean height was measured from the above-ground stem base to the top leaf without the spike from a total of 135 to 162 rice plants (three pots). Mean fresh weight, biovolume, dry weight, nitrogen content, and seed yield of plants per pot were calculated as the averages of the three replicate pots. Root biovolume was measured using water displacement. Plant dry weight was measured after drying in an oven for 2 days at 80°C. Total nitrogen content was measured by the Kjeldahl method, and total grain yield was calculated as the mean seed weight times the total seed yield per pot.
Extraction and analysis of phytohormones from rice tissues.
High-performance liquid chromatography (11, 19) was used to measure indoleacetic acid (IAA) and gibberellin (GA) extracted from all root and shoot tissues (including three to four leaves and leaf sheaths) of gnotobiotic cultured rice after 40 days of incubation with rhizobia.
RESULTS AND DISCUSSION
Microscopy of rice tissues colonized with gfp-tagged Sinorhizobium meliloti 1021.
Roots examined at six dpi contained fluorescent green cells of gfp-tagged S. meliloti 1021 preferentially colonized around lateral root junctions (Fig. 2A), where they gained entry into the root interior between displaced epidermal cells, a finding consistent with previous studies indicating this as the major site of colonization on epidermal root surfaces and primary host infection in cereals by rhizobia (8, 27, 28, 37, 38). By 10 to 14 dpi, the fluorescent bacteria had spread to intercellular spaces and within lysed plant cells in neighboring regions of epidermal (including root hairs; Fig. 2B), cortical, and vascular root tissues (Fig. 2C). By 21 dpi, some fluorescent rhizobia had disseminated upward to aerenchyma and vascular tissue within leaf sheaths above the stem base and within leaves. GFP-tagged S. meliloti 1021 heavily colonized these above-ground internal tissues by 35 dpi (Fig. 2D to G). These observations were similar using the two rice varieties cv. Zhonghua 8 and cv Nipponbare.
FIG. 2.
Confocal laser scanning micrographs of gfp-tagged cells of wild-type S. meliloti 1021 colonized within healthy rice tissues. (A) Lateral root emergence; (B) lysed root hair; (C) cross-section of the tap root; (D and G) cross-sections of the leaf sheath above the stem base; (E and F) within leaves. Bars: 50 μm in panels A to E and 20 μm in panels F to G.
CMEIAS image analysis of nine Z-series of confocal fluorescence optisections representing three tissue locations indicated a high in situ local abundance of the internal gfp-tagged bacteria, with a range of 3.66 to 20.13% microbial cover and a local population density ranging from 2.61 × 108 to 9.03 × 1010 bacteria per cm3 of infected plant tissue (Table 2).
TABLE 2.
In situ local abundance of gfp-tagged S. meliloti 1021 cells within confocal optisections of infected plant tissues in ricea
| Tissue location | % Microbial cover | Local population density (bacteria/cm3) |
|---|---|---|
| Root | 3.66-19.73 | 1.52 × 109-9.03 × 1010 |
| Stem base | 5.61-19.15 | 2.61 × 108-1.37 × 1010 |
| Lower leaves | 8.30-20.13 | 7.69 × 109-8.95 × 1010 |
Plant roots were inoculated and grown gnotobiotically for 35 days before examination by computer-assisted microscopy and CMEIAS image analysis.
Enumeration of culturable endophytic rhizobia within rice tissues.
Four different plating experiments verified this endophytic colonization strategy of disseminating migration from primary host root infection to aerial plant parts and extended it to include gfp-tagged derivatives of four species of wild-type rhizobia (Sinorhizobium meliloti 1021, Azorhizobium caulinodans ORS 571, Rhizobium leguminosarum [bv. viciae] USDA 2370, Mesorhizobium huakui 93, and Sinorhizobium meliloti USDA 1002). For each plating experiment, 10 colonies were picked at random after enumeration and verified that they exhibited green autofluorescence and were able to nodulate their respective legume hosts in gnotobiotic culture.
