Abstract
We have compared the proteomic profiles of L. lactis subsp. cremoris NCDO763 growing in the synthetic medium M17Lac, skim milk microfiltrate (SMM), and skim milk. SMM was used as a simple model medium to reproduce the initial phase of growth of L. lactis in milk. To widen the analysis of the cytoplasmic proteome, we used two different gel systems (pH ranges of 4 to 7 and 4.5 to 5.5), and the proteins associated with the cell envelopes were also studied by two-dimensional electrophoresis. In the course of the study, we analyzed about 800 spots and identified 330 proteins by mass spectrometry. We observed that the levels of more than 50 and 30 proteins were significantly increased upon growth in SMM and milk, respectively. The large redeployment of protein synthesis was essentially associated with an activation of pathways involved in the metabolism of nitrogenous compounds: peptidolytic and peptide transport systems, amino acid biosynthesis and interconversion, and de novo biosynthesis of purines. We also showed that enzymes involved in reactions feeding the purine biosynthetic pathway in one-carbon units and amino acids have an increased level in SMM and milk. The analysis of the proteomic data suggested that the glutamine synthetase (GS) would play a pivotal role in the adaptation to SMM and milk. The analysis of glnA expression during growth in milk and the construction of a glnA-defective mutant confirmed that GS is an essential enzyme for the development of L. lactis in dairy media. This analysis thus provides a proteomic signature of L. lactis, a model lactic acid bacterium, growing in its technological environment.
The bacterium Lactococcus lactis is the main source of mesophilic starters used for the manufacture of fermented dairy products, and strong research efforts have been dedicated in the past 20 years to the isolation and description of functions required for proper development in milk (7, 22, 24). Dairy lactococci present half a dozen amino acid auxotrophies, whereas milk is not an abundant source of free amino acids (34). Also, the hydrolysis of caseins by a cell-wall-attached protease (PrtP) is required to achieve a final biomass of approximately 2 × 109 CFU/ml (3, 24). A limited number of the resulting peptides are internalized by the oligopeptide transport system (OppA) and degraded to amino acids by a pool of cytoplasmic peptidases (23). Logically, the activities of both PrtP and OppA have been demonstrated to be crucial for optimal growth of lactococci in milk (3, 39). Another essential property of dairy lactococci is their capacity to internalize lactose by use of a phosphotransferase system (LacEF) and to degrade lactose-6-phosphate by the tagatose pathway (7). The genes encoding the lactose phosphotransferase system (lacEF), the phospho-β-galactosidase (lacG), and the enzymes of the tagatose phosphate pathway (lacABCD) are organized in an operon that is also located on the protease plasmid (3). Besides the capacity to use casein and lactose efficiently, a small number of enzymes have been reported as being essential or important for proper growth in milk. These proteins are involved in the metabolism of amino acids such as AspC, BcaT, and AraT (4, 8, 41) or in the synthesis of their precursors such as the pyruvate carboxylase (PycA) (41).
The sequencing of the genome of Lactococcus lactis paved the way for a global analysis of its adaptation to various environments (2). Numerous reports have shown interest in the proteomic approach to describe metabolic adaptation to changing environments (see, for example, references 32, 38, and 40), and our recent analysis of the cytosolic proteomes of two strains of L. lactis indicated that analysis by two-dimensional electrophoresis (2-DE) seemed to be well adapted to monitor changes in the concentrations of metabolic enzymes (15). The aim of the present study was to characterize the proteome profile of L. lactis during its development in milk. For this purpose, we have compared the proteomes of the dairy strain L. lactis subsp. cremoris NCDO763 cultivated in three media: (i) M17Lac; (ii) skim milk microfiltrate (SMM), from which the micellar caseins have been removed and which was used to simulate the initial phase of development in milk; and (iii) skim milk (milk). We observed in-depth modifications of the proteome pattern for bacteria cultivated in SMM or milk. The modifications were essentially associated with pathways involved in the supply of amino acids and purine nucleotides. The proteomic analysis attracted our attention to the role of glutamine synthetase (GS), and further genetic experiments highlighted the key role of this enzyme in the development of L. lactis in dairy environments.
MATERIALS AND METHODS
Bacterial strains, cultures, and media.
The strain Lactococcus lactis subsp. cremoris NCDO763 was cultivated in M17Lac (Difco, Sparks, MD), in SMM (see below), or in reconstituted skim milk (10% milk powder; NIZO, Ede, The Netherlands). The three media were buffered with 75 mM β-glycerophosphate at pH 7.2. Six other strains (L. lactis subsp. lactis C10, IL-1403, NCDO2118, and NCDO2110 and L. lactis subsp. cremoris AM2 and HP) were used in the course of the study to monitor the capacity of the species to grow in SMM. The SMM, a gift of the Laboratoire des Sciences et Technologie du Lait et de l'Oeuf (INRA Rennes, France), was produced by cross flow microfiltration of cow skim milk on ceramic membrane (pore size, 0.1 μm) (36). The resulting SMM is a yellow, crystal-clear solution whose composition is close to that of sweet SMM (9). The elimination of caseins during the filtration procedure allows direct measurement of the cell density by turbidimetry. The medium was filter sterilized (0.22 μm) and stored in the dark at 4°C. Under these conditions the SMM was stable for at least 6 months.
Overnight cultures of L. lactis NCDO763 grown in M17Lac, SMM, or milk were used to inoculate (1/100) 400 ml of M17 broth containing 5 g/liter lactose (M17Lac), SMM, or milk in 0.5-liter Erlenmeyer flasks. Cells were batch cultivated at 30°C without shaking, and cultures were stopped in exponential phase at a cell density of to 8 ×108 ± 0.5 ×108 CFU/ml. Trisodium citrate (1 M) was added at a final concentration of 0.25 M to the cell suspensions, which were maintained at 4°C for 5 min under these conditions before centrifugation. This treatment, adapted from that described previously (16), limits the casein precipitation in the milk and was applied to the three media. The bacteria were harvested by centrifugation (10,000 × g, 10 min, 4°C). The cell pellet was washed twice with ice-cold 200 mM Na-phosphate, pH 6.4, and resuspended in 3.5 ml of 20 mM Na-phosphate buffer, pH 6.4, 1 mM EDTA.
Cytoplasmic protein extract preparation.
The protein extract was prepared essentially as previously described (15). A modification of the previous protocol was the addition of a cocktail of protease inhibitors diluted 20 times (P8465; Sigma-Aldrich, St. Louis, MO) in the phosphate-buffered cell suspension. The cell suspension (approximately 35 units of optical density at 600 nm [OD600]/ml) was transferred to the precooled chamber of a BASIC Z cell disrupter (Celld, Warwickshire, United Kingdom) and was subjected to a pressure of 2,500 bars. The suspension was centrifuged at 5,000 × g for 15 min at 4°C to remove unbroken cells and large cellular debris. The supernatant was collected and centrifuged at 220,000 × g for 30 min at 4°C. The total protein concentration (2.5 ± 0.5 mg/ml) in the resulting supernatant (cytosolic fraction) was determined with the Coomassie protein assay reagent (Pierce, Rockford, IL), using bovine serum albumin as a standard. The cytosolic fraction was aliquoted and stored frozen at −20°C.
CEA protein extract preparation.
The transparent pellet obtained after ultracentrifugation (see above) was resuspended in 0.5 ml 50 mM Na-phosphate buffer, pH 6.4, 1 mM EDTA and sonicated to facilitate the resuspension. The concentration of the resulting cell envelope-associated (CEA) protein sample was 2 ± 0.5 mg/ml. For the same number of CFU, the amount of protein in this fraction was thus approximately 10 times less than that in the cytoplasmic extract. To remove lipids, the CEA protein extract was treated with methanol-chloroform as described previously (43). In brief, the solution was first treated with a mixture of methanol and chloroform (3:1), vortexed, and centrifuged (12,000 × g, 4°C, 2 min), and 4 volumes of water were then added. The upper aqueous phase was removed after centrifugation (12,000 × g, 4°C, 2 min), and 3 volumes of methanol were added to precipitate the proteins.
