Abstract
4"-Oxo-avermectin is a key intermediate in the manufacture of the agriculturally important insecticide emamectin benzoate from the natural product avermectin. Seventeen biocatalytically active Streptomyces strains with the ability to oxidize avermectin to 4"-oxo-avermectin in a regioselective manner have been discovered in a screen of 3,334 microorganisms. The enzymes responsible for this oxidation reaction in these biocatalytically active strains were found to be cytochrome P450 monooxygenases (CYPs) and were termed Ema1 to Ema17. The genes for Ema1 to Ema17 have been cloned, sequenced, and compared to reveal a new subfamily of CYPs. Ema1 to Ema16 have been overexpressed in Escherichia coli and purified as His-tagged recombinant proteins, and their basic enzyme kinetic parameters have been determined.
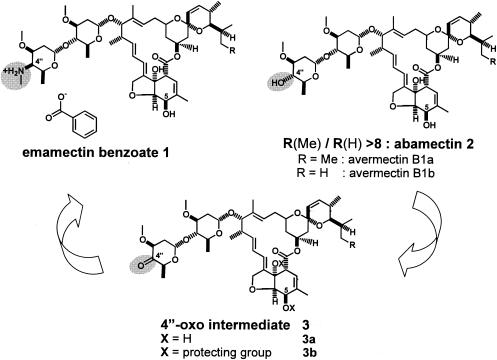
Emamectin benzoate (MK-244) (Fig. 1, compound 1) is an extremely potent insecticide that controls many agriculturally important pests such as thrips, leaf miners, and worm pests including the beet armyworm, corn earworm, cutworms, and diamondback moth. It is chemically synthesized from the natural product mixture abamectin 2 in a series of four steps (11). To achieve widespread commercial exploitation of emamectin benzoate 1, the synthetic conversion from abamectin 2 needs to be economical. A critical step in the synthesis described by Cvetovich et al. (11) is the oxidation of the 4"-carbinol group of abamectin 2 to the 4"-oxo intermediate 3. To our knowledge, all attempts to obtain an economical and safe regioselective oxidation process from 2 to the 4"-oxo intermediate 3a have failed so far, due to the high reactivity of the allylic secondary alcohol in position 5.
FIG. 1.
(Bio)synthetic conversion of abamectin to emamectin benzoate.
The problem of regioselective oxidation was circumvented by selectively protecting the hydroxyl groups of the macrolactone prior to the oxidation to the 4"-oxo intermediate 3b (11). Obviously, the protection groups need to be removed en route to emamectin benzoate 1. This protection-deprotection strategy leads to high production costs. Therefore, with a more economical production process in mind, it seemed worthwhile to resume the search for a suitable reagent capable of oxidizing abamectin 2 regioselectively to the nonprotected 4"-oxo intermediate 3a, and this time we planned to focus on microbial oxidation systems.
Microbial oxidations have been well established in a number of industrial processes (1, 30) with one of the earliest examples being the biological production of L-ascorbic acid (27). l-Sorbose, the former starting material for l-ascorbic acid, was later elegantly synthesized by Anderson et al. from d-glucose, involving a regioselective oxidation of the intermediate d-sorbitol by a genetically modified biocatalyst (3). Microbial oxidation of 1-methyl-pyranosides was found to yield several of the corresponding 4-oxo derivatives (31). However, none of the hexoses tested were oxidized, perhaps because the Acetobacter enzyme system employed in this study cannot cope with large substituents in position 3 and/or 5. Therefore, it seemed unlikely that Acetobacter would oxidize the outer oleandrose of the di-oleandrose side chain of abamectin 2.
Bernaerts and de Ley have shown that some disaccharides can be converted to their 3-oxo-derivatives by bacterial oxidation (6); more recently, Noll-Borchers and Buchholz elaborated on practical aspects of this reaction, using isomaltulose as a substrate and resting cells of Agrobacterium tumefaciens as a biocatalyst (25). The authors reported yields of 3-oxo-isomaltulose of up to 90% at a substrate concentration of 50 g/liter and a biocatalyst concentration (bacterial wet weight) of 50 g/liter.
Although none of the microbial biocatalytic systems described in the literature seemed to provide a solution to our problem, their excellent regioselectivity and remarkable practicability encouraged us to search on a broad basis for new microorganisms that would oxidize abamectin 2 in the desired regioselective manner. The present article describes the search for enzymes capable of oxidizing the 4"-position of avermectin; the identification of 17 Streptomyces strains that catalyze this biotransformation; the discovery that the biocatalytically active enzymes are bacterial P450 monooxygenases; the cloning, sequencing, and analysis of the corresponding genes (ema1 to ema17); and the isolation and biochemical characterization of 16 His-tagged biocatalytically active Ema enzymes. The accompanying article (20) describes the expression of the genes for Ema1 and Ema2 in Streptomyces and Pseudomonas hosts and the optimization of the whole-cell biocatalytic conversion reaction by using different promoters, vectors, gene versions, and cloned ferredoxins and ferredoxin reductases.
MATERIALS AND METHODS
Analytical procedures.
Chromatographic analyses of avermectin and its derivatives were carried out on a Merck-Hitachi HPLC using a 125- by 4-mm Kromasil C18 column (100 Å, 5 μm; Macherey-Nagel Inc., Easton, PA), using a linear gradient from 50% CH3CN to 100% CH3CN within 20 min at a flow rate of 1.5 ml/min. Compounds were detected at 243 nm. The retention times of avermectin derivatives under these conditions were: 10.0 min, 4"-oxo-avermectin B1a ketohydrate and desoleandrosyl-avermectin B1a; 13.0 min, avermectin B1a; 15.8 min, 5-oxo-avermectin B1a; and 16.1 min, 4"-oxo-avermectin B1a.
Microbial screens.
For assays with liquid cultures, starter cultures of Actinomycetes organisms were grown in Erlenmeyer flasks with one baffle for 3 days at 28°C in 100 ml ISP-2 medium (Difco, Becton Dickinson, Sparks, MD) with shaking at 120 rpm. Main cultures were grown for 3 days in 100 ml ISP-2 medium supplemented with technical-grade abamectin (final concentration, 1 g/liter). (The stock solution is 20 g/liter abamectin in dimethyl sulfoxide [DMSO]-Tween-40 = 1:1. Abamectin is the natural mixture of 85% avermectin B1a and 15% avermectin B1b as produced by Streptomyces avermitilis). Fungal cultures were cultivated similarly but at 25°C with shaking at 200 rpm in Erlenmeyer flasks without baffles. Gram-negative bacteria were cultivated in Luria broth (Difco, Becton Dickinson) for 24 h for both the starter and the main cultures. Ten milliliters of each main culture was extracted with 10 ml methyl-t-butyl ether (MTBE), and the ether phases were collected and evaporated in vacuo. The residues were redissolved in 1.2 ml acetonitrile, and the formation of avermectin derivatives was followed by high-performance liquid chromatography (HPLC).
