Abstract
Gram-positive bacteria of the genus Exiguobacterium have been repeatedly isolated from Siberian permafrost ranging in age from 20,000 to 2 to 3 million years and have been sporadically recovered from markedly diverse habitats, including microbial mats in Lake Fryxell (Antarctic), surface water, and food-processing environments. However, there is currently no information on genomic diversity of this microorganism or on the physiological strategies that have allowed its survival under prolonged freezing in the permafrost. Analysis of the genome sequence of the most ancient available Exiguobacterium isolate (Exiguobacterium sp. strain 255-15, from 2 to 3 million-year-old Siberian permafrost) revealed numerous putative transposase sequences, primarily of the IS200/IS605, IS30, and IS3 families, with four transposase families identified. Several of the transposase genes appeared to be part of insertion sequences. Southern blots with different transposase probes yielded high-resolution genomic fingerprints which differentiated the different permafrost isolates from each other and from the Exiguobacterium spp. type strains which have been derived from diverse surface habitats. Each of the Exiguobacterium sp. strain 255-15 transposases that were used as probes had highly conserved homologs in the genome of other Exiguobacterium strains, both from permafrost and from modern sites. These findings suggest that, prior to their entrapment in permafrost, Exiguobacterium isolates had acquired transposases and that conserved transposases are present in Exiguobacterium spp., which now can be isolated from various modern surface habitats.
Permafrost is a unique soil structure that embodies constantly frozen buried soils and sediments and underlies an estimated 20 to 25% of the earth's land surface (14). Permafrost reaches a thickness of more than 700 to 1,000 m in North America and Eurasia and also occurs in the ice-free regions of Antarctica and Greenland and in alpine regions. The permafrost dates back to as long as 2 to 3 million years in northeastern Siberia and is probably even older in Antarctica. The mean annual temperature of permafrost is between −10 and −12°C in the Arctic and between −18 and −27°C in the Antarctic. Permafrost sediments are firmly cemented by ice with content ranging from 12 to 70%, depending on the presence of ice wedges. The chemical composition of permanently frozen ground differs in different areas and depths in accordance with their origin (for a review, see reference 9).
Extensive studies have demonstrated that bacterial cells within these special soils survive for thousands of years and in deeper strata for up to several millions of years, with the age of the embedded cells corresponding to the age of the surrounding permafrost soil structure (9, 16, 21, 25). These bacteria pose quite unique opportunities for exploring the genomic organization of ancient, preserved microorganisms and the genetic relatedness between the permafrost microorganisms and their modern relatives from surface habitats. From an evolutionary point of view, permafrost bacteria have been naturally preserved in the frozen sediments in the absence of gene flow from any outside biota and represent ancestors of several bacterial species that can be isolated from modern surface habitats.
During previous work, we isolated representatives of several different taxa of gram-positive and gram-negative bacteria from Siberian permafrost core samples of markedly differing ages (10,000 to 2 to 3 million years) (24). Bacteria of the genus Exiguobacterium were unusual in being isolated from permafrost cores representing all available age ranges, including cores dated to 20,000 years (two strains), 600,000 years (one strain), and 2 to 3 million years (one strain) (T. A. Vishnivetskaya, S. Kathariou, and J. M. Tiedje, unpublished).
Exiguobacterium spp. are asporogenous gram-positive bacteria in the order Firmicutes (low-GC gram-positives). On the basis of the small-subunit rRNA sequences, Exiguobacterium has been clustered together with the round-spore-forming bacilli of the genus Bacillus and with the asporogenous taxa Caryophanon, Kurthia, and Planococcus (6, 7). To date, several Exiguobacterium species isolated from contemporary natural and food-processing environments have been described, including E. aurantiacum (type strain DSM6208, from alkaline potato processing plant effluent), E. undae (type strain DSM14481, from garden pond water in Germany), E. antarcticum (type strain DSM14480, from a microbial mat in Lake Fryxell in the Antarctic), and E. acetylicum (type strain DSM20416, from creamery plant waste) (3, 6-8).
Although a comparative analysis of physiological characteristics and of the phylogenetic relationships among these four Exiguobacterium spp. type strains has been described (8), there is currently no information on genomic diversity in this organism or on molecular subtyping tools that can be used to facilitate ecological and other studies. Such information and tools would be invaluable, not only because of the organism's remarkable ability to survive under subzero conditions in the permafrost but also because Exiguobacterium is emerging as a bacterium with impressive diversity in geographic distribution and habitats from which it has been isolated: in addition to the diverse origins of the type strains listed above, Exiguobacterium has been recently isolated from a glacial ice core sample in Greenland (13), the stem of potato plants in Austria (15), as well as hot springs in Colorado, in Yellowstone National Park, and in Southern India (Ganeshpuri, northeast of Bombay) (10; R. F. Ramaley, unpublished data).