2-DE.
A volume of cytosolic fraction corresponding to 300 μg of protein was thawed on ice and precipitated with 60% (vol/vol) methanol. The protein pellet was resuspended in 500 μl of isoelectric focusing (IEF) buffer 1, consisting of 7 M urea, 2 M thiourea, 4% CHAPS{3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 100 mM dithiothreitol or 4 mM tributylphosphine (for basic gels), and 0.5% pH 4 to 7 or 4.5 to 5.5 immobilized pH gradient (IPG) buffer (Amersham-Pharmacia Biotec, Uppsala, Sweden). The sample was loaded on 24-cm pH 4 to 7 or pH 4.5 to 5.5 IPG strip (Bio-Rad, Hercules, CA) which was rehydrated at 50 V for 12 h. IEF was carried out for 60,000 V · h at a maximum of 8,000 V, using the Protean II IEF cell (Bio-Rad). For CEA protein extract, the protein pellet (250 μg/gel) obtained after methanol precipitation (see above) was resuspended in IEF buffer 2, consisting of 1% ASB14 (sulfobetaine), 4 mM tributylphosphine, 7 M urea, 2 M thiourea, and 0.5% pH 4 to 7 IPG buffer.
After completion of IEF, the IPG strip was positioned on sodium dodecyl sulfate-polyacrylamide gels, using 1% low-melting-point agarose in 150 mM Tris-HCl, pH 8.8. Second-dimension electrophoresis was performed on 12% polyacrylamide gels (24 by 20 by 0.1 cm) in 25 mM Tris, 192 mM glycine, 0.1% sodium dodecyl sulfate, pH 8.3, using the Ettan-Dalt II apparatus. Electrophoresis was run at 1 W/gel for 16 h at 4°C. The gels were stained with BioSafe colloidal Coomassie blue (Bio-Rad) for 1 h and destained with three successive washes in deionized water. The set of images and the information associated with the spots can be retrieved by downloading the PARIS software (http://www.inra.fr/bia/J/imaste/paris/) (42).
Image acquisition and analysis of 2-DE.
Gel images were generated using an Epson Expresssion 1640XL scanner controlled by Silver Fast software. Image files were recorded at 256 gray levels. Image manipulation and analysis were performed with Image Master 4.01 software (Amersham-Pharmacia). Comparative analysis was performed by analyzing images from two independent cultures for the three conditions. Two gels (from two independent cultures) were analyzed for each growth condition. The normalized intensity (NI) of each spot was calculated as the ratio of its intensity in pixel units versus the sum of the intensities of all the spots in the gel. The average NI was calculated for each spot from the two independent cultures and used to calculate the ratio (fold change) of expression level of the corresponding protein between the different experimental conditions (SMM versus M17Lac and milk versus M17Lac). A protein was included in the list of up- or down-regulated proteins according to the following criteria: (i) a minimum of a twofold change of NI between two conditions was observed in the set of duplicate gels, and (ii) the difference was statistically significant (Student's t test, P < 0.05). For the 25 proteins which were not detected in M17Lac but appeared as novel spots in SMM or milk, the statistical test could not be applied and we used as the sole criterion the detection of the spot in the two independent SMM or milk cultures (two gels). A ratio was calculated by using as a denominator the normalized volume of the smallest spot detected on M17Lac 2-DE. This provided an approximate minimum fold change for these proteins.
Protein identification by peptide mass fingerprinting and N-terminal sequencing.
Proteins were identified by mass spectrometry as described previously (15). The analysis of the masses of tryptic peptides was performed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry with a Voyager DE STR instrument (Applied Biosystems, Framingham, MA). Database searches were conducted with the MS-Fit software (http://prospector.ucsf.edu) either on an L. lactis-specific database containing protein sequences deduced from the genome sequences of L. lactis IL-1403 (2) and SK11 (DOE Joint Genome Institute, http://genome.jgi-psf.org/microbial/) or on GENPEPT (ftp://ftp.ncifcrf.gov/pub/genpept/).
Construction of L. lactis TIL520 (glnA::pTIL520) and TIL521 (glnA::luxAB).
DNA manipulations were carried out essentially as described previously (35). Restriction enzymes (from Eurogentec, Liége, Belgium, or Roche Molecular Biochemicals, Indianapolis, IN.), T4 DNA ligase (Epicenter, Madison, WI.), and Taq DNA polymerase (Qbiogen, Illkirch, France) were used as recommended by the manufacturers. Oligonucleotides were purchased from Invitrogen SARL (Cergy-Pontoise, France). Plasmids were extracted by using a QIAprep Spin miniprep kit (QIAGEN S.A., Courtaboeuf, France). PCRs were performed with a GeneAmp 2400 PCR system (Perkin-Elmer, Boston, MA). DNA sequences were determined with an Applied Biosystems 370A automated DNA sequencer and with ABI PRISM dye terminator cycle sequencing and dye primer cycle sequencing kits (Perkin-Elmer). Preparation of competent cells and electrotransformation of L. lactis were carried out as described previously (17).
L. lactis NCDO763 was used as a parental strain. The inactivation of glnA was performed by single-crossover integration of an internal glnA fragment cloned into the nonreplicative vector pRV300 (28). An internal 521-base-pair internal fragment of glnA was PCR amplified with primers 5′-GGGGTACCCCGCTTCAAGAGCTTTAAC-3′ (KpnI site underlined) and 5′-GGAATTCCATGAGTTAACAGTTGGG-3′ (EcoRI site underlined). The fragment was digested with EcoRI-KpnI and cloned into the EcoRI-KpnI-linearized pRV300 vector. The resulting plasmid (pTIL520) was produced in Escherichia coli TG1 and used to transform L. lactis NCDO763. Erythromycin (Em)-resistant (5 μg · ml−1) clones were selected, and the integration of pRV520 into the chromosomal glnA gene was verified by PCR. The resulting strain was named TIL520. As an additional control for the disruption of glnA, the absence of the protein was verified by 2-DE. The organization of the glnRA locus makes the possibility of a polar effect of the insertion unlikely: sequence analysis downstream of glnA predicted a Rho-independent terminator, and the following gene, ywiE, is orientated in the opposite direction.
Transcriptional fusion of glnA with luxAB of Vibrio harveii was constructed as follows. A 1.2-kb fragment spanning the 5′ end of glnA was generated by PCR from the L. lactis NCDO763 chromosomal DNA with the PtpL-SphI-F (GTCGACGCATGCATTGTTTTTGTCTGTTTAGGAAATA) and GlnA-XbaI-R (GCTCTAGAAAGTGTCAAGGTCTGGATAAAGATA) primers. The amplified region contains the 5′-end fragment of the ptpL gene, the glnR coding sequence, and 231 bp from the glnA coding sequence corresponding to the first 77 codons. This fragment was cloned into vector pORINeWLux (pJIM2374), which is nonreplicative in L. lactis (6), to give pTIL521. Plasmid pTIL521 was produced using E. coli TG1 repA+ in the presence of 150 μg · ml−1 Em. L. lactis NCDO763 competent cells were transformed by electroporation with pIL521. The integrants were selected at 30°C on M17Lac containing 5 μg · ml−1 Em. The correct chromosomal integration of pIL521 was tested by PCR. Integration into the expected location was confirmed for one clone, called TIL521, which was selected for further studies. In TIL521, the lux genes are under the control of the glnRA promoter. The strain still possesses a fully functional glnRA operon, and its growth in M17Lac, SMM, or milk was found to be indistinguishable from that of the wild-type strain.