In the first experiment, all five gfp-tagged derivatives of wild-type rhizobia could be recovered from within surface-sterilized roots (104.3 to 106.1 CFU/gfw [gram fresh weight]) and leaf sheaths (103.0 to 105.4 CFU/gfw) above the stem base after inoculation with a mixed inoculum containing 5 × 107 cells of each strain and incubation for 15 days in gnotobiotic culture. None of these gfp-tagged test strains could be cultured from the surfaces of leaf sheaths and leaves before surface sterilization, ruling out bacterial contamination or ascending migration by external routes (e.g., via water splash or mechanical transfer mechanisms).
Only gfp-tagged Azorhizobium caulinodans ORS571 was recovered from within surface-sterilized leaves (103.26 CFU/gfw) at 15 dpi, indicating that this test strain of rhizobia was efficient in endophytic ascending migration within rice, a finding consistent with its ability to infect other nonlegume plants (33). The second plating experiment conducted at 15 dpi indicated that a higher inoculum (108 cells/plant) of S. meliloti 1021 could compensate for its slower rate of endophytic ascending migration into rice leaf sheaths and leaves.
The third plating experiment examined the population dynamics of gfp-tagged A. caulinodans ORS571 and S. meliloti 1021 within various rice tissues after inoculation of their roots in gnotobiotic culture. The results with A. caulinodans ORS571 (Fig. 3) were similar to those with S. meliloti 1021 (not shown), indicating transient growth followed by maintenance of persistent or slightly declining populations on root surfaces and within leaf sheaths and leaves.
FIG. 3.
Population dynamics of gfp-tagged A. caulinodans ORS571 in various rice tissues after inoculation of roots and growth in gnotobiotic culture. Tissue samples were: surface-sterilized roots (r2) and roots (r1), leaf sheaths (s), and leaves (l) that had not been surface sterilized. gfp-tagged and antibiotic-resistant bacteria could not be cultured from the surfaces of the latter two sources of sampled tissues. The datum points and bars are means and standard errors of the mean from three replicates at each sampling time.
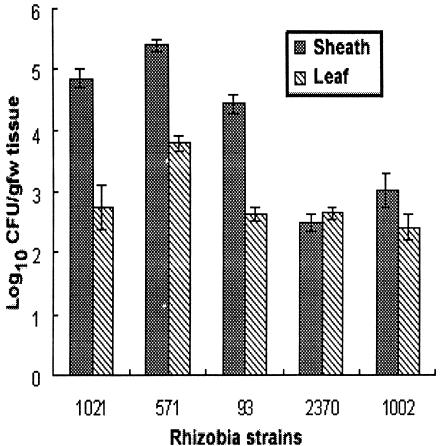
The fourth plating experiment examined the persistence of viable populations of endophytic rhizobia within rice plants grown in pots outdoors for a more extended period of its life cycle. All five gfp-tagged rhizobial test strains inoculated into the rhizosphere of rice were recovered from within surface-sterilized leaf sheaths and in the third to fifth leaves of these plants even at the mature heading stage in reproductive development (Fig. 4). No colonies of the gfp-tagged inoculant strains were recovered from surface-sterilized and macerated flag or ear leaves that developed at higher locations on the mature rice plants.
FIG. 4.
Culturable population densities of gfp-tagged S. meliloti 1021, A caulinodans ORS571, M. huakuii 93, R. leguminosarum USDA 2370, and S. meliloti 1002 within large tissue samples of leaf sheaths and leaves of rice plants grown in open potted soil plus vermiculite (1:1 ratio) and harvested at 125 dpi. The data reported are the means ± the standard errors of the mean from three tissue sample replicates plated on TY media.