For assays with agar surface cultures, all microorganisms were grown for 7 days at 28°C in standard 24-well plates on ISP-2 medium supplemented with 0.1 g/liter (final concentration) of technical-grade abamectin solution (see above). Each well was extracted with 1.5 ml acetonitrile, and the formation of avermectin derivatives was followed by HPLC.
Feasibility study for the production of 4"-oxo-avermectin.
Starter cultures of Streptomyces tubercidicus strain I-1529 were grown in ISP-2 medium at 28°C with shaking at 120 rpm for 3 days, and 2,000 ml of starter culture was used to inoculate 40 liters of growth medium (10 g/liter glucose, 10 g/liter malt extract, 10g/liter Pharmamedia, 1 g/liter meat extract, and 3 g/liter yeast extract) in a 50-liter Braun Biostat UD fermentor. The fermentor was set to the following parameters: stirring at 200 to 300 rpm, guided by pO2 set to a minimum of 25%, aeration at 0.75 vol/vol/min (30 liters/min), temperature 28°C. Mycelia were harvested by centrifugation after 2 days of growth; the typical yield was 70 to 80 g (wet cell)/liter.
A total of 88.0 g abamectin was dissolved in 2.5 liters of DMSO:Tween 40 at a 1:1 ratio, and 25-ml aliquots were distributed into 100 3-liter Erlenmeyer flasks, each containing 1 liter of potassium phosphate buffer (70 mM; pH 6.0). A total of 100 g (wet weight) of fresh cells was added to each flask (total volume, 1.125 liters containing 880 mg abamectin/liter = 750 mg avermectin/liter), and the reaction mixtures were incubated at room temperature with shaking at 120 rpm for 96 h. The reaction mixtures were centrifuged, and the supernatants were pooled and extracted three times with 1 volume equivalent of MTBE. The pooled MTBE phases were dried over anhydrous Na2SO4 and concentrated in vacuo to yield extract S.
The centrifuged cells were extracted in 800-g aliquots in 2-liter Erlenmeyer flasks. Approximately 90 g of diatomaceous earth (Hyflo Supercell; VWR, Darmstadt, Germany) and 1.2 liters of acetone were added to each Erlenmeyer flask. The mixtures were homogenized by being stirred for 1 h, and the resulting pulp was filtered and washed with acetone until colorless elution was achieved, to provide filtrate C1 and filter cake C1.
Filtrate C1 was concentrated in vacuo on the rotary evaporator until all acetone had been removed. The resulting aqueous phase was extracted three times with toluene, and the combined toluene phases were dried over anhydrous Na2SO4 and evaporated in vacuo to yield extract C1.
Filter cake C1 was extracted with 1.5 liters of toluene by being stirred for 1 h. The resulting pulp was filtered and washed with toluene until colorless elution was achieved, to yield filtrate C2 and filter cake C2. Filter cake C2 was discarded. Filtrate C2 was concentrated in vacuo to yield extract C2.
The extracts S, C1, and C2 were pooled, dried at high vacuum, purified on silica gel (Silica 32-63, 60 Å; Chemie Brunschwig AG, Basel, Switzerland), and eluted with ethyl acetate-hexane (at a 3:2 ratio) to yield 10.5 g of 4"-oxo-avermectin (14.0% isolated yield).
Bacterial strains and plasmids.
Streptomyces lividans ZX7 (17) was received from the John Innes Centre (Norwich, United Kingdom). Biocatalytically active Streptomyces strains and their strain collection sources are listed in Table 1. Escherichia coli DH10B was used for routine cloning. E. coli BL21(DE3) (Stratagene, La Jolla, CA) was used for expressing His-tagged proteins.
TABLE 1.
Biocatalytically active strains and CYP enzymes identified in this studya
| CYP identified
|
Producer organism
|
||||
|---|---|---|---|---|---|
| Common name | Assigned name | GenBank no. | Species | Strain | Strain collection |
| Ema1 | CYP107Z12 | AY545995 | S. tubercidicus | R-922 | SYN |
| Cyp229 | CYP105B2 | AY549203 | S. tubercidicus | R-922 | SYN |
| Cyp230 | CYP105S1 | AY549204 | S. tubercidicus | R-922 | SYN |
| CypLC | CYP107L4 | AY549202 | S. tubercidicus | R-922 | SYN |
| Ema2 | CYP107Z10 | AY549181 | S. tubercidicus | I-1529 | SYN |
| Cyp233 | CYP105D8 | AY549200 | S. tubercidicus | I-1529 | SYN |
| Cyp234 | CYP105S2 | AY549201 | S. tubercidicus | I-1529 | SYN |
| CypEA | CYP147C1 | AY549197 | S. tubercidicus | I-1529 | SYN |
| CypLA | CYP107L3 | AY549198 | S. tubercidicus | I-1529 | SYN |
| CypLB | CYP185A1 | AY549199 | S. tubercidicus | I-1529 | SYN |
| Ema3 | CYP107Z2v2 | AY549182 | Streptomyces rimosus subsp. paromyceticus | 1053 | SYN |
| Ema4 | CYP107Z5v3 | AY549183 | S. lydicus | R-401 | SYN |
| Ema5 | CYP107Z6 | AY549184 | Streptomyces sp. | I-1525 | SYN |
| Ema6 | CYP107Z5v2 | AY549185 | Streptomyces saraceticus | DSM-40241 | DSMZ |
| Ema7 | CYP107Z3 | AY549186 | Streptomyces sp. | IHS-0435 | SYN |
| Ema8 | CYP107Z2v1 | AY549187 | Streptomyces albofaciens | C-0083 | SYN |
| Ema9 | CYP107Z11 | AY549188 | Streptomyces platensis | NRAA-7479 | SYN |
| Ema10 | CYP107Z5v3 | AY549189 | Streptomyces kasugaensis | A/96 | SYN |
| Ema11 | CYP107Z1 | AY549190 | S. rimosus subsp. paromyceticus | R-2374 | SYN |
| Ema12 | CYP107Z9 | AY549191 | S. tubercidicus | NRAA-7027 | SYN |
| Ema13 | CYP107Z8 | AY549192 | S. platensis | Tue-3077 | SYN |
| Ema14 | CYP107Z10 | AY549193 | S. platensis | I-1548 | SYN |
| Ema15 | CYP107Z5v1 | AY549194 | S. lydicus | NRRL-2433 | NRRL |
| Ema16 | CYP107Z4 | AY549195 | S. lydicus | NRAB-0114 | SYN |
| Ema17 | CYP107Z7 | AY549196 | S. tubercidicus | DSM-40261 | DSMZ |
Strain collections: DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany); NRRL, Northern Regional Research Laboratory (Peoria, IL); SYN, Syngenta in-house strain collection. CYP identified: common names, GenBank numbers, and assigned CYP nomenclature names (David R. Nelson, CYP Nomenclature Committee) of the CYP enzymes whose encoding genes were isolated from the given strain in this study.