Measurement of luciferase activity.
To measure luciferase activity, 1 ml of a culture in milk was withdrawn at the indicated times and rapidly mixed with 5 μl 95% nonyl aldehyde (30 μM final concentration) (Sigma-Aldrich, Steinheim, Germany). The emission of light was immediately measured in a Luminoskan II luminometer (Labsystems, Helsinki, Finland). In parallel, growth was monitored with a spectrophotometer at a wavelength of 600 nm, and the emitted light value was standardized to the optical density (ULU/OD). The experiments were repeated twice with similar results.
RESULTS AND DISCUSSION
Growth of L. lactis in M17Lac, SMM, and milk.
The present study focused on strain NCDO763, the parental strain of the model strain MG1363, which is widely used in the scientific community studying lactic acid bacteria (LAB) (27). The adaptation of L. lactis to dairy media was studied by comparing the proteomic profiles of cells cultivated in M17Lac, SMM, and skim milk. M17 is the reference synthetic medium for the bacterium and is thought to keep the biosynthetic activities of dairy lactococci at their lowest (37). SMM is obtained from skim milk by a microfiltration procedure which eliminates the micellar caseins (see Materials and Methods). This medium supports the growth of dairy strains of L. lactis without additives. It was used to mimic the initial phase of development of L. lactis in milk, where the bacterial growth is independent of casein hydrolysis (18, 31).
Compared to L. lactis cultivated in M17Lac, L. lactis cultivated in milk and SMM displayed the steepest acidification curves (data not shown). To eliminate the pH factor as a possible cause of proteome variation, we choose to work with buffered media. In these conditions, buffered SMM has the capacity to sustain growth of L. lactis NCDO763 to a cell density of 1.2 × 109 CFU/ml. This corresponds to approximately two generations less than for bacteria growing in milk (4.7 × 109 CFU/ml).
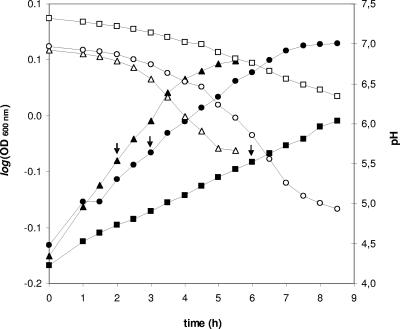
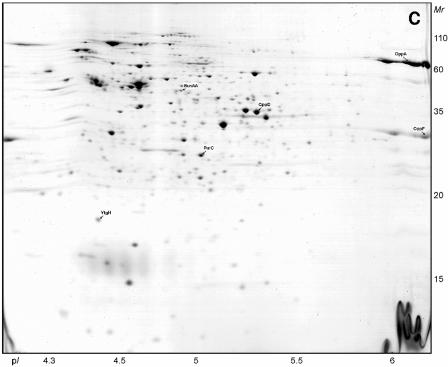
We monitored the growth and acidification of L. lactis NCDO763 in the three buffered media (Fig. 1). In these conditions, the growth rate in milk (0.68 h−1) is between those observed in M17Lac (0.94 h−1) and SMM (0.34 h−1). For the proteomic analysis, the cells were harvested in the exponential phase at a similar cell density (8 × 108 ± 0.5 × 108 CFU/ml) and pH (6.8 ± 0.1).
FIG. 1.
Growth and acidification curves of L. lactis NCDO763 in M17Lac, SMM, and milk. The evolution of OD600 (filled symbols) and pH (open symbols) in M17Lac (triangles), SMM (squares), and milk (circles) was measured. The arrows indicate the times of sampling of cells for proteomic analysis.
One of the difficulties in working with milk is that the caseins, which are present at a high concentration (25 g/liter), tend to sediment with the bacteria during harvest. To bypass this problem, we incubated the three cultures for 5 min in the presence of 0.25 M trisodium citrate (see Materials and Methods). In control experiments, we found that this treatment was without consequence for the 2-DE profiles of M17Lac- or SMM-grown bacteria (data not shown).
Proteomic profiles of L. lactis cultivated in M17Lac and dairy media.
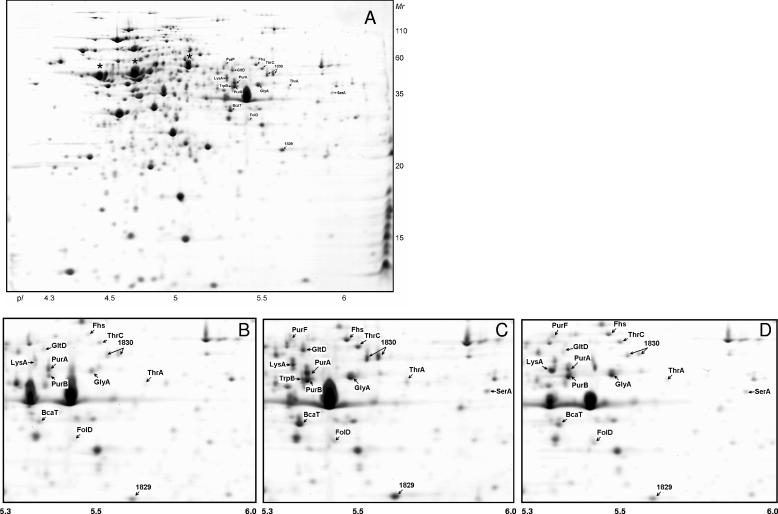
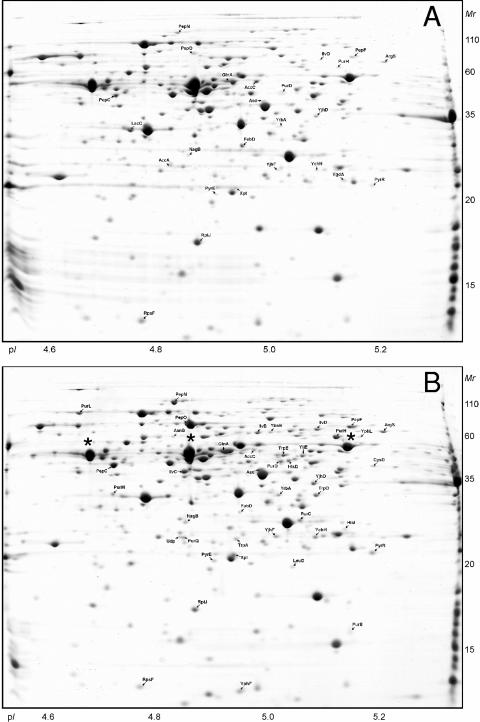
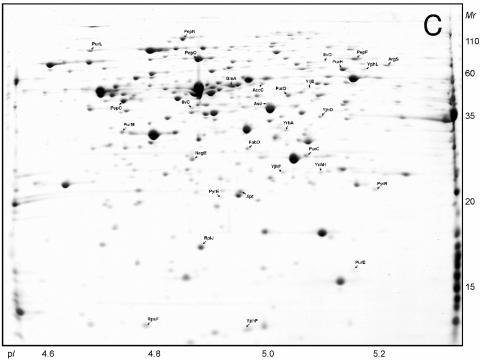
The proteins synthesized by L. lactis NCDO763 during growth in the three media were analyzed by 2-DE. As previously reported (15), the proteome of L. lactis analyzed in the pH 4 to 7 range shows some areas with a strong spot density (Fig. 2). To improve the gel resolution and increase the number of proteins detected, we performed 2-DE with a narrow pH gradient (pH 4.5 to 5.5) (Fig. 3). We also studied the proteins that can be solubilized from the cell fraction obtained after ultracentrifugation (CEA extract [see Materials and Methods]) (Fig. 4). This sample is enriched in proteins preferentially located at the cytosol-membrane interface and allowed us to identify proteins associated with membrane-located cellular processes.