Random picks of isolated rhizobial colonies from the above-ground surface-sterilized tissues of the inoculated rice plants were able to nodulate the homologous host legume again, and individual cells fluoresced green (indicative of gfp expression) when examined by fluorescence microscopy (not shown). Furthermore, PCR analysis also detected the gfp and 16S rRNA genes from colonies of the same rhizobial inoculant strains isolated from within these above-ground rice tissues. This detection of the same genetic markers (plus antibiotic resistance) in isolates obtained from the aerial plant tissues fulfills the criteria of Koch's postulates indicating that this disseminating colonization pattern in rice can be accomplished by several species of rhizobia.
These results of population studies in planta are consistent with a dynamic process of disseminating, endophytic colonization by rhizobia in rice that begins on and then within roots, followed by their ascending migration from the root interior up into the leaf sheath and leaves where they persist and transiently grow, respectively.
The significantly higher population densities of rhizobia in planta when calculated from in situ image analysis compared to viable plate counts raises some pertinent issues. First, it illustrates the effect of significantly reducing the sample “noise” (i.e., uninfected host tissue) when calculating in situ local population density by direct microscopy/image analysis in contrast to viable plate counts. Second, it raises the interesting possibility that the rhizobia remain active but some may transform to a nonculturable state within the rice tissues. Such transformations are profound during bacteroid formation in the Rhizobium-legume root-nodule symbiosis. The magnitude of these population dynamics in rice are predictably influenced by an expanding growth of plant tissue inaccessible to the rhizobia, survival/(in)compatibility of the endophytic rhizobia amid plant defenses and other stress responses in planta, and maintenance of metabolic activity (e.g., gfp expression and phytohormone production or induction) despite concurrent loss of culturability (3, 18, 27, 29, 30). More studies, similar to the one that used transmission electron microscopy to define the “endophytic” state of the rhizobia within the rice root cells (29), are needed to establish how close to an “endosymbiotic” state and for how long do the rhizobia associate with the above-ground rice plant tissues.
Influence of rhizobial inoculation on growth physiology of rice.
The ability of the gfp-tagged rhizobia to influence the vegetative growth, photosynthetic activity, and reproductive capacity of rice was assessed on plants grown outdoors in potted soil under ambient conditions. All of the plants inoculated with certain strains grew better than did the uninoculated control plants. Among the five test strains, Azorhizobium caulinodans ORS 571, Sinorhizobium meliloti 1021, and Mesorhizobium huakui 93 enhanced rice growth more than did the other two test strains (Table 3). Consistent with previous studies (2-5, 15, 17, 22, 23, 26-27, 29, 37, 38), inoculation of rice with these test strains of gfp-tagged rhizobia evoked positive growth responses such as significantly increased root volume, shoot dry weight, shoot height, shoot N content, and grain yield compared to uninoculated control plants (Table 3). Likewise, rice plants inoculated with certain test strains had significantly increased surface areas of flag leaves (considered to have the highest photosynthetic efficiency), net photosynthetic rate, stomatal conductance, transpiration velocity, and water utilization efficiency (Table 4), indicating that rhizobial inoculation of rice can also evoke physiological responses resulting in increased photosynthetic capacity and resistance to drought, even when the bacteria cannot be cultured from within these same plant tissues.
TABLE 3.
Growth responses of potted rice plants 160 days after inoculation with various gfp-tagged strains of wild-type rhizobiaa
| Rhizobium test strain | Total plant root vol/pot (cm3) ± SE | Shoot dry wt/pot (g) ± SE | Plant height/shoot (cm) ± SE | Total shoot N/pot (g) ± SE | % Shoot N/pot ± SE | Grain yield/pot (g) ± SE |
|---|---|---|---|---|---|---|
| Ac-ORS571 | 210 ± 36A | 63 ± 2A | 79.59 ± 5.67A | 0.7020 ± 0.0368AB | 1.11 ± 0.03B | 86 ± 5A |
| Sm-1021 | 180 ± 26A | 67 ± 5A | 81.67 ± 8.29A | 0.8235 ± 0.0653A | 1.23 ± 0.08AB | 86 ± 4A |
| Sm-1002 | 168 ± 8AB | 52 ± 4BC | 75.70 ± 5.88B | 0.6805 ± 0.0932AB | 1.30 ± 0.07AB | 61 ± 4B |
| Rl-2370 | 175 ± 23A | 61 ± 8AB | 76.35 ± 6.84B | 0.8569 ± 0.1239A | 1.40 ± 0.07A | 64 ± 9B |
| Mh-93 | 193 ± 16A | 67 ± 4A | 75.87 ± 8.00B | 0.7962 ± 0.0148A | 1.19 ± 0.02B | 77 ± 5A |
| Control | 130 ± 10B | 47 ± 6C | 66.06 ± 9.32C | 0.6047 ± 0.0900B | 1.28 ± 0.08AB | 51 ± 4C |
Data are represented as means per pot from three pot replicates, each containing 15 transplanted plants per strain. Values followed by a different superscript capital letter are significantly different at the 95% confidence level according to Duncan's multiple-range test.