The Streptomyces expression vector pTBK (34) carries the thiostrepton-inducible promoter PtipA (23), the φC31-derived integrase (18), and the tsr thiostrepton resistance and the neo (aphII) kanamycin resistance genes (17). Plasmid pGM160 (24) was also used for gene library construction in Streptomyces. Plasmid pET28a (Novagen-EMD Biosciences, Madison, WI) was used for expressing His-tagged proteins.
General culture conditions and molecular genetic procedures.
E. coli strains were grown on 2× tryptone-yeast or LB medium with the appropriate antibiotics. Streptomyces strains were maintained on ISP-2 agar, supplemented with antibiotics as appropriate. Liquid Streptomyces cultures for DNA isolation were grown in YEME (17) or in peptone-yeast-glucose medium. Transformed Streptomyces strains for avermectin conversion assays were grown on ISP-2 agar plates with 1 g/liter (final concentration) technical-grade abamectin, supplemented with 50 μg/ml thiostrepton (pGM160 derivatives) or 50 μg/ml kanamycin (pTBK derivatives) for selection and 5 μg/ml thiostrepton for induction of the PtipA promoter (pTBK derivatives), as appropriate. E. coli transformations and recombinant DNA procedures followed standard protocols (29). DNA sequence analysis was performed using the dideoxynucleotide chain termination method on Applied Biosystems model 377 sequencers. The individual DNA sequencing runs were assembled using Sequencher, version 4.0 (Gene Codes Corp., Ann Arbor, MI), and the sequence was analyzed using the Vector NTI suite, version 7 (InforMax, Inc., Frederick, MD). S. lividans protoplasts were transformed as previously described (17).
Cloning procedures.
A genomic DNA library of S. tubercidicus I-1529 was constructed in the Streptomyces plasmid pGM160 (24) by partially cloning Sau3AI-digested genomic DNA of 7 to 9 kb in size. The S. tubercidicus I-1529 expression library was constructed by partially cloning Sau3AI-digested genomic DNA of approximately 4 kb in size into pTBK. Both clone banks were amplified in E. coli and transformed into S. lividans ZX7 for screening.
PCR amplification of cytochrome P450 monooxygenase (CYP)-encoding DNA fragments followed the protocol of Hyun et al. (15). The primers used are listed in Table 2. The 350-bp PCR-amplified fragments were cloned into pCR2.1-TOPO TA (Invitrogen, Carlsbad, CA). Approximately 150 individual clones were sequenced and found to represent eight unique CYP gene fragments from strain R-922 and seven unique fragments from I-1529. The DNA fragments cloned by PCR were used as hybridization probes against cosmid libraries of strains R-922 and I-1529 raised in SuperCos I (Stratagene, La Jolla, CA). Two unique CYP genes from strain R-922 (cyp229 and cyp230) and two genes from I-1529 (cyp233 and cyp234) were subcloned and completely sequenced.
TABLE 2.
PCR primers used in this study
| Primer | Primer sequence and the amino acid sequence to which it was designeda | Degeneracy |
|---|---|---|
| O2-binding domain primers (5′ to 3′) | ||
| OBD1 | I A G H E T T | 8 |
| ATC GCS GGS CAC GAG ACS AC | ||
| OBD2 | V A G H E T T | 16 |
| GTS GCS GGS CAC GAG ACS AC | ||
| OBD3 | L A G H E T T | 16 |
| CTS GCS GGS CAC GAG ACS AC | ||
| OBD4 | L L L I A G H E T | 32 |
| TS CTS CTS ATC GCS GGS CAC GAG ACb | ||
| Heme-binding domain primers (3′ to 5′) | ||
| HBD1 | H Q C L G Q N L A | 8 |
| GTG GTC ACG GAS CCS TGC TTG GAS CGb | ||
| HBD2 | F G H G V H Q C | 8 |
| AAG CCS GTG CCS CAS GTG GTC ACG | ||
| HBD3 | F G F G V H Q C | 8 |
| AAG GCS AAG CCS CAS GTG GTC ACG | ||
| HBD4 | F G H G I H Q C | 4 |
| AAG CCS GTG CCS TAG GTG GTC ACG | ||
| HBD5 | F G H G V H F C | 8 |
| AAG CCS GTG CCS CAS GTG AAG ACG | ||
| Ema1 forward primers (5′ to 3′) | ||
| 2aF | P G E D N V M | 64 |
| 5′-CCS GGS GAR CCS AAY GTS ATG-3′ | ||
| 2bF | A L I T D P F | 32 |
| 5′-GCS CTS ATY ACS GAC CCS TTC-3′ | ||
| 3F | F M D D S P V W | 32 |
| 5′-TTC ATG GAC GAC WSS CCS GTS TGG-3′ | ||
| 1F | L N Y D A P D H | 32 |
| 5′-CTS AAY TAY GAC GCS CCS GAC CAC-3′ | ||
| 7F | V E Q I A D A L | 32 |
| 5′-GTS GAR CAG ATY GCS GAC GCS CTS-3′ | ||
| Ema1 reverse primer (3′ to 5′) 5R | D L I S M D P D | 64 |
| 3′-CTG GAS TAR WSS TAC CTG GGS CTG-5′ |
The amino acid sequence is shown in the top line and the corresponding nucleotide sequence is shown below in the second line. Ambiguity codes: Y = C or T, R = A or G, S = C or G, W = A or T.
Primers described by Hyun et al. (15).
The I-1529-derived cypEA, cypLA, and cypLB and the R-922-derived cypLC genes were identified in the same cosmid libraries, using the epoF CYP gene (21) as a heterologous probe.
Degenerate PCR primers (Table 2) were designed against peptide sequences obtained from the wild-type Ema1 enzyme purified from S. tubercidicus strain R-922 (details of the enzyme purification procedures will be published elsewhere) and used with genomic DNA of S. tubercidicus R-922 as a template. Each of the five forward primers (2aF, 2bF, 3F, 1F, and 7F) was paired with the reverse primer (5R). In each reaction, a DNA fragment of the expected size was produced. The 580- and 600-bp PCR fragments generated by using primers 2bF and 5R and 2aF and 5R, respectively, were cloned into pCR-Blunt II-TOPO (Invitrogen, Carlsbad, CA). The cloned 600- and 580-bp fragments were identical in the 580 bp of sequence that they were expected to share. The 600-bp PCR fragment produced using primers 2aF and 5R was used as a hybridization probe against the cosmid library of strain R-922. The complete coding sequence of the ema1 gene was identified.