FIG. 2.
(A) Two-dimensional gels (pH 4 to 7) of the cytoplasmic proteins of L. lactis NCDO763 grown in SMM. (B to D) The sector of interest of the 2-DE of the cytoplasmic proteins of L. lactis NCDO763 cultivated in M17Lac (B), SMM (C), and milk (D). The proteins which are more abundant in SMM or milk are indicated. The same three proteins (from left to right, enolase, EF-Tu, and pyruvate kinase) are marked with an asterisk in panel A and in Fig. 3B.
FIG. 3.
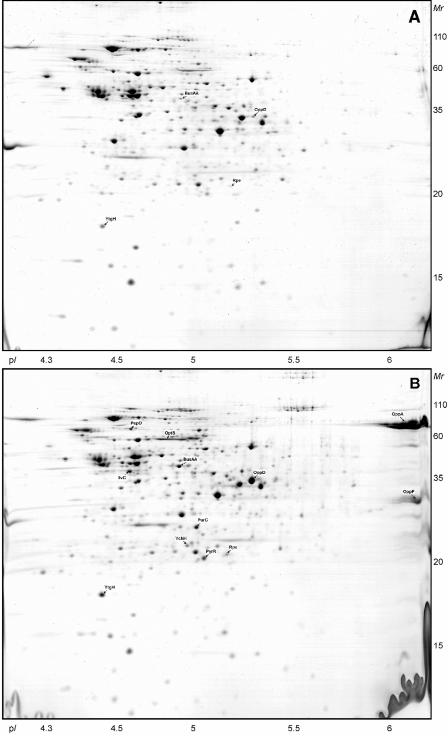
Two-dimensional gels (pH 4.5 to 5.5) of the cytoplasmic proteins of L. lactis NCDO763 grown in M17Lac (A), SMM (B), and milk (C). The proteins which are differentially regulated in SMM or milk compared to M17Lac are indicated. The same three proteins (from left to right, enolase, EF-Tu, and pyruvate kinase) are marked with an asterisk in panel A and in Fig. 2A.
FIG. 4.
Two-dimensional gels (pH 6 to 11) of the CEA proteins of L. lactis NCDO763 grown in M17Lac (A), SMM (B), and milk (C). The proteins which are differentially regulated in SMM or milk compared to M17Lac are indicated.
Overall, about 900 spots, corresponding to more than 330 distinct proteins, have been identified and quantitatively analyzed in the present study. This corresponds to 25% of the theoretical acidic proteome deduced from the genome sequence of strain IL-1403. The list of the proteins identified and the corresponding images are in Table S1 and Fig. S1 and S2 in the supplemental material.
Representative images of the 2-DE gels analyzed in the present work are presented in Fig. 2 to 4. We found that the levels of 31 proteins increased upon growth in milk. In SMM the same group of proteins was up-regulated and contained 28 additional proteins. The changes in the proteome profiles are summarized in Table 1. In the following sections, we discuss the main positive variations associated with the cultures in SMM and milk.
TABLE 1.
Proteome modifications during growth of L. lactis NCDO763 in dairy media
| Protein function and spot no. | Gela | Gene | Fold change in:b
|
Protein | |
|---|---|---|---|---|---|
| SMM | Milk | ||||
| Up-regulated proteins | |||||
| Nucleotide metabolism | |||||
| 158 | 4-7 | purA | 3.0 | — | Adenylosuccinate synthase (EC 6.3.4.4) |
| 386 | 4-7 | purB | 4.0 | 4.2 | Adenylosuccinate lyase (EC 4.3.2.2) |
| 257 | 4.5-5.5 | purC | >14 | >14 | SAICAR synthetase (EC 6.3.2.6) |
| 160 | 4.5-5.5 | purD | 2.9 | 3.2 | GAR synthase (EC 6.3.4.3) |
| 337 | 4.5-5.5 | purE | >3 | >3 | Phosphoribosylaminoimidazole carboxylase (EC 4.1.1.21) |
| 98 | 4-7 | purF | >6.8 | >5.3 | Phosphoribosylpyrophosphate amidotransferase (EC 2.4.2.14) |
| 95 | 4.5-5.5 | purH | 23.3 | 24.8 | Bifunctional purine biosynthesis protein (EC 2.1.2.3) (EC 3.5.4.10) |
| 30 | 4.5-5.5 | purL | >19 | >17 | Phosphoribosylformylglycinamidine synthase II (EC 6.3.5.3) |
| 223 | 4.5-5.5 | purM | >9 | >11 | AIR synthetase (EC 6.3.1.1) |
| 290 | 4.5-5.5 | purQ | >8 | — | Phosphoribosylformylglycinamidine synthase I (EC 6.3.5.3) |
| 356 | 4.5-5.5 | yphF (PurS) | >18 | >15 | Phosphoribosylformylglycinamidine (FGAM) synthase PurS |
| 315 | 4.5-5.5 | pyrE | 4.8 | — | Orotate phosphoribosyltransferase (EC 2.4.2.10) |
| 309 | 4.5-5.5 | pyrR | 7.8 | 2.3 | Pyrimidine operon regulator |
| 288 | 4.5-5.5 | udp | >4 | — | Uridine phosphorylase |
| 311 | 4.5-5.5 | xpt | 3.2 | 2.7 | Xanthine phosphoribosyltransferase (EC 2.4.2.) |
| Amino acid and nitrogen metabolism | |||||
| 183 | 4.5-5.5 | asd | 3.0 | — | Aspartate-semialdehyde dehydrogenase (EC 1.2.1.11) |
| 91 | 4.5-5.5 | asnB | >3 | — | Asparagine synthetase B |
| 229 | 4-7 | bcaT | 4.8 | 3.1 | Branched-chain amino acid aminotransferase (EC 2.6.1.42) |
| 176 | 4.5-5.5 | cysD | >5 | — | O-Acetylhomoserine sulfhydrylase |
| 124 | 4.5-5.5 | glnA | 3.4 | 2.5 | Glutamine synthetase (EC 6.3.1.2) |
| 113 | 4-7 | gltd | 9.8 | 3.3 | Glutamate synthase (NADPH) small chain (EC 1.4.1.13) |
| 166 | 4-7 | glyA | 4.6 | 3.8 | Serine hydroxymethyltransferase |
| 157 | 4.5-5.5 | hisD | >12 | — | Histidinol dehydrogenase (EC 1.1.1.23) |
| 272 | 4.5-5.5 | hisl | >3 | — | Phosphoribosyl-AMP cyclohydrolase |
| 82 | 4.5-5.5 | ilvB | >18 | — | Acetolactate synthase large subunit (EC 4.1.3.18) |
| 187 | 4.5-5.5 | ilvC | >39 | >3 | Ketol-acid reductoisomerase (EC 1.1.1.86) |
| 71 | 4.5-5.5 | ilvD | 8.2 | — | Dihydroxy-acid dehydratase (EC:4.2.1.9) |
| 321 | 4.5-5.5 | leuD | >3 | — | 3-Isopropylmalate dehydratase small subunit (EC 4.2.1.33) |
| 139 | 4-7 | lysA | 4.4 | 3.3 | Diaminopimelate decarboxylase (EC 4.1.1.20) |
| 191 | 4-7 | serA | >6 | >2 | d-3-Phosphoglycerate dehydrogenase (EC 1.1.1.95) |
| 178 | 4-7 | thrA | 6.1 | — | Aspartokinase (EC 2.7.2.4) |
| 111 | 4-7 | thrC | 4.0 | — | Threonine synthase (EC 4.2.99.2) |
| 292 | 4.5-5.5 | trpA | >8 | — | Tryptophan synthase alpha subunit (EC 4.2.1.20) |
| 387 | 4-7 | trpB | >13 | — | Tryptophan synthase beta chain (EC 4.2.1.20) |
| 226 | 4.5-5.5 | trpD | >20 | — | Anthranilate phosphoribosyltransferase (EC 2.4.2.18) |
| 136 | 4.5-5.5 | trpE | >22 | — | Anthranilate synthase component I (EC:4.1.3.27) |
| 287 | 4.5-5.5 | ychH | 8.1 | 2.5 | 2,3,4,5-Tetrahydropyridine-2,6-dicarboxylate |
| N-Succinyltransferase (EC 2.3.1.117) | |||||
| Peptidases | |||||
| 174 | 4.5-5.5 | pepC | 3.0 | 2.0 | Aminopeptidase C |
| 66 | 4.5-5.5 | pepF | 4.1 | 3.6 | Oligoendopeptidase F |
| 12 | 4.5-5.5 | pepN | 4.1 | 3.1 | Aminopeptidase N |
| 55 | 4.5-5.5 | pepO | 19.2 | 9.3 | Neutral endopeptidase |
| Carbohydrate metabolism | |||||