TABLE 4.
Parameters of photosynthetic rate, stomatal conductance, CO2 concentration, transpiration velocity, water utilization efficiency, and morphological change in flag leavesa
| Rhizobium test strain | Net photosynthetic rate (μmol of CO2 m−2 s−1) ± SE | Stomatal conductance (μmol of H2O m−2 s−1) ± SE | CO2 concn (μmol mol−1) within intercellular leaf spaces ± SE | Transpiration velocity (μmol of H2O m−2 s−1) ± SE | Water utilization efficiency (net photosynthetic rate/transpiration velocity) ± SE | Area (cm2) of flag leaf (S = 0.75 × length × width) ± SE | Length/width of flag leaves ± SE |
|---|---|---|---|---|---|---|---|
| Ac-ORS571 | 16.4223 ± 1.3903A | 0.2402 ± 0.0225A | 231.5825 ± 6.8692B | 4.5299 ± 0.3397A | 3.6264 ± 0.1715BC | 17.6437 ± 4.9422ABC | 14.8490 ± 3.0377D |
| Sm-1021 | 14.9850 ± 1.6388B | 0.2180 ± 0.0374AB | 221.3119 ± 11.5020B | 3.7224 ± 0.3272BC | 4.0222 ± 0.1922AB | 20.0250 ± 3.9161A | 16.1600 ± 1.6222CD |
| Sm-1002 | 13.7036 ± 0.7259B | 0.1941 ± 0.0173B | 225.3235 ± 19.0891B | 3.3079 ± 0.2186C | 4.1549 ± 0.3220A | 19.5789 ± 4.4695AB | 18.3021 ± 2.0242B |
| Rl-2370 | 13.8499 ± 0.3768B | 0.2408 ± 0.0403A | 261.7970 ± 15.5635A | 4.1640 ± 0.4180AB | 3.3584 ± 0.4119C | 18.9789 ± 4.4902AB | 17.6614 ± 2.6436BC |
| Mh-93 | 13.8633 ± 0.7615B | 0.2187 ± 0.0169AB | 252.8500 ± 8.0899A | 4.3762 ± 0.2911AB | 3.1778 ± 0.2521CD | 16.7857 ± 3.4302BC | 16.4579 ± 2.6239CD |
| Control | 10.2302 ± 1.0323C | 0.1899 ± 0.0324B | 262.6345 ± 24.6411A | 3.8537 ± 0.9159BC | 2.7688 ± 0.6929D | 15.2362 ± 4.0063C | 21.1538 ± 2.1794A |
Data are represented as means per pot from three pot replicates, each containing 15 transplanted plants per strain. Values followed by a different superscript capital letter are significantly different at the 95% confidence level according to Duncan's multiple-range test.
These results extend the list of benefits to rice after inoculation of their rhizospheres with certain strains of rhizobia by further revealing various additional indexes of growth physiology that are enhanced by this endophytic association. We plan to conduct additional studies (e.g., in situ host gene expression activated by bacterial quorum sensing cell-to-cell communication events plus proteomic/metabolomic analysis of the plant-microbe interactions) to explain why the magnitude of plant growth responses to some inoculant test strains varied significantly.