The ema1 gene was used as a heterologous probe in Southern hybridizations against several restriction digests of genomic DNA from the remaining 16 biocatalytically active Streptomyces strains. For each strain, there was only one strongly hybridizing DNA fragment in each digest, suggesting that other P450 genes were not detected under the high-stringency hybridization conditions. DNA in narrow size ranges that included the sizes of the ema1-hybridizing fragments was used to generate genomic sublibraries raised in pBluescript II KS(+) (Stratagene, La Jolla, CA). Colony hybridizations with the ema1 gene were used to identify clones containing ema1-homologous DNA fragments. The nucleotide sequence of the cloned DNA in each ema1-homologous clone was determined.
To express the Ema proteins with C-terminal His tags, the corresponding genes were cloned into NcoI-XhoI digested pET28a. Each individual ema gene was PCR amplified using primers that introduced an NcoI-compatible PciI site at the 5′ end and an XhoI site at the 3′ end of each open reading frame. The newly introduced PciI sites (ACATGT; start codon in boldface) replaced the start codons, while the XhoI sites (CTCGAG) replaced the stop codons of the native ema genes. This cloning strategy positions the start codon of each individual ema gene at an optimal distance from the ribosome-binding site on pET28a and appends a Leu-Glu linker, followed by a His tag, onto the C termini of each Ema protein.
Expression and purification of 16 His-tagged Ema proteins.
Transformed E. coli BL21(DE3) cells were grown at 37°C in 1 liter of LB broth supplemented with 5 mg/liter kanamycin. At an optical density at 600 nm of 0.5, 0.5 mM (final concentration) δ-amino-levulinic acid was added. At an optical density at 600 nm of 0.9, expression was induced by the addition of 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the culture was incubated at 28°C for 8 h. The cells were harvested by centrifugation, washed with 50 mM K2HPO4-KH2PO4 at pH 7.0, and used directly for enzyme isolation. The cell pellet obtained from 1 liter of culture was resuspended in 15 ml resuspension buffer (50 mM K2HPO4-KH2PO4 [pH 7.0], 5 mM benzamidine hydrochloride, 0.5 mM Pefabloc SC, 5 mM MgCl2, and 10% [vol/vol] glycerol), followed by disruption of the cells with a French pressure cell (SLM Instruments, Rochester, NY). The extract was clarified by centrifugation at 39,800 × g at 4°C for 45 min, and the supernatant was filtered with a 0.45-μm Millex-HV PVDF Durapore syringe filter (Millipore, Billerica, MA). The sample was loaded onto a HiTrap Chelating HP Affinity column (Amersham Bioscience, Piscataway, NJ) equilibrated with 0.5 M NaCl in 25 mM K2HPO4-KH2PO4, pH 7.2. Elution was performed with a linear gradient of 0 to 0.5 M imidazole in 25 mM K2HPO4-KH2PO4 (pH 7.2) and 0.5 M NaCl. The red-colored Ema-containing fractions eluted at 0.2 to 0.25 M imidazole. After concentration and buffer exchange on a 10-kDa Ultrafree-4 centrifugal filter unit (Millipore, Billerica, MA), the concentrated fractions were separated on a HiTrap Q anion-exchange column (Amersham Bioscience, Piscataway, NJ) equilibrated with 25 mM Tris-HCl, pH 7.5. Elution was performed with a linear gradient of 0.1 to 1 M KCl in 25 mM Tris-HCl, pH 7.5. The red-colored Ema-containing fractions eluted around 0.45 M KCl, and these fractions were pooled and concentrated. The purity of each Ema enzyme preparation exceeded 95%, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. Protein concentrations were determined by the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA) (7), with bovine gamma globulin (Bio-Rad Laboratories) as a standard. Glycerol was added to a final concentration of 50% to enhance storage stability at −20°C.
Kinetic characterization of purified His-tagged enzymes.
The activity of the purified His-tagged Ema enzymes was measured spectrophotometrically, following the consumption of NADPH at 340 nm at 30°C for 5 to 10 min. The assay mixture contained 50 to 300 μg of the enzyme dissolved in 50 mM K2HPO4-KH2PO4, pH 7.0, supplemented with 3.5 μM ferredoxin from Spinacia oleracea (Fluka, Buchs, Switzerland) and 40 mU/ml ferredoxin-NADP+-reductase from Spinacia oleracea (Sigma, St. Louis, MO). A total of 1 μl of the substrate avermectin B1a was added as a solution in isopropanol to final concentrations varying between 1.4 and 46 μM. Following preincubation of the mixture at 30°C for 5 min, the reaction was started by addition of NADPH to a final concentration of 0.2 mM. Kinetic parameters (Km and Vmax) were determined by hyperbolic regression using Hyper32 software (http://homepage.ntlworld.com/john.easterby/hyper32.html). The turnover numbers (kcat) of the purified enzymes were determined with the calculated average molecular weights of the respective enzymes.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the CYP genes identified in this study are listed in Table 1.
RESULTS
Screen for microorganisms capable of oxidizing the 4"-carbinol of avermectin.
Liquid cultures of 473 microbial strains (207 Actinomycetes bacteria, 203 non-Actinomycetes bacteria, and 63 fungi), and agar surface cultures of an additional 2,861 strains (2,039 Actinomycetes bacteria, 40 non-Actinomycetes bacteria, and 782 fungi) were screened for their ability to oxidize avermectin as described in Materials and Methods. Among the total of 3,334 screened microorganisms, 17 actinomycete strains were found to oxidize the 4"-carbinol of avermectin in a regioselective manner (Table 1).
Feasibility study for the production of 4"-oxo-avermectin.
Since the final objective of the present study was to develop an industrial-scale process for the regioselective oxidation of avermectin, it was necessary to demonstrate early on that the biocatalytic reaction could be scaled up and that it would be compatible with the constraints of industrial downstream processing. Therefore, preparation of 10 g of 4"-oxo-avermectin from avermectin was attempted. Two S. tubercidicus strains, I-1529 and R-922 (Table 1), showed a relatively high activity in pilot-scale biocatalytic reactions (results not shown), and strain I-1529 was chosen for the feasibility study. Resting cells of strain I-1529 successfully converted 16% of the avermectin substrate into 4"-oxo-avermectin, as measured by HPLC, in the course of 96 h at avermectin concentrations of 0.75 g/liter as described in Materials and Methods. After purification on a silica gel, 10.5 g of >95% pure 4"-oxo-avermectin was isolated, providing a 14.0% overall isolated yield.