| 263 | 4.5-5.5 | nagB | 3.9 | 6.8 | Glucosamine-6-phosphate isomerase (EC 5.3.1.10) |
| 20 | CEA | rpe | 10.2 | — | Ribulose-phosphate 3-epimerase (EC 5.1.3.1) |
| Folic acid and derivative biosynthesis | |||||
| 99 | 4-7 | fhs | 16.4 | 8.5 | Formyltetrahydrofolate synthetase (EC 6.3.4.3) |
| 248 | 4-7 | folD | 3.6 | 3.1 | Tetrahydrofolate dehydrogenase/cyclohydrolase (EC 1.5.1.5; 3.5.4.9) |
| tRNA synthetases | |||||
| 76 | 4.5-5.5 | argS | 3.0 | 2.1 | Arginyl-tRNA synthetase (EC 6.1.1.19) |
| ABC transport systems | |||||
| 3 | CEA | oppA | >380 | >281 | Oligopeptide transport, substrate binding protein |
| 12 | CEA | oppD | 23.5 | 12.2 | Oligopeptide transport, ATP binding protein |
| 15 | CEA | oppF | >45 | >65 | Oligopeptide transport, ATP binding protein |
| 79 | 4.5-5.5 | ybaB | >6 | — | Unknown substrate, ATP binding protein |
| 6 | CEA | optS | >42 | — | Oligopeptide transport, substrate binding protein |
| 8 | CEA | busAA | 4.0 | — | Betaine transport, ATP binding protein |
| Unknown function | |||||
| 210 | 4.5-5.5 | yjhD | 3.0 | — | Conserved protein |
| 284 | 4.5-5.5 | yjhF | 4.9 | — | Phosphoglycerate mutase paralog |
| 141 | 4.5-5.5 | yljE | >15 | — | RNA methyltransferase, TrmA family |
| 225 | 4.5-5.5 | yrbA | 3.4 | 2.0 | Oxidoreductase |
| 24 | CEA | ytgH | 3.1 | — | Adaptations to atypical conditions, similar to gls24 |
| 126 | 4-7 | 1830-SK11 | 5.7 | — | Putative oxydreductase |
| 302 | 4-7 | 1829-SK11 | 3.1 | — | Similar to product of gene 1829 of L. lactis SK11 |
| Down-regulated | |||||
| Ribosomal proteins | |||||
| 332 | 4.5-5.5 | rplJ | 0.5 | 0.5 | 50S ribosomal protein L10 |
| 354 | 4.5-5.5 | rpsF | 0.5 | 0.5 | 30S ribosomal protein S6 |
| Fatty acid biosynthesis | |||||
| 411 | 4.5-5.5 | accA | NDc | ND | Acetyl coenzyme A carboxylase carboxyl transferase subunit alpha (EC 6.4.1.2) |
| 116 | 4.5-5.5 | accC | 0.5 | 0.4 | Biotin carboxylase (EC 6.3.4.14) |
| 250 | 4.5-5.5 | fabD | 0.2 | 0.4 | Malonyl coenzyme A-acyl carrier protein transacylase (EC 2.3.1.39) |
| Miscellaneous | |||||
| 416 | 4.5-5.5 | lacC/fruK | ND | ND | Fructose-1-phosphate kinase |
| Unknown function | |||||
| 408 | 4.5-5.5 | ygdA | ND | ND | Hypothetical protein |
pH range of 2-DE in which the corresponding protein was identified. CEA pH range, 4 to 7.
Fold change was calculated as the mean of the normalized volume of the spot detected on SMM or milk divided by the mean of the normalized volume of the spot detected M17Lac. “Greater than” indicates that the spot was not detected on M17Lac, the value was calculated by using the lowest normalized volume as a denominator; —, the fold change between milk and M17Lac was not significant.
ND, not determined. The corresponding spot was not detected on SMM or milk.
Increases of the peptide hydrolytic and transport capacities.
As noted above, the development of L. lactis in milk is dependent upon transport of peptides followed by their intracellular hydrolysis. Twelve cytosolic peptidases (PepA, PepC, PepDB, PepF, PepM, PepN, PepP, PepQ, PepO, PepO2, PepT, and PepV) have been identified on 2-DE; only four of them were found to be up-regulated in SMM and milk (Fig. 3 and Table 1). Three of these peptidases (PepC, PepN, and PepO) are encoded by genes which are repressed by the addition of a Casitone peptide source in the medium (13) and belong to the CodY regulon in L. lactis (14). The increase of PepF is interesting in that the control of pepF expression is independent of CodY (13). This could indicate either that an alternative mode of regulation of genes involved in nutritional peptide catabolism exists in L. lactis or that PepF activity is not dedicated to nitrogen utilization. The latter hypothesis is reminiscent of the finding that a Bacillus subtilis PepF ortholog, YjbG, is involved in the degradation of peptides having signaling properties (19).
We observed the synthesis of two PepO-like endopeptidases by the strain NCDO763, which differed in their pIs and peptide mass fingerprints (see Fig. S1 in the supplemental material). One protein, which increased massively during growth on SMM, has been identified as that encoded by the pepO gene, which is part of the oppDFBCA-pepO operon in strain MG1363 (39). The second PepO-like protein (PepO2) was produced at similar levels in the three media. Its tryptic fingerprint was mapped to the protein deduced from the pepO2 gene (accession no. AAF67832) from strain MG1363, which is also present in the genome of L. lactis SK11 (http://genome.jgipsf.org/draft_microbes/laccr/laccr.home.html; gene 113). These data show that the two PepO-encoding genes are functional in L. lactis NCDO763 and are not controlled in a similar way. It is thus likely that the duplication of the pepO gene corresponds to an adaptation to milk.
The gels of the CEA extracts obtained from cells cultivated on SMM and milk displayed a strong up-regulation of the three soluble components (OppA, OppF, and OppD) of the oligopeptide transport machinery Opp (Fig. 5). These proteins were detected only on the 2-DE gels of the CEA extracts. The implementation of this protocol thus helped us to analyze the cell transport capacity.
FIG. 5.
Activity of the glnA promoter in milk. Luciferase activity (circles) and pH (triangles) were measured during growth of strain TIL521 (glnA::luxAB, GS+) in milk (open symbols) or in milk supplemented with 10 mM glutamine (filled symbols) at 30°C. The level of luciferase activity in M17Lac was 75 ± 25 ULU/OD. The OD measurements are identical under both conditions; for clarity, they are not presented.