Influence of endophytic rhizobia on levels of growth-regulating phytohormones in rice tissues.
Rice-adapted isolates of rhizobia have previously been shown to produce the growth-regulating phytohormones IAA and GA in pure culture and increase IAA levels accumulated externally in root exudate of gnotobiotically cultured rice plants (2, 3, 38). Our finding that endophytic rhizobia disseminate from below-ground into above-ground rice tissues prompted us to examine whether the levels of growth-regulating phytohormones within these tissues are influenced by the internal, ascending rhizobia. High-pressure liquid chromatography analyses indicated elevated levels of IAA and GA3 extracted from the leaf sheaths and leaves of rice plants whose rhizospheres were inoculated with S. meliloti 1021 and A. caulinodans ORS571, respectively (Table 5). We predict that this rhizobium-induced elevation of the levels of these growth-stimulating phytohormones within the above-ground rice tissues contributes to the underlying mechanism(s) allowing certain strains of these bacteria to enhance vegetative and reproductive growth of cereals in general (2-4, 17, 22, 23, 37, 38).
TABLE 5.
High-pressure liquid chromatography analysis of phytohormones accumulated within rice tissues 20 dpi with rhizobia in gnotobiotic culture (values based on three replicates)a
| Inoculant test strain | Mean GA3 (mg/100 gfw) ± SE
|
Mean IAA (mg/100 gfw) ± SE
|
||
|---|---|---|---|---|
| Roots | Leaf sheaths and leaves | Roots | Leaf sheaths and leaves | |
| S. meliloti 1021 | 1.13 ± 0.46A | 2.79 ± 0.69B | 0.06 | 0.55 ± 0.06A |
| A. caulinodans ORS571 | 0.75 ± 0.32B | 7.90 ± 1.57A | 0.07 | 0.41 ± 0.08B |
| Uninoculated control | 0.46 ± 0.16B | 2.37 ± 0.43B | 0 | 0.24 ± 0.13C |
IAA was detected in only one of three plant root replicates inoculated with S. meliloti 1021. Values followed by a different superscript capital letter are significantly different at the 95% confidence level according to Duncan's multiple-range test.
One of us (F.B.D.) recently developed new computing tools to examine the connectivity between various parameters that can be measured by image analysis, e.g., local clustered density of bacteria, with their in situ distribution, and to apply this information to geostatistical tests for spatial dependence by modeling the patterns of microbial distribution during their colonization of surfaces (8). In cases where the local density of the bacteria fulfills the criterion of spatial autocorrelation (statistical connectivity to spatial location), the resultant “best-fit” semivariogram model can be applied to statistically defendable, predictive kriging interpolation methods to map the influence this regionalized variable has on colonization by neighboring microbes over the continuous spatial domain of interest, even in areas not sampled (25, 31). We applied this powerful geostatistical approach to analyze the local density of endophytic rhizobia colonized within the leaf sheath of rice above the stem base and found it to be spatially dependent and autocorrelated for neighboring microbes separated by a distance of up to approximately 80 μm. This maximal separation distance defines the spatial scale whereby “bacteria can influence their nearest neighbors' spatial distribution.” Interestingly, this maximal radius of influence is remarkably similar in length to the maximal “in situ calling distances” (∼78 μm) that allow individual cells of Pseudomonas rhizobacteria to communicate with each other via N-acyl homoserine lactone signal molecules during their colonization of plant roots (14). A kriging interpolation map was produced from the geostatistical model that best fits the autocorrelation data on these density-dependent activities affecting rhizobial colonization patterns over the same spatial domain within the leaf sheath of rice (Fig. 5A and B). This pseudocolored kriging map provides a vivid prediction of diffusion gradients of potent bioactive metabolites (e.g., phytohormones and communication signal molecules) produced by these endophytic bacteria in situ, based on the assumption that each bacterium contributes equally to their production (either directly or indirectly) in planta, and hence, gradients would be stronger in areas surrounding higher local densities of organisms.