Recently, the determination of the genomic sequence of Exiguobacterium sp. strain 255-15, isolated from one of the most ancient permafrost sediments (dated to 2 to 3 million years) has been undertaken in the context of the Joint Genome Institute Microbial Sequencing program (http://spider.jgi -psf.org/JGI_microbial/html/). Our analysis of the available genome sequence data revealed the presence of a large number of putative transposases, primarily of the IS200/IS605, IS30, and IS3 families. The putative transposase genes from this ancient permafrost strain were found to have highly conserved homologs in the genome of several other Exiguobacterium spp., not only from permafrost but also from modern environments. Strain specificity of the hybridization patterns obtained with transposase probes suggests the potential of these genes for the molecular subtyping for this microorganism.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains investigated in the present study and their sources are listed in Table 1. Permafrost strains 7-3 and 5138 had been isolated in earlier studies using direct plating of soil suspensions on Trypticase soy agar (Difco). Strains 190-11 and 255-15 had been isolated during these studies after incubation of natural permafrost sediments for 2 or 12 weeks, respectively, at 4°C, and subsequent plating on Trypticase soy agar (16, 24). Four strains, namely, E. aurantiacum DSM6208, E. undae DSM14481, E. antarcticum DSM14480, and E. acetylicum DSM20416, were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ GmbH; Braunschweig, Germany). Unless otherwise indicated, bacteria were grown in Trypticase soy broth with 0.7% yeast extract at 24°C overnight in standing cultures. For selected experiments, bacteria were also grown in the same medium at 4°C for 1 week. Bacterial cells were preserved at −70°C in Trypticase soy broth supplemented with 20% glycerol.
TABLE 1.
Strains of the genus Exiguobacterium included in this study
| Straina | Source | Reference(s) |
|---|---|---|
| Exiguobacterium sp. strains | ||
| 7-3 and 5138 | Permafrost sample (20,000 years old) from 8 m of borehole 6Kh-Yu, Khomus-Yuryakh River, northeast Siberia, Russia, 68°19′N, 154°58′E; August 1989 | 22 |
| 190-11 | Permafrost sample (600,000 years old) from 5.5 m of borehole 5/94, middle part of Bol'shaya Chykochya River, right bank, northeast Siberia, Russia, 69°10′N, 158°4′E; July 1994 | 24 |
| 255-15 | Permafrost sample (2 to 3 million years old) from 43.6 m of borehole 2/94, middle part of Bol'shaya Chykochya River, right bank, northeast Siberia, Russia, 69°10′N, 158°4′E; July 1994 | 24 |
| E. aurantiacum DSM 6208* | Alkaline effluent from sodium hydroxide peeling of potato in processing factories | 6 |
| E. antarcticum DSM 14480* | Microbial mat from the shallow, moated area of Lake Fryxell, McMurdo Dry Valleys region, Antarctica, 77°36′S, 162°6′E; February 1999 | 3,8 |
| E. undae DSM 14481* | Garden pond in Wolfenbuttel, Lower Saxony, Germany; June 2001 | 8 |
| E. acetylicum DSM 20416* | Creamery waste | 7 |
Strains followed by asterisks were obtained from DSMZ GmbH, Braunschweig, Germany.
Repeated freeze-thawing.
Bacteria grown at either 24 or 4°C were frozen in 1-ml volumes at −20°C in cryogenic vials (Nalge-Nunc International Corp.). Repeated freeze-thawing (from −20 to 24°C) was performed every 2 days. After each complete thawing at 24°C the vials were returned to −20°C and maintained at that temperature for 48 h until the next thawing cycle.
Extraction of genomic DNA.
Bacterial cells were harvested by centrifugation of 7 ml of an overnight culture and resuspended in 360 μl of lysis buffer (20 mM Tris-HCl [pH 8.0], 2 mM EDTA, 1.2% Triton X-100) with lysozyme (40 mg/ml). After incubation for 1 h at 37°C, 50 μl of proteinase K (20 mg/ml) (QIAGEN, Valencia, CA) and 400 μl of buffer AL supplied by QIAGEN (catalog no. 19075) were added, followed by incubation for 30 min at 70°C. Further steps for DNA isolation were as described in the DNeasy Tissue Handbook (QIAGEN) for gram-positive bacteria.
16S rRNA gene amplification and sequencing.