Initial attempts to clone the genetic determinant for the biocatalytic activity.
The isolation of the genetic determinant for the biocatalytic activity was first attempted by constructing DNA libraries from S. tubercidicus I-1529 in the biocatalytically naïve strain S. lividans ZX7 and screening the transformants for their ability to oxidize avermectin to 4"-oxo-avermectin. Over 5,000 clones were screened from a library raised in pGM160 (24), and >11,000 clones were assayed from an expression library utilizing the PtipA promoter resident on pTBK. No catalytically competent clones were identified, and this approach was abandoned.
Next, based on preliminary biochemical results suggesting that the biocatalytic enzyme from strain R-922 is a P450 monooxygenase, we tried to isolate diverse CYP genes from strains S. tubercidicus R-922 and I-1529. The CYP-encoding genes cyp229 and cyp230 (both from strain R-922) and also cyp233 and cyp234 (both from I-1529) (Table 1) were isolated using degenerate PCR primers (Table 2) designed to bind to the conserved oxygen-binding and heme-binding domains of CYPs (15). Three additional CYPs from strain I-1529 (expressed by cypLA, cypLB, and cypEA), and one from strain R-922 (expressed by cypLC) were also identified in cosmid libraries of these strains using the epoF CYP gene (13, 21) as a hybridization probe (Table 1). None of these CYP genes, however, encoded biocatalytically active enzymes upon expressing them in S. lividans (results not shown).
Cloning of ema1 from strain R-922.
The biocatalytic CYP enzyme from S. tubercidicus R-922, termed Ema1, was purified and its amino acid sequence was partially determined (results will be published elsewhere). PCR primers were designed by reverse translation from some of the determined amino acid sequences (Table 2). PCR products generated with these primers using S. tubercidicus R-922 genomic DNA as a template were cloned and sequenced, and the deduced amino acid sequences showed a perfect match with the amino acid sequences of tryptic peptides derived from the purified Ema1 CYP (data not shown). This result strongly suggested that the PCR amplicons isolated were derived from the gene that encodes the biocatalytic CYP enzyme.
The PCR products were used as probes against a cosmid library of S. tubercidicus R-922 to identify the full-length gene encoding Ema1. The ema1 gene was subcloned and sequenced from the identified cosmids, and its deduced amino acid sequence matched perfectly all the identified peptide fragments from Ema1. The ema1 gene was also cloned into the Streptomyces expression vector pTBK behind the PtipA promoter and assayed in S. lividans ZX7 as a host to reveal biocatalytic activity, as described in the accompanying article (20). The closest match in the databases to the deduced amino acid sequence of Ema1 is a CYP107-family P450 monooxygenase from the carbomycin biosynthetic gene cluster of S. thermotolerans (GenBank accession number 1076117) (4), whose identity with Ema1 is 49%.
Cloning and sequencing of the genes encoding Ema2 to Ema17.
The genes encoding the biocatalytic CYPs from the remaining 16 biotransforming strains were isolated using the ema1 gene as a heterologous probe as described in Materials and Methods.
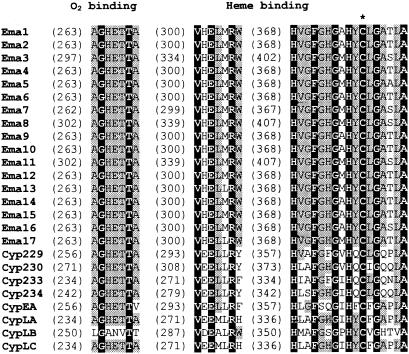
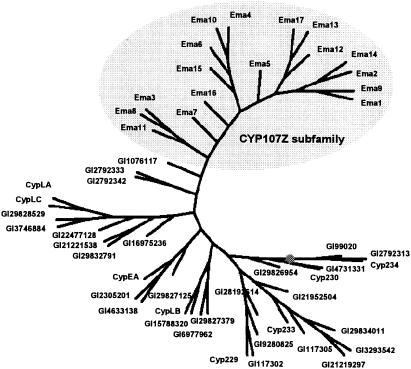
The deduced amino acid sequences of the 17 Ema proteins showed 60 to 100% mutual identity. Ema4 and Ema10 and Ema2 and Ema14 were found to be identical, respectively, despite originating from strains that were classified in the strain collections under different species names. The corresponding genes for Ema4 and Ema10 are also 100% identical at the nucleotide level, while those of Ema2 and Ema14 differ by a single nucleotide. The 17 Ema proteins and also the eight unrelated CYP enzymes that we identified from strains R-922 and I-1529 all harbor the strongly conserved threonine in the putative I-helix proposed to be involved in O2 binding (10). They all also feature the E/DxxR motif in the putative K helix and the Cys pocket with the invariable cysteine in the β-bulge preceding the L helix, both involved in heme binding (Fig. 2) (8, 14). A comparison of the 17 Ema proteins and the 8 unrelated CYP proteins identified from strains R-922 and I-1529 with their nearest databank homologs revealed that the Ema CYP enzymes formed a distinct subfamily of the CYP107 family within the large CYP protein superfamily (Fig. 3). They were most similar to the carbomycin CYP (GenBank accession number 1076117) mentioned earlier and to two distinct CYPs from the rifamycin biosynthetic gene cluster of Amycolatopsis mediterranei (GenBank accession numbers 2792333 and 2792342, respectively).
FIG. 2.
Multiple sequence alignment of conserved motifs in CYP enzymes identified in this study. The Cys pocket and the E/DxxR motif in the K helix, both involved in heme binding, and the O2-binding pocket sequences are shown. Amino acids conserved in all the CYPs listed are shown in white boldface letters on a black background, and amino acids conserved in at least 18 out of the 25 CYPs listed are identified by gray backgrounds. The Cys residue that coordinates the heme is indicated by an asterisk.
FIG. 3.
Comparison of CYP sequences. A multiple sequence alignment was generated with the AlignX program of Vector NTI 7 using the 17 Ema and 8 Cyp proteins identified in this study, and their nearest homologs in the databanks. The alignment was graphically rendered as a half-rooted tree with Phylodraw, version 0.8 (http://pearl.cs.pusan.ac.kr/phylodraw/) using the neighbor-joining matrix. Proteins labeled Ema are the biocatalytically active CYPs, and proteins labeled Cyp are the CYP homologs identified in this study. All other CYPs are labeled with their GenBank number. The root node is labeled with a dot. The newly established CYP107Z subfamily is highlighted by a gray oval.
Cloning, expression, and purification of 16 His-tagged Ema enzymes.