FIG. 5.
Distribution of gfp-tagged S. meliloti 1021 cells within a section of the rice leaf sheath (A) and a two-dimensional kriging map of their geostatistically autocorrelated, local spatial density (B).
Summary and conclusions.
It is clear from this and earlier studies that rhizobia possess traits that allow them to associate beneficially with a wide diversity of host plants, not only in the classic legume root-nodule symbiosis but also in the endophytic association with cereal plants. This study shows that various species and strains of rhizobia can not only use “crack entry” at lateral root emergence as the means to gain access into and colonize the interior of rice roots but also utilize a dynamic infection process that permits them to migrate endophytically upward into the stem base, leaf base, leaf sheaths, and some leaves of the plant. There they grow transiently to high local populations that are metabolically active, and some rhizobia persist throughout the vegetative and into the reproductive phases of development. Such intimate interactions elevate phytohormone levels within the rice tissues, and also a variety of beneficial responses ensue, impacting significantly on the growth physiology of the rice plant. These new findings indicate that the natural, endophytic Rhizobium-rice association is far more complex, inclusive, dynamic, and invasive than previously thought, therefore heightening its status as an exciting experimental research model of beneficial plant-bacterium interactions and its potential value for future exploitation as a biofertilizer strategy in sustainable agriculture to produce the world's most important cereal crops for human nutrition.
Acknowledgments
We thank Y. M. Liu. for technical assistance.
Portions of this work were supported by National Key Program no. 2001CB108903 to Y.-X.J., the Center for Microbial Ecology and Long-Term Ecological Research programs at Michigan State University for F.B.D., and project BIO5-001-015 of the US-Egypt Science and Technology Program to F.B.D. and Y.G.Y.
REFERENCES
- 1.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 2.Biswas, J. C., J. K. Ladha, and F. B. Dazzo. 2000. Rhizobia inoculation improves nutrient uptake and growth in lowland rice. Soil Sci. Soc. Am. J. 64:1644-1650. [Google Scholar]
- 3.Biswas, J. C., J. K. Ladha, F. B. Dazzo, Y. G., Yanni, and B. G. Rolfe. 2000. Rhizobial inoculation influences seedling vigor and yield of rice. Agron. J. 92:880-886. [Google Scholar]
- 4.Chabot, R. H., H. Antoun, J. Kloepper, and C. Beauchamp. 1996. Root colonization of maize and lettuce by bioluminescent Rhizobium leguminosarum biovar phaseolus. Appl. Environ. Microbiol. 62:2767-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaintreuil, C., E. Giraud, Y. Prin, J. B. Lorquin, M. Gillis, P. de Laudie, and B. Dreyfus. 2000. Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl. Environ. Microbiol. 66:5437-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi, F., S. H. Shen, H. P. Cheng, Y. X. Jing, and F. B. Dazzo. 2004. The migration of rhizobia inside rice roots, p. 40, abstr. L40. In Y. Wang (ed.), Proceedings of the 14th International Congress on Nitrogen Fixation. International Congress on Nitrogen Fixation, Beijing, China.
- 8.Dazzo, F. B. 2004. Applications of quantitative microscopy in studies of plant surface microbiology, p. 503-550. In A. Varma, L. Abbott, D. Werner, and R. Hampp (ed.), Plant surface microbiology. Springer-Verlag, Berlin, Germany.