The amount of the oligopeptide binding component OppA is in large excess over those of the two ATP binding modules OppD and OppF. These proteins have been proposed to be encoded by a single transcriptional unit (39), and if this is the case, a posttranscriptional mechanism could modulate their cellular concentration. However, a second putative promoter was detected upstream of the oppA-pepO genes (39). The activation of the second promoter in milk could explain the high level of the OppA protein compared to OppD and OppF.
The peptides initially present in milk have been shown to be essential for the development of strains of L. lactis that are deficient in casein hydrolysis (18). In SMM, these peptides are likely to be a crucial source of amino acids. The analysis of the proteome in SMM shows that L. lactis has deployed the capacity to internalize and degrade these peptides through the OppA and cytoplasmic proteolytic systems, respectively.
Interestingly, we observed the synthesis of the OptS protein in cells cultivated in SMM. OptS is a putative substrate binding protein of a second oligopeptide ABC transporter, Opt (25). The operon encoding the Opp system (opp-pepO) belongs to the CodY regulon; the marked variation in expression of the OptS and OppA proteins during growth in milk suggests that a different regulatory mechanism drives the synthesis of OptS. It also indicates that the Opt system is probably not involved in casein-derived oligopeptide transport. In strain IL-1403, OptS was also not implicated in nutrient acquisition (25). The role of the system is thus currently unknown; it could function as an accessory nutrient acquisition system that is synthesized only under especially demanding conditions (such as in SMM), or an alternative hypothesis would be that it plays a part in the transport of peptides having intracellular signaling functions (26).
Changes in amino acid-synthesizing and -interconverting enzymes.
The concentration of free amino acids in milk is low, and the composition of the pool is unbalanced (18). Twenty-two enzymes involved in biosynthesis or interconversion of amino acids were found to be synthesized at a higher level during growth in SMM (Table 1), among which are aspartate kinase (ThrA) and aspartate-beta-semialdehyde dehydrogenase (Asd). These enzymes lie at the first branch point in the aspartate metabolic pathway that leads to the formation of l-aspartate 4-semialdehyde, the precursor of the amino acids lysine, isoleucine, methionine, and threonine. The amounts of several enzymes implicated in the later stages of the synthesis of these amino acids were found to increase in SMM: the threonine synthase (TrhC) responsible for the last step of threonine biosynthesis; YchH and diaminopimelate decarboxylase (LysA), which participate in the biosynthesis of lysine from aspartate-semialdehyde; and O-acetylhomoserine (thiol)-lyase (CysD), which is responsible for the synthesis of methionine. We also observed in SMM the synthesis of four proteins (TrpABDE) involved in the de novo synthesis of the aromatic amino acid tryptophan. Previous studies of the expression of the tryptophan biosynthetic operon in L. lactis have shown that it was activated by tryptophan depletion and general amino acid starvation (33); these two conditions are met in SMM. Four enzymes of the branched-chain amino acid (BCAA) biosynthetic pathway in SMM were produced in SMM: the large subunit of the acetolactate synthase IlvB, the ketol-acid reductoisomerase (IlvC), and the dihydroxy-acid dehydratase (IlvD) (Fig. 1). The ilv genes are located in the 12-kb operon which includes 10 structural genes involved in the biosynthesis or transport of BCAA, whose expression is triggered by BCAA starvation (11). The BCAA-specific aminotransferase BcaT, which is active in L. lactis NCDO763 (44), was also overproduced in SMM. The proteomic data obtained in SMM indicate that L. lactis NCDO763 can produce all the proteins necessary for Ile/Val biosynthesis from pyruvate, while the dairy strains of L. lactis are known to be auxotrophic for BCAA (34). Rather than a deficiency in enzyme production, the proteomic analysis suggests that the auxotrophy must be caused by a deficit in the activity of one or more of these products.
In milk-cultivated cells (Fig. 2C and 3C), the amounts of most the enzymes described above returned to the levels detected in M17Lac. This indicates that most of the amino acid starvation encountered in SMM is relieved by the casein hydrolysis taking place in milk. An exception is for BcaT, whose amount remained at a high level in milk. It has been previously shown that a bcaT araT double mutant is severely impaired for growth in milk, and it was postulated that BcaT, by maintaining a low level of intracellular BCAAs, relieves the repression of CodY-regulated genes during growth in milk (4). The proteomic analysis supports this idea and further shows that at the protein level, BcaT is regulated differently from the other enzymes of the BCAA biosynthetic pathway. The control of the expression of both bcaT and the leu-ilv operon is dependent upon BCAA availability; however, while bcaT belongs to the CodY regulon, the mechanism in play for leu-ilv has not been identified.
The initial phase of growth of L. lactis in milk is known to be dependent upon the small amount of free amino acids and peptides as a nitrogen source (18, 31), and a similar observation was recently made for another dairy species, Streptococcus thermophilus (29). The analysis of the proteome of L. lactis growing in SMM shows the synthesis of several enzymes involved in amino acid synthesis, whereas most of them are absent in the milk proteome profile. These results suggest that the amino acid-synthesizing abilities are involved in the initial phase of growth in milk, when it is important to out-compete eventual food-borne pathogens or spoilage microorganisms.
Activation of the purine nucleotide biosynthetic pathway.
Milk is poor in purine bases, while it contains a substantial amount of the pyrimidine precursor orotate (1); growth in this medium thus requires a functional purine biosynthetic pathway. L. lactis cultivated on SMM and milk synthesized high levels of 11 enzymes directly involved in the de novo synthesis of purine bases (Table 1). In parallel, the concentration of GlyA (serine methylase) increased significantly in SMM and milk. GlyA catalyzes the transformation of serine to glycine, an important step for the supply of methyl groups to diverse biosynthetic pathways. We also observed the up-regulation of formyl-tetrahydrofolate (THF) synthetase (Fhs) and THF-dehydrogenase/cyclohydrolase (FolD) in dairy media. These proteins are involved in the formation of folic acid derivatives that supply one-carbon units to various anabolic pathways. In L. lactis and B. subtilis, the promoter regions of the genes glyA and fhs contain the regulatory sequence (Pur box) characteristic of PurR-activated genes (1).
A recent proteomic analysis of L. lactis MG1363 cultivated under purine starvation conditions led to the identification of six up-regulated enzymes (PurS, PurE, PurM, PurL, GlyA, and Fhs), which are all listed in Table 1 (1). The present work thus extends the characterization of the experimental purine stimulon in L. lactis. We also observed that the amounts of three proteins that are directly related to the purine metabolism, i.e., adenylosuccinate lyase (PurB), xanthine phosphoribosyl transferase (Xpt), and FolD, increased in dairy media. The corresponding genes, however, do not show a canonical Pur box regulatory region, either because the Pur box is degenerate (folD and xpt) or because it does not have adequate spacing with the −10 region (folD, xpt, and purB) (1). The recent identification of a putative riboswitch G box structure that is responsive to the intracellular guanine concentration in the 5′ untranslated region of the L. lactis xpt gene (30) suggests that an mRNA-mediated modulation of gene expression could be at work in L. lactis growing in dairy media.
A major characteristic of the milk proteome profile is the production of enzymes involved directly or indirectly in the purine biosynthetic pathway. Several important steps towards the understanding of the regulation of expression of the corresponding genes in L. lactis have been accomplished (1, 20, 21, 30), and it is important to continue in this direction.
Glutamine synthetase is an essential enzyme for growth of L. lactis in milk.