Sixteen of the 17 ema genes (ema1 to ema16) were cloned into the expression vector pET28a, followed by transformation of the constructs into E. coli BL21(DE3). The 16 corresponding Ema enzymes were expressed as their C-terminally His-tagged versions, and the proteins were purified to near-homogeneity by a two-step purification procedure, yielding >95% pure, red-colored, biocatalytically active CYPs, as described in Materials and Methods.
Kinetic characterization of 16 His-tagged Ema enzymes.
The kinetic parameters of the purified His-tagged enzymes were determined spectrophotometrically in the presence of ferredoxin and ferredoxin-NADP+-reductase from spinach.
The Vmax of the purified His-tagged Ema enzymes varied between 0.001 and 0.060 μmol min−1 mg−1 Ema CYP protein, with Ema1 showing the highest specific activity (Table 3). The calculated turnover numbers were 0.048 s−1 for Ema1, followed by 0.042 s−1 for Ema13 and 0.034 s−1 for Ema4 and Ema10. The Km values of the purified enzymes were in the micromolar range.
TABLE 3.
Measured kinetic parameters of Ema1 to Ema16a
| CYP | Km (μM) | Vmax (μmol min−1 mg−1) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Ema1 | 4.3 ± 2.3 | 0.060 ± 0.009 | 0.048 | 11.1 × 103 |
| Ema2/14 | 3.7 ± 3.1 | 0.014 ± 0.003 | 0.011 | 3.1 × 103 |
| Ema3 | 6.9 ± 8.4 | 0.013 ± 0.005 | 0.011 | 1.6 × 103 |
| Ema4/10 | 4.4 ± 2.6 | 0.043 ± 0.007 | 0.034 | 7.8 × 103 |
| Ema5 | 2.2 ± 4.1 | 0.023 ± 0.008 | 0.018 | 8.2 × 103 |
| Ema6 | 1.1 ± 0.7 | 0.014 ± 0.002 | 0.011 | 10.3 × 103 |
| Ema7 | 2.3 ± 2.8 | 0.005 ± 0.001 | 0.004 | 1.7 × 103 |
| Ema8 | 2.6 ± 1.4 | 0.007 ± 0.001 | 0.006 | 2.4 × 103 |
| Ema9 | 4.3 ± 2.3 | 0.021 ± 0.003 | 0.017 | 3.9 × 103 |
| Ema11 | 0.001 ± 0.001 | 0.001 | ||
| Ema12 | 2.7 ± 2.8 | 0.012 ± 0.003 | 0.010 | 3.5 × 103 |
| Ema13 | 6.5 ± 3.3 | 0.053 ± 0.006 | 0.042 | 6.6 × 103 |
| Ema15 | 2.6 ± 1.7 | 0.034 ± 0.005 | 0.027 | 10.4 × 103 |
| Ema16 | 3.8 ± 3.6 | 0.008 ± 0.002 | 0.006 | 1.7 × 103 |
The values were determined as described in Materials and Methods. Ema2/Ema14 and Ema4/Ema10 are identical by their respective amino acid sequences.
DISCUSSION
In this work, we identified 17 Streptomyces strains that are able to biotransform avermectin to 4"-oxo-avermectin, a key intermediate towards the synthesis of the commercial insecticide emamectin benzoate. We identified the biocatalytic enzymes from these strains as cytochrome P450 monooxygenases. Purification of the CYP Ema1 from S. tubercidicus R-922 (details will be published elsewhere) allowed us to clone the corresponding gene by using PCR primers based on the deduced peptide sequences of the purified wild-type enzyme. The genes encoding the biocatalytic CYPs from the remaining 16 strains were also cloned, sequenced, and compared to ema1.
Our first attempt to accomplish the regioselective biological oxidation of avermectin was to screen commercially available redoxactive enzymes, including dehydrogenases, oxidases, and laccases (data not shown). Unfortunately, only laccase AB from Agaricus bisporus was able to oxidize avermectin; despite all our efforts, the oxidation proved not to be regiospecific. Thus, we decided to screen microbial strains available in culture collections for their ability to regioselectively oxidize the 4"-carbinol of avermectin.
To establish a high-throughput microbial screen, several obstacles had to be overcome. Avermectin is very poorly soluble in water (33), so a biocompatible solvent system had to be found for the solubilization of sufficiently high concentrations of avermectin that allow the detection and structure determination of microbial oxidation products. The system we developed uses DMSO and Tween 40 as cosolvents and allows the solubilization of avermectin up to concentrations as high as 4 g/liter (results not shown). After the initial discovery of the biocatalytically competent strains S. tubercidicus I-1529 and R922, we have also tested more water-miscible derivatives of avermectin in the biocatalytic reaction, using both resting cells and (later) purified Ema1 (results not shown). Since the conversion was no higher for any of these polar avermectin derivatives than for avermectin, all further tests were carried out with unmodified avermectin.
Next, a protocol for the reproducible extraction of avermectin from reaction mixtures containing the microorganisms or their cell extracts had to be established. The solvent had to be able to extract a sufficiently high amount of avermectin and its derivatives without the extraction of high amounts of polar medium components such as sugars and amino acids. Ethyl acetate was a very efficient extractant, but unfortunately, it also extracted a lot of polar entities (results not shown). Toluene and MTBE both showed very good extraction characteristics. Further, an analytical method had to be implemented for the separation and quantification of the two oxidized avermectin isomers, 4"-oxo-avermectin and 5-oxo-avermectin, which have almost identical physical properties. We were able to achieve a baseline separation of these isomers by using Kromasil RP-18 material and a 20-min acetonitrile-water gradient. Finally, standard growth and reaction conditions had to be established for the different classes of microorganisms. Our initial microbial screening protocol involved liquid cultures, but this proved too labor intensive and low-throughput. Thus, a protocol employing microorganisms growing on avermectin-containing agar medium was established. A total of 3,334 microbial strains were screened, and 17 strains, all Streptomycetes bacteria, were identified to be capable of oxidizing the 4"-carbinol of avermectin. These 17 strains all belong to the Streptomyces lydicus and S. tubercidicus clusters based on their biochemical characteristics and 16S rRNA gene sequences (analyses conducted at the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) (Table 1).