- 9.Ditta, G., S. Stanfiels, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, Y., A. L. Iniguez, B. M. Ahmer, and E. W. Triplett. 2003. Kinetics and strain specificity of rhizosphere and endophytic colonization by enteric bacteria in seedlings of Medicago sativa and Medicago truncatula. Appl. Environ. Microbiol. 69:1783-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durley, H. C., T. Kannangara, and G. M. Simpson. 1982. Leaf analysis for abscisic, phaseic, and 3-indolyacetic acids by high-performance liquid chromatography. J. Chromatogr. 236:181-188. [Google Scholar]
- 12.Endre, G., A. Kereszt, Z. Kevei, S. Mihacea, P. Kalo, and G. B. Kiss. 2002. A receptor kinase gene regulating symbiotic nodule development. Nature 417:962-966. [DOI] [PubMed] [Google Scholar]
- 13.Gage, D. J., T. Bobo, and S. R. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 178:7159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gantner, S., M. Schmid, C. Durr, R. Schulegger, A. Steidle, P. Hutzler, C. Langebartels, L. Eberl, A. Hartmann, and F. B. Dazzo. 2004. In situ calling distances and high population-independent rhizobacterial cell-to-cell communication, p. 95, abstr. PAS7/30. In A. Hartmann (ed.), International Congress Rhizosphere 2004: perspectives and challenges—a tribute to Lorenz Hiltner. International Congress Rhizosphere, Munich, Germany.
- 15.Gutierrez-Zamora, M., and E. Martinez-Romero. 2001. Natural endophytic association between Rhizobium etli and maize (Zea mays). J. Biotechnol. 91:117-126. [DOI] [PubMed] [Google Scholar]
- 16.Gyaneshwar, P., E. K. James, N. Mathan, P. M. Reddy, B. Reinhold-Hurek, and J. K. Ladha. 2001. Endophytic colonization of rice by a diazotrophic strain of Serratia marcescens. J. Bacteriol. 183:2634-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilali, A., D. Prevost, W. Broughton, and H. Antoun. 2001. Effets de l'inoculation avec des souches de Rhizobium leguminosarum biovar trifolii sur la croissance der ble dans deux sols der Maroc. Can. J. Microbiol. 41:590-593. [PubMed] [Google Scholar]
- 18.Iniguez, A. L., Y. Dong, H. Carter, B. Almer, J. Stone, and E. W. Triplett. 2005. Regulation of endophytic bacterial colonization by plant defenses. Mol. Plant-Microbe Interact. 18:169-178. [DOI] [PubMed] [Google Scholar]
- 19.Li, J.-C., S. Jing, X.-I. Zhao, G. Wang, H.-F. Yu, and Y.-J. Ren. 1994. Separation and determination of three kinds of plant hormone by high-performance liquid chromatography. Chinese J. Anal. Chem. 22:801-804. [Google Scholar]
- 20.Liu, J., F. B. Dazzo, B. Yu, O. Glagoleva, and A. Jain. 2001. CMEIAS: a computer-aided system for the image analysis of bacterial morphotypes in microbial communities. Microb. Ecol. 41:173-194. [DOI] [PubMed] [Google Scholar]
- 21.Long, S. R. 2001. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 125:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupway, N., G. Clayton, K. Hanson, W. Rice, and V. Bierderbeck. 2004. Endophytic rhizobia in barley, wheat, and canola roots. Can. J. Plant Sci. 84:37-45. [Google Scholar]
- 23.V. Matiru, M. A. Jaffer, and F. D. Dakora. 2000. Rhizobial colonization of roots of African landraces of sorghum and millet and the effects of sorghum growth and P nutrition, p. 99-100. In Proceedings of the 4th Congress of the African Association for Biological Nitrogen Fixation: imperatives for BNF research and application in Africa for the 21st century. African Association for Biological Nitrogen Fixation, Nairobi, Kenya.
- 24.Mundt, J. O., and N. F. Hinkle. 1976. Bacteria within ovules and seeds. Appl. Environ. Microbiol. 32:694-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer, M. W. 2003. Scale detection using semivariograms and autocorrelograms, p. 129-144. In S. Gergel, and M. Turner (ed.), Learning landscape ecology. Springer, New York, N.Y.