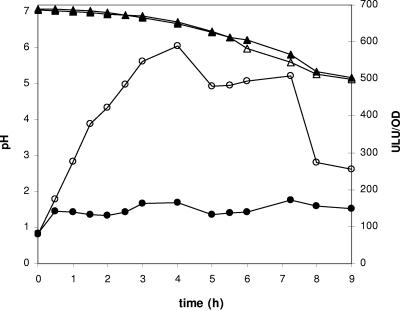
Differently from most of the enzymes involved in amino acid biosynthesis, which are increased in SMM but are down-regulated in milk, glutamine synthetase, encoded by the glnA gene, was maintained at a high level in milk-cultivated cells (Table 1; Fig. 3). By carrying out the ATP-dependent formation of glutamine from ammonia and glutamate, GS conveys nitrogen for amino acid and nucleotide biosynthesis. To determine if an increased expression of glnA was at the origin of the up-regulation of GS in milk, we generated a mutant strain that carries a glnA::luxAB transcriptional fusion (strain TIL521). The expression of glnA was then assessed by measuring luciferase activity in milk (Fig. 5). We observed a fivefold increase of luciferase activity during the first 4 hours of culture, after which it remained constant before starting to decrease at the entry into stationary phase (Fig. 5). The complementation of milk with 10 mM glutamine in the medium abolished this pattern, and the luciferase production was maintained at a low and constant level during the culture (Fig. 5), similar to that measured in M17Lac (data not shown). These results show that transcriptional activation mediated at least in part by glutamine starvation explains GS up-regulation in milk. Furthermore, the high level of activity measured throughout the development in milk suggests that the degradation of caseins does not fulfill the glutamine requirements.
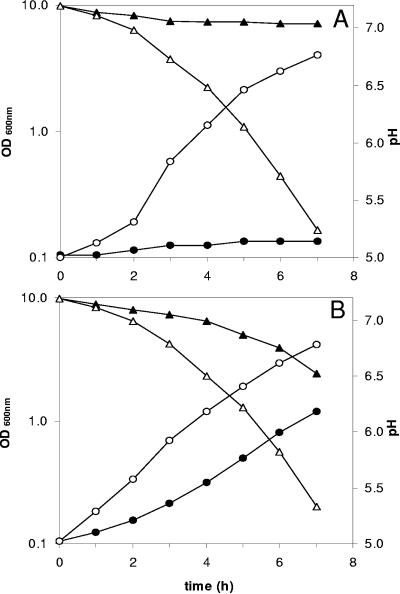
The de novo synthesis of purine bases requires the neosynthesis of 5-phospho-d-ribosylamine. The amino group of this compound is provided by glutamine, which is undetectable in SMM or milk (data not shown). The high intracellular concentration of glutamate, which is by far the most abundant free amino acid in milk (>350 μM), and the highest level of synthesis of GS revealed by the proteome analysis suggest a key role of the enzyme for the growth of L. lactis in dairy media. To confirm this hypothesis, we constructed a mutant strain (strain TIL520) in which the glnA gene was inactivated (see Materials and Methods). While the development of TIL520 in M17Lac was slightly affected (a 20% reduction in the maximal growth rate), the growth of TIL520 was totally impaired in SMM (data not shown) and milk (Fig. 6A), a phenotype that can be partially restored by the addition of glutamine to the medium (Fig. 6B). This result demonstrates that the production of glutamine through the activity of GS is a key aspect of the metabolic adaptation of L. lactis to milk.
FIG. 6.
Development of L. lactis NCDO763 (wild type) and L. lactis TIL520 (glnA::pTIL520) in milk. The growth of wild-type L. lactis NCDO763 (open symbols) and TIL520 (filled symbols) was monitored by measuring pH (triangles) and OD600 (circles) in milk (A) and milk complemented with 5 mM glutamine (B).
In B. subtilis, the expression of glnA is under the dual control of two transcriptional repressors, GlnR and TnrA (10). The situation in L. lactis is somewhat different in that no obvious TnrA homolog can be found in the genome of the dairy strain IL-1403. Given the importance of GS activity for development in milk, the control of its synthesis will certainly deserve further investigation. From this perspective, it would be of a special interest to compare the regulatory mechanisms of glnA expression and GS activity in dairy and nondairy strains of L. lactis.
The explanation of certain characteristics of the proteomic profile of L. lactis in milk would require further studies. This is the case for the strong increase in the concentration of the glucosamine-6-phosphate deaminase, NagB, in both SMM and milk (Table 1; Fig. 3). NagB catalyzes the conversion of the amino sugar glucosamine-6 phosphate into the glycolytic substrate fructose-6-phosphate, and ammonia. Bovine milk contains oligosaccharides, glycoproteins, and glycolipids, which are potential sources of N-acetylglucosamine (12). The amino sugar is fermentable by L. lactis (7), and it can thus be transported into the cell and further modified into glucosamine-6 phosphate, the substrate of NagB. This observation thus opens the possibility that L. lactis can use N-acetylglucosamine as an alternative carbon source in milk, leading to an intracellular production of ammonia that could be used for the biosynthesis of nitrogenous molecules or cytoplasmic pH homeostasis.
Concluding remarks.
By comparing the proteomic profiles of L. lactis cultivated in M17Lac broth, milk microfiltrate, and milk, the present work provides a global view of the adaptation to milk by a model dairy lactic acid bacterium. We observed that 10% (milk) to 20% (milk microfiltrate) of the proteins which can be identified on 2-DE are up-regulated in these media compared to in M17Lac. In the future, the comparison of proteomic patterns of strains possessing various technological abilities should give some clues to understand their properties. A difficulty would reside in the important protein polymorphism between strains (15), which makes a direct comparison of the 2-DE images of different strains challenging if not impossible. However, the systemic identification of all milk-variable proteins in a large number of strains is a realistic objective. Such a program could provide novel information for the selection of strains.
In the cheese industry L. lactis is widely used as a starter, and the acidification of the medium after its active development in milk is a major property required for technological strains. However, the metabolic activity of resting cells at acidic pH is also likely to participate in the next steps of the cheese making. From this perspective, a proteomic analysis of L. lactis along the ripening process would be of interest for understanding its role in this process.
In the group of lactic acid bacteria, Lactococcus, Streptococcus, and Lactobacillus are the predominant genera used for lactic acid food fermentation and particularly for the production of dairy products. We have already observed that the proteomic profiles of dairy LAB obtained after growth in broth show some similarities (reference 5 and unpublished results). It will be of interest to compare the strategies of adaptation to milk among dairy LAB. Once again, the comparison of proteome profiles would be an inexpensive and versatile tool for this purpose.
ADDENDUM IN PROOF
Since the submission of this paper, Monnet et al. (Appl. Environ. Microbiol. 71:3376-3378, 2005) have reported the essentiality of glutamine synthetase for the growth in milk of Streptococcus thermophilus, another dairy organism.
Supplementary Material
Acknowledgments
We thank Pierre Schuck and Jacques Faucant of the Laboratoire des Sciences et Technologie du Lait et de l'Oeuf (INRA, Rennes, France) for the generous gift of milk microfiltrate.