The 16S rRNA genes were amplified with Bacteria-specific primers 8F and 1492R, targeted to E. coli 16S rRNA positions 8 to 27 (5′-AGA GTT TGA TCC TGG CTC AG-3′) and positions 1512 to 1492 (5′-ACG GTT ACC TTG TTA CGA CTT-3′), respectively. The thermal PCR profile was as follows: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 1 min, primer annealing at 54°C for 1 min, and elongation at 72°C for 1.5 min. The final elongation step was 7 min at 72°C.
The 16S rRNA gene of Exiguobacterium strain 5138 was sequenced at the University of North Carolina at Chapel Hill (UNC-CH) Genome Analysis Facility on a 3730 DNA Analyzer (Applied Biosystems). Primers 8F and 1492R (see above), and additional internal primers targeted to E. coli positions 342 to 357 (5′-CTA CGG G(AG)(GC) GCA GCA G-3′) and positions 1100 to 1115 (5′-AGG GTT GCG CTC GTT G-3′) were used to obtain overlapping DNA fragments.
Transposase coding sequences and primer design.
Genome sequence data of Exiguobacterium strain 255-15 are available in a number of contigs (http://genome.ornl.gov/microbial/exig/) and have been deposited in the NCBI database (accession number NZ_AADW00000000, contigs NZ_AADW01000001 to AADW01000138). Transposase genes were found in genome of strain 255-15 by a keyword search through the orthologous groups of proteins (COGs) tab-delimited datafile (http://genome.ornl.gov/microbial/exig/10feb04/cogs_summary). Nucleotide and amino acid sequences were obtained with annotated database files and Artemis_v5 (Software Foundation, Inc., Boston, MA) (18). Primers were designed by using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi/) (17) and were purchased from QIAGEN. Primer sequences are listed in Table 2.
TABLE 2.
Primer pairs, positions of amplified fragments, and thermal cycling conditions used in this study
| Primera | Accession no. | Sequences (5′ to 3′) | Positions of amplified fragments on chromosomal DNA of strain 255-15b | Thermal cycling steps used in PCR
|
Product size (bp) | ||
|---|---|---|---|---|---|---|---|
| Denaturation | Annealing | Elongation | |||||
| or0823F | AADW01000010 | TTCGAAAGAAATCCCAATGC | Contig196 position 14723. . .15621 | 95°C, 60 s | 52°C, 60 s | 72°C, 90 s | 899 |
| or0823R | AADW01000010 | GACATGTCCATTCACGCAAG | |||||
| or1938F | AADW01000039 | GAAACTGAACCCTGGCGTAA | Contig178 position 18387. . .17483 | 95°C, 60 s | 54°C, 60 s | 72°C, 90 s | 905 |
| or1938R | AADW01000039 | CACCGCAAGTACATGTCCAC | |||||
| or1018F | AADW01000014 | TGACGAGCAAGAATGTCCAG | Contig184 position 36355. . .37270 | 95°C, 60 s | 56°C, 60 s | 72°C, 90 s | 916 |
| or1018R | AADW01000014 | CCATCAACCACAGGTGCATA | |||||
| or1017F | AADW01000014 | CCACCGTTTCTCTGATCCAT | Contig184 position 35835. . .36169 | 95°C, 60 s | 54°C, 60 s | 72°C, 50 s | 335 |
| or1017R | AADW01000014 | TGTTTCGCTCGACACATTTC | |||||
| or0882F | AADW01000011 | AAATACAGACGCAAAGTGATCG | Contig194 position 32455. . .32763, Contig203 position 20830. . .21138 | 95°C, 60 s | 54°C, 60 s | 72°C, 50 s | 309 |
| or0882R | AADW01000011 | GTGTCTGATGACGTCGCTTG | |||||
| or0411F | AADW01000004 | GGAATCTGCATCCTCGAAAA | Contig112 position 10037. . .10706, | 95°C, 60 s | 54°C, 60 s | 72°C, 70 s | 708 |
| or0411R | AADW01000004 | GTGCAACACTTCTTCCGTGA | Contig172 position 16196. . .16903, | ||||
| Contig184 position 4718. . .5425, | |||||||
| Contig196 position 24844. . .25551, | |||||||
| Contig200 position 7001. . .7708, | |||||||
| Contig203 position 160319. . .161026 | |||||||
| or0173F | AADW01000001 | TCGCTCTTCGAGCTTTCAGT | Contig141 position 9091. . .9677, Contig193 position 38093. . .38679, Contig185 position 12469. . .11921, Contig197 position 59520. . .60106, Contig203 position 58478. . .59064 | 95°C, 60 s | 54°C, 60 s | 72°C, 70 s | 587 |
| or0173R | AADW01000001 | CGTGACCTCCTCTCGATCTC | |||||
| or0174F | AADW01000001 | TTGCGGTGAACGGAATCTAT | Contig141 position 9877. . .10532, Contig199 position 213. . .869, Contig203 57622. . .58278 | 95°C, 60 s | 54°C, 60 s | 72°C, 70 s | 657 |
| or0174R | AADW01000001 | CCAAGCCGTTTAATCGTTGT | |||||
| or1723F | AADW01000031 | CATTCGCCGTTCATCATCTT | Contig81 position 3707. . .4110, Contig176 position 30272. . .30675, Contig184 53340. . .53743, Contig199 position 55267. . .55670 | 95°C, 60 s | 54°C, 60 s | 72°C, 50 s | 404 |
| or1723R | AADW01000031 | ATATCGCTCGACGGATTTTC | |||||
The primer names include gene identification number of the ORF selected for primer design; the accession number of the contig harboring ORF is indicated in column 2.