- 26.Peng, S. B., J. C. Biswas, J. K. Ladha, P. G. Yaneshwar, and Y. Chen. 2002. Influence of rhizobial inoculation on photosynthesis and grain yield of rice. Agron. J. 94:925-929. [Google Scholar]
- 27.Prayitno, J., J. Stefaniak, J. McIver, J. Weinman, F. B. Dazzo, J. K. Ladha, W. Baraquio, Y. G. Yanni, and B. G. Rolfe. 1999. Interactions of rice seedlings with nitrogen-fixing bacteria isolated from rice roots. Austr. J. Plant Physiol. 26:521-535. [Google Scholar]
- 28.Reddy, C. K., and F. B. Dazzo. 2004. Computer-assisted segmentation of bacteria in color micrographs. Microsc. Anal. 18:5-7. [Google Scholar]
- 29.Reddy, P. M., J. K. Ladha, R. So, R. Hernandez, F. B. Dazzo, O. R. Angeles, M. C. Ramos, and F. de Bruijn. 1997. Rhizobial communication with rice: induction of phenotypic changes, mode of invasion, and extent of colonization in roots. Plant Soil 194:81-99. [Google Scholar]
- 30.Reinhold-Hurek, B., and T. Hurek. 1998. Life in grasses: diazotrophic endophytes. Trends Microbiol. 6:139-144. [DOI] [PubMed] [Google Scholar]
- 31.Robertson, G. P. 2004. GS+ geostatistics for the environmental sciences. Gamma Design Software, Plainwell, Mich.
- 32.Stolzfus, R., So, J. R., P. P. Malarvithi, J. K. Ladha, and F. J. de Bruijn. 1997. Isolation of endophytic bacteria from rice and assessment of their potential for supplying rice with biologically fixed nitrogen. Plant Soil 194:25-36. [Google Scholar]
- 33.Stone, P. J., K. J. O'Callaghan, M. R. Davey, and E. C. Cocking. 2001. Azorhizobium caulinodans ORS571 colonizes the xylem of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 14:93-97. [DOI] [PubMed] [Google Scholar]
- 34.Stracke, S., G. Kistner, S. Yoshida, L. Mulder, S. Sato, R. Kaneko, S. Tabata, N. Sandal, J. Stougaard, K. Szczyglowski, and M. Parniske. 2002. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417:959-962. [DOI] [PubMed] [Google Scholar]
- 35.Verkhusha, V. V., I. M. Kuznetsova, O. V. Stepanenko, A. G. Zaraisky, M. M. Shavlovsky, K. K. Turoverov, and V. N. Uversky. 2003. High stability of Discosoma DsRed as compared to Aequorea EGFP. Biochemistry 42: 7879-7884. [DOI] [PubMed] [Google Scholar]
- 36.Weidner, S., A. Puhler, and H. Kuster. 2003. Genomics insight into symbiotic nitrogen fixation. Cur. Opin. Biotechnol. 14:200-205. [DOI] [PubMed] [Google Scholar]
- 37.Yanni, Y. G., R. Y. Rizk, V. Corich, A. Squartini, K. Ninke, S. Philip-Hollingsworth, G. Orgambide, F. de Bruijn, R. Stoltzfus, D. Buckley, T. Schmidt, P. F. Mateos, J. K. Ladha, and F. B. Dazzo. 1997. Natural endophytic association between Rhizobium leguminosarum bv. trifolii and rice roots and assessment of its potential to promote rice growth. Plant Soil 194:99-114. [Google Scholar]
- 38.Yanni, Y. G., R. Y. Rizk, F. Abd El-Fattah, A. Squartini, V. Corich, A. Giacomini, F. de Bruijn, J. Rademaker, J. Maya-Flores, P. Ostrom, M. Vega-Hernandez, R. I. Hollingsworth, E. Martinez-Molina, P. Mateos, E. Velazquez, J. Wopereis, E. Triplett, M. Umali-Garcia, J. A. Anarna, B. G. Rolfe, J. K. Ladha, J. Hill, R. Mujoo, P. K. Ng, and F. B. Dazzo. 2001. The beneficial plant growth-promoting association of Rhizobium leguminosarum biovar trifolii with rice roots. Austr. J. Plant Physiol. 62:845-870. [Google Scholar]