We acknowledge the support of the ACI-Bioinformatique IMASTE of the French Ministry of Research and Technology.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Beyer, N. H., P. Roepstorff, K. Hammer, and M. Kilstrup. 2003. Proteome analysis of the purine stimulon from Lactococcus lactis. Proteomics 3:786-797. [DOI] [PubMed] [Google Scholar]
- 2.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruinenberg, P. G., P. Vos, and W. M. De Vos. 1992. Proteinase overproduction in Lactococcus lactis strains: regulation and effect on growth and acidification in milk. Appl. Environ. Microbiol. 58:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambellon, E., and M. Yvon. 2003. CodY-regulated aminotransferases AraT and BcaT play a major role in the growth of Lactococcus lactis in milk by regulating the intracellular pool of amino acids Appl. Environ. Microbiol. 69:3061-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champomier-Vergès M.-C., E. Maguin, M.-Y. Mistou, P. Anglade, and J.-F. Chich. 2002. Lactic acid bacteria and proteomics: current knowledge and perspectives. J. Chromatogr. B 329:329-342. [DOI] [PubMed] [Google Scholar]
- 6.Delorme, C., S. D. Ehrlich, and P. Renault. 1999. Regulation of expression of the Lactococcus lactis histidine operon. J. Bacteriol. 181:2026-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vos, W. M. 1996. Metabolic engineering of sugar catabolism in lactic acid bacteria. Antonie Leeuwenhoek 70:223-242. [DOI] [PubMed] [Google Scholar]
- 8.Dudley, E., and J. Steele. 2001. Lactococcus lactis LM0230 contains a single aminotransferase involved in aspartate biosynthesis, which is essential for growth in milk. Microbiology 147:215-224. [DOI] [PubMed] [Google Scholar]
- 9.Fauquant, J., J.-L. Maubois, and A. Pierre. 1988. Microfiltration du lait sur membrane minérale. Tech. Lait 1028:21-23. [Google Scholar]
- 10.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol. Microbiol. 32:223-232. [DOI] [PubMed] [Google Scholar]
- 11.Godon, J. J., C. Delorme, J. Bardowski, M. C. Chopin, S. D. Ehrlich, and P. Renault. 1993. Gene inactivation in Lactococcus lactis: branched-chain amino acid biosynthesis. J. Bacteriol. 175:4383-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopal, P. K., and H. S. Gill. 2000. Oligosaccharides and glycoconjugates in bovine milk and colostrum. Br. J. Nutr. 84(Suppl. 1):S69-S74. [DOI] [PubMed] [Google Scholar]
- 13.Guedon, E., P. Renault, S. D. Ehrlich, and C. Delorme. 2001. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 183:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227-1239. [DOI] [PubMed] [Google Scholar]
- 15.Guillot, A., C. Gitton, P. Anglade, and M. Y. Mistou. 2003. Proteomic analysis of Lactococcus lactis, a lactic acid bacterium. Proteomics 3:337-354. [DOI] [PubMed] [Google Scholar]
- 16.Guimont, C., M.-A. Chopard, J.-L. Gaillard, and J.-F. Chamba. 2002. Comparative study of the protein composition of three strains of Streptococcus thermophilus grown either in M17 medium or in milk. Lait 82:645-656. [Google Scholar]
- 17.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 18.Juillard, V., D. Le Bars, E. R. Kunji, W. N. Konings, J. C. Gripon, and J. Richard. 1995. Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth in milk. Appl. Environ. Microbiol. 61:3024-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanamaru, K., S. Stephenson, and M. Perego. 2002. Overexpression of the PepF oligopeptidase inhibits sporulation initiation in Bacillus subtilis. J. Bacteriol. 184:43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilstrup, M., S. G. Jessing, S. B. Wichmand-Jorgensen, M. Madsen, and D. Nilsson. 1998. Activation control of pur gene expression in Lactococcus lactis: proposal for a consensus activator binding sequence based on deletion analysis and site-directed mutagenesis of purC and purD promoter regions. J. Bacteriol. 180:3900-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilstrup, M., and J. Martinussen. 1998. A transcriptional activator, homologous to the Bacillus subtilis PurR repressor, is required for expression of purine biosynthetic genes in Lactococcus lactis. J. Bacteriol. 180:3907-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konings, W. N., J. Kok, O. P. Kuipers, and B. Poolman. 2000. Lactic acid bacteria: the bugs of the new millennium. Curr. Opin. Microbiol. 3:276-282. [DOI] [PubMed] [Google Scholar]
- 23.Kunji, E. R., G. Fang, C. M. Jeronimus-Stratingh, A. P. Bruins, B. Poolman, and W. N. Konings. 1998. Reconstruction of the proteolytic pathway for use of beta-casein by Lactococcus lactis. Mol. Microbiol. 27:1107-1118. [DOI] [PubMed] [Google Scholar]
- 24.Kunji, E. R., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 25.Lamarque, M., P. Charbonnel, D. Aubel, J. C. Piard, D. Atlan, and V. Juillard. 2004. A multifunction ABC transporter (Opt) contributes to diversity of peptide uptake specificity within the genus Lactococcus. J. Bacteriol. 186:6492-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazazzera, B. A. 2001. The intracellular function of extracellular signaling peptides. Peptides 22:1519-1527. [DOI] [PubMed] [Google Scholar]
- 27.Le Bourgeois, P., M. L. Daveran-Mingot, and P. Ritzenthaler. 2000. Genome plasticity among related Lactococcus strains: identification of genetic events associated with macrorestriction polymorphisms. J. Bacteriol. 182:2481-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leloup, L., S. D. Ehrlich, M. Zagorec, and F. Morel-Deville. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letort, C., M. Nardi, P. Garault, V. Monnet, and V. Juillard. 2002. Casein utilization by Streptococcus thermophilus results in a diauxic growth in milk. Appl. Environ. Microbiol. 68:3162-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandal, M., B. Boese, J. E. Barrick, W. C. Winkler, and R. R. Breaker. 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113:577-586. [DOI] [PubMed] [Google Scholar]
- 31.Niven, G. W., D. J. Knight, and F. Mulholland. 1998. Changes in the concentrations of free amino acids in milk during growth of Lactococcus lactis indicate biphasic nitrogen metabolism. J. Dairy Res. 65:101-107. [DOI] [PubMed] [Google Scholar]
- 32.Palmfeldt, J., F. Levander, B. Hahn-Hagerdal, and P. James. 2004. Acidic proteome of growing and resting Lactococcus lactis metabolizing maltose. Proteomics 4:3881-3898. [DOI] [PubMed] [Google Scholar]
- 33.Raya, R., J. Bardowski, P. S. Andersen, S. D. Ehrlich, and A. Chopin. 1998. Multiple transcriptional control of the Lactococcus lactis trp operon. J. Bacteriol. 180:3174-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiter, B., and J. D. Oram. 1962. Nutritional studies on cheese starters. Dairy Res. 29:63-77. [Google Scholar]
- 35.Romeo, Y., D. Obis, J. Bouvier, A. Guillot, A. Fourcans, I. Bouvier, C. Gutierrez, and M. Y. Mistou. 2003. Osmoregulation in Lactococcus lactis: BusR, a transcriptional repressor of the glycine betaine uptake system BusA. Mol. Microbiol. 47:1135-1147. [DOI] [PubMed] [Google Scholar]
- 36.Saboya, L. V., and J. L. Maubois. 2000. Current development of microfiltration technology in the dairy industry. Lait 80:541-553. [Google Scholar]
- 37.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tobisch, S., D. Zuhlke, J. Bernhardt, J. Stulke, and M. Hecker. 1999. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J. Bacteriol. 181:6996-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tynkkynen, S., G. Buist, E. Kunji, J. Kok, B. Poolman, G. Venema, and A. Haandrikman. 1993. Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis. J. Bacteriol. 175:7523-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vido, K., D. Le Bars, M. Y. Mistou, P. Anglade, A. Gruss, and P. Gaudu. 2004. Proteome analyses of heme-dependent respiration in Lactococcus lactis: involvement of the proteolytic system. J. Bacteriol. 186:1648-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, H., W. Yu, T. Coolbear, D. O'Sullivan, and L. L. McKay. 1998. A deficiency in aspartate biosynthesis in Lactococcus lactis subsp. lactis C2 causes slow milk coagulation. Appl. Environ. Microbiol. 64:1673-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, J., C. Caron, M. Y. Mistou, C. Gitton, and A. Trubuil. 2004. PARIS: a proteomic analysis and resources indexation system. Bioinformatics 20:133-135. [DOI] [PubMed] [Google Scholar]
- 43.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]
- 44.Yvon, M., E. Chambellon, A. Bolotin, and F. Roudot-Algaron. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl. Environ. Microbiol. 66:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.