Four different classes of enzymes are known to oxidize secondary alcohols to the corresponding ketones: dehydrogenases, oxidases, laccases, and P450 monooxygenases (12, 22). Based on the observed high regiospecificity of the biotransformations with the identified Streptomyces strains, we ruled out the possibility that the corresponding enzymes are laccases: these multicopper oxidoreductases generally exhibit low specificity (28). While in vitro enzyme assays using crude cell extracts to characterize the biocatalytic enzymes as oxidases or dehydrogenases were unsuccessful, supplementations of the reactions with ferredoxin and ferredoxin-NADP+-reductase, together with NADPH as a source for electrons, allowed the desired oxidation reaction to be observed. This indicated that the enzyme that catalyzes the biotransformation is a P450 monooxygenase and that the oxidation at the 4"-carbinol involves an interesting regioselective hydroxylation reaction. Thus, the Ema CYP enzymes would hydroxylate the 4" secondary alcohol, followed by the collapse of the resultant 4"-gem-diol to the corresponding ketone by dehydration. The 4"-oxo-avermectin hydrate was observed by HPLC, characterized by liquid chromatography-mass spectrometry and was shown to equilibrate with the 4"-ketone in water-containing solvents (results not shown).
Our initial attempts to clone the genetic determinant for the biocatalytic activity from S. tubercidicus I-1529 were designed to detect the heterologous expression of Ema2 CYP in S. lividans ZX7. In spite of screening >16,000 colonies, we were not able to isolate any biocatalytically competent transformants. Interestingly, our later attempts to express the cloned ema2 in S. lividans ZX7 in different expression vectors also failed to yield biocatalytically competent cells unless a suitable ferredoxin-encoding gene was coexpressed in the same cells (20). We also cloned five CYP-homologous genes from strain I-1529 and three from strain R-922, but none of these encoded biocatalytically active CYP enzymes. Streptomyces strains produce many different CYP enzymes, with the two Streptomyces strains whose genome is completely sequenced, Streptomyces coelicolor A3 (2) and S. avermitilis MA-4680, harboring 18 and 33 CYP-encoding genes, respectively (5, 16, 19), so our failure to clone the biocatalytic CYP via this approach is probably not surprising.
The biocatalytic CYP was then purified from S. tubercidicus R-922 and characterized as a new soluble CYP requiring ferredoxin, ferredoxin reductase, and NADPH for electron supply (results not shown).
The ema1 gene of S. tubercidicus strain R-922 was cloned based on amino acid sequence information gathered from the purified Ema1 enzyme (results not shown). The remaining 16 CYP enzymes that are able to catalyze the oxidation of the 4"-carbinol of avermectin were cloned by their homology to Ema1. These 17 enzymes are highly similar to each other, with identities exceeding 60%. They are clearly distinct, however, from other CYP enzymes in the databanks (Fig. 3), with their closest homologue, a CYP from the carbomycin biosynthetic gene cluster (CYP107C1, GenBank accession number 1076117), displaying identities to the Ema CYPs of <50%. This places the Ema proteins in the CYP107 family populated by many Streptomyces CYPs involved in xenobiotic and secondary metabolism but into the newly established subfamily of CYP107Z (David R. Nelson, personal communication). CypLA and CypLC of the eight non-Ema CYPs identified in this study from S. tubercidicus strains I-1529 and R-922 are also members of the CYP107 family but belong to a different subfamily (CYP107L). The remaining six non-Ema CYPs cloned from strains I-1529 and R-922 are members of different CYP families (Table 1; Fig. 3).
Sixteen of the 17 Ema enzymes were overproduced as their His-tagged versions in E. coli BL21(DE3). After purification, their ability to regioselectively oxidize avermectin in aqueous solution was shown, and the kinetic parameters of the enzymes were determined. The highest specific activity was observed with Ema1, showing a Vmax of 0.060 μmol/min/mg and a turnover of 0.048 s−1. In comparison, the three best-characterized bacterial CYP enzymes described in the literature show much higher turnover numbers. The turnover number of CYP BM-3, the fastest-ever reported for an NADPH-dependent P450 enzyme, is 53 s−1 for the oxidation of arachidonic acid and 27 s−1 for palmitic acid (9). The turnover of cytochrome P450cam for the hydroxylation of camphor is also in the range of 27 s−1 (26), whereas EryF, the CYP responsible for the hydroxylation of 6-deoxyerythronolide B to erythronolide B in Saccharopolyspora erythraea, shows a turnover number of 0.88 s−1 for its natural substrate (2, 32). Since avermectin B1a is not the natural substrate for the Ema enzymes, this substrate might not interact optimally with the active site of these CYPs, leading to the observed lower turnover numbers.
As mentioned earlier, the substrate avermectin B1a is barely water soluble. To obtain consistent kinetic data, avermectin B1a had to be added to the assay mixture dissolved in isopropanol. Although no immediate precipitation was observed, we are aware that the substrate might not have formed a proper solution under these assay conditions.
Heterologous expression of the cloned ema1 gene in a S. lividans host led to the regioselective oxidation of avermectin to 4"-oxo-avermectin. Although the initial yields of this whole-cell biocatalytic reaction were low, optimization of expression parameters and provision of electron supply components increased the bioconversion yields dramatically, as described in the accompanying paper (20). Chemical synthesis of 4"-oxo-avermectin from avermectin involves several steps and uses protecting group chemistry, which makes this conversion time consuming and expensive. A one-step biocatalytic oxidation as performed by the new bacterial CYPs described in this work might overcome these limitations and open new ways for the industrial scale synthesis of emamectin from avermectin.
Acknowledgments
We acknowledge the excellent technical assistance of Mario Jörg, Leonhard Hagmann, Tammo Winkler, Andreas Stämpfli, Marion Petrzika-Kitzka, Roland Dahinden, Snezana Neff, Traugott Schüz, Hansjürg Widmer, Amber Gaudreau, Sandy Emhart, and Keri Cavanaugh; DNA sequencing by Ann Hu and Makoto Ono; and protein sequencing by Daniel Hess (Friedrich Miescher Institute, Basel, Switzerland). We are grateful to David R. Nelson (University of Tennessee, Memphis, TN) for assigning CYP names and to David C. Lamb (University of Wales Aberystwyth, Aberytswyth, United Kingdom) for sharing then-unpublished information on CYPs from S. avermitilis.
This work was supported by Syngenta Biotechnology, Inc. (Research Triangle Park, NC), and Syngenta Crop Protection AG (Basel, Switzerland).
REFERENCES
- 1.Adachi, O., D. Moonmangmee, H. Toyama, M. Yamada, E. Shinagawa, and K. Matsushita. 2003. New developments in oxidative fermentation. Appl. Microbiol. Biotechnol. 60:643-653. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J. F., and C. R. Hutchinson. 1992. Characterization of Saccharopolyspora erythraea cytochrome P-450 genes and enzymes, including 6-deoxyerythronolide B hydroxylase. J. Bacteriol. 174:725-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, S., C. B. Marks, R. Lazarus, J. Miller, K. Stafford, J. Seymour, D. Light, W. Rastetter, and D. Estell. 1985. Production of 2-keto-l-gulonate, an intermediate in L-ascorbate synthesis, by a genetically modified Erwinia herbicola. Science 230:144-149. [DOI] [PubMed] [Google Scholar]
- 4.Arisawa, A., H. Tsunekawa, K. Okamura, and R. Okamoto. 1995. Nucleotide sequence analysis of the carbomycin biosynthetic genes including the 3-O-acyltransferase gene from Streptomyces thermotolerans. Biosci. Biotechnol. Biochem. 59:582-588. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 6.Bernaerts, M. J., and J. de Ley. 1958. 3-Ketoglycosides, new intermediates in the bacterial catabolism of disaccharides. Biochim. Biophys. Acta 30: 661-662. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Buchatskii, A. G., K. Y. Kazachenko, and A. A. Alexandrov. 2001. Cytochrome P450 pattern revision. J. Biomol. Struct. Dyn. 19:273-277. [DOI] [PubMed] [Google Scholar]
- 9.Capdevila, J. H., S. Wei, C. Helvig, J. R. Falck, Y. Belosludtsev, G. Truan, S. E. Graham-Lorence, and J. A. Peterson. 1996. The highly stereoselective oxidation of polyunsaturated fatty acids by cytochrome P450BM-3. J. Biol. Chem. 271:22663-22671. [DOI] [PubMed] [Google Scholar]
- 10.Cupp-Vickery, J. R., and T. L. Poulos. 1995. Structure of the cytochrome P450ERYF involved in erythromycin biosynthesis. Nat. Struct. Biol. 2: 144-153. [DOI] [PubMed] [Google Scholar]
- 11.Cvetovich, R. J., D. H. Kelly, L. M. DiMichele, R. F. Shuman, and E. J. J. Grabowski. 1994. Syntheses of 4"-epi-amino-4"-deoxyavermectins B1. J. Org. Chem. 59:7704-7708. [Google Scholar]
- 12.Drauz, K., and H. Waldmann. 1995. Enzyme catalysis in organic synthesis: a comprehensive handbook, vol. II. VCH, Weinheim, Germany.
- 13.Gerth, K., H. Steinmetz, G. Höfle, and H. Reichenbach. 2001. Studies on the biosynthesis of epothilones: the PKS and epothilone C/D monooxygenase. J. Antibiot. 54:144-148. [DOI] [PubMed] [Google Scholar]
- 14.Hasemann, C. A., R. G. Kurumbail, S. S. Boddupalli, J. A. Peterson, and J. Deisenhofer. 1995. Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure 3:41-62. [DOI] [PubMed] [Google Scholar]
- 15.Hyun, C.-G., J.-M. Kim, S.-K. Hong, and J.-W. Suh. 1998. An efficient approach for cloning P450 hydroxylase genes from Actinomycetes. J. Microbiol. Biotechnol. 8:295-299. [Google Scholar]
- 16.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 17.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 18.Kuhstoss, S., and R. N. Rao. 1991. Analysis of the integration function of the streptomycete bacteriophage φC31. J. Mol. Biol. 222:897-908. [DOI] [PubMed] [Google Scholar]
- 19.Lamb, D. C., T. Skaug, H.-L. Song, C. J. Jackson, L. M. Podust, M. R. Waterman, D. B. Kell, D. E. Kelly, and S. L. Kelly. 2002. The cytochrome P450 complement (CYPome) of Streptomyces coelicolor A3(2). J. Biol. Chem. 277:24000-24005. [DOI] [PubMed] [Google Scholar]
- 20.Molnár, I., D. S. Hill, R. Zirkle, P. E. Hammer, F. Gross, T. G. Buckel, V. Jungmann, J. P. Pachlatko, and J. M. Ligon. 2005. Biocatalytic conversion of avermectin to 4"-oxo-avermectin: heterologous expression of the ema1 cytochrome P450 monooxygenase. Appl. Environ. Microbiol. 71:6977-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molnár, I., T. Schupp, M. Ono, R. Zirkle, M. Milnamow, B. Nowak-Thompson, N. Engel, C. Toupet, A. Stratmann, D. D. Cyr, J. Gorlach, J. M. Mayo, A. Hu, S. Goff, J. Schmid, and J. M. Ligon. 2000. The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chem. Biol. 7:97-109. [DOI] [PubMed] [Google Scholar]
- 22.Morita, M., and Y. Watanabe. 1977. A secondary alcohol oxidase: a component of a polyvinyl alcohol degrading enzyme preparation. Agric. Biol. Chem. 41:1535-1537. [Google Scholar]
- 23.Murakami, T., T. G. Holt, and C. J. Thompson. 1989. Thiostrepton-induced gene expression in Streptomyces lividans. J. Bacteriol. 171:1459-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muth, G., B. Nussbaumer, W. Wohlleben, and A. Puehler. 1989. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol. Gen. Genet. 219:341-348. [Google Scholar]
- 25.Noll-Borchers, M., and K. Buchholz. 1993. Kinetics and yields of 3-keto-isomaltulose by microbial oxidation of isomaltulose. Biotechnol. Lett. 15:139-144. [Google Scholar]
- 26.Peterson, J. A., and D. M. Mock. 1975. Metabolic control of cytochrome P 450cam. Adv. Exp. Med. Biol. 58:311-324. [PubMed] [Google Scholar]
- 27.Reichstein, T., and A. Grussner. 1934. A high-yield synthesis of l-ascorbic acid (vitamin C). Helv. Chim. Acta 17:311-328. [Google Scholar]
- 28.Reinhammar, B., and B. G. Malmström. 1981. “Blue” copper-containing oxidases, p. 109-149. In T. G. Spiro (ed.), Copper proteins. Wiley, New York, N.Y.
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schmid, A., J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts, and B. Witholt. 2001. Industrial biocatalysis today and tomorrow. Nature 409: 258-268. [DOI] [PubMed] [Google Scholar]
- 31.Schnarr, G. W., and W. A. Szarek. 1978. Oxidation of methyl glycopyranosides by Acetobacter suboxydans. Can. J. Chem. 56:1752-1757. [Google Scholar]
- 32.Shafiee, A., and C. R. Hutchinson. 1987. Macrolide antibiotic biosynthesis: isolation and properties of two forms of 6-deoxyerythronolide B hydroxylase from Saccharopolyspora erythraea (Streptomyces erythreus). Biochemistry 26:6204-6210. [DOI] [PubMed] [Google Scholar]
- 33.Tomlin, C. 2003. The pesticide manual, 13th ed. British Crop Protection Council Publications, Surrey, United Kingdom.
- 34.Zirkle, R., J. M. Ligon, and I. Molnár. 2004. Heterologous production of the antifungal polyketide antibiotic soraphen A of Sorangium cellulosum So ce26 in Streptomyces lividans. Microbiology 150:2761-2774. [DOI] [PubMed] [Google Scholar]