ORFs used for probe construction are indicated in boldface.
DNA probe construction and Southern blots.
Fragments of transposase genes were amplified by PCR. The reaction volume (100 μl) contained 1× PCR buffer, 2.5 mM concentrations of each deoxynucleotide triphosphate, 0.2 μM final concentrations of each primer, 2.5 U of TaKaRa ExTaq polymerase (TaKaRa Bio, Inc., Japan), and 20 ng of genomic DNA of strain 255-15 as a template. Cycling conditions were defined for each primer pair: an initial denaturation at 95°C for 5 min, followed by 30 cycles (1 min at 95°C and 1 min at 52 to 56°C, depending on primer melting temperature, and 50 to 90 s at 72°C, depending on length of amplified fragment, followed by a final extension for 7 min at 72°C) (see Table 2 for details). The PCR products were purified by using QIAquick spin columns (QIAGEN) and labeled with digoxigenin-dUTP (Roche Diagnostics Corp.) according to the manufacturer's instructions.
For Southern blots, genomic DNA (1 μg) was digested with EcoRI (New England Biolabs, Beverly, MA) overnight at 37°C, resolved on an 0.8% agarose gel, transferred to a nylon membrane (Osmonics, Inc.), and hybridized overnight with digoxigenin-labeled probes at 42°C in standard buffer (Roche) with 50% formamide. Membrane was washed at 42°C twice for 10 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer with 0.1% sodium dodecyl sulfate and then twice for 20 min in 0.5× SSC buffer with 0.1% sodium dodecyl sulfate. Hybridizing bands were detected by chemiluminescence using CSPD (Roche) according to the instructions of the manufacturer.
Phylogenetic analyses.
Sequences were initially aligned with the most similar sequences in the small subunit rRNA database using the algorithm in the Ribosomal Database Project (5). Additional 16S rRNA sequences from closely related bacteria were retrieved from GenBank after identification by BLAST (1). Multiple sequence alignment was done by using the program CLUSTAL X (23). Phylogenetic 16S rRNA analyses were performed by the maximum-likelihood (19) and neighbor-joining (20) methods. Bootstrap values were based on 100 trees generated by using the program MEGA3 (11).
Nucleotide sequence accession number.
The 16S rRNA gene sequence of Exiguobacterium sp. strain 5138 was deposited in NCBI database under accession number AY831656.
RESULTS
Phylogenetic analysis of Exiguobacterium spp. from permafrost.
Sequences of 16S rRNA genes from the eight Exiguobacterium isolates in our collection and other Exiguobacterium spp. formed two distinct clusters. One cluster consisted of isolates mostly from freshwaters or Siberian permafrost, whereas the members of the other cluster were primarily from slightly alkaline and marine environments (Fig. 1). All studied Exiguobacterium strains except strain E. aurantiacum DSM6208 were grouped together in the first cluster with sequence identity range of 95.6 to 99.7%. Permafrost isolates 7-3 and 255-15 were closely (99.7%) related to each other, and they both had 99.0% sequence identity to E. antarcticum DSM 14480, whereas permafrost strains 190-11 and 5138 had 98.3 to 99.6% 16S rRNA sequence identity with each other and with E. undae DSM 14481, E. oxidotolerans T-2-2, and Exiguobacterium sp. strain ARCTIC-P28 (Fig. 1). 16S rRNA gene sequence identity between E. aurantiacum DSM6208 and the other seven Exiguobacterium spp. isolates characterized in the present study ranged from 92.4 to 95.5%, with highest similarity to isolates 7-3 and 255-15.
FIG. 1.
Phylogenetic relationships of Siberian permafrost isolates and selected relatives. Strains studied are shown in boldface. The sequence of Arthrobacter globiformis DSM 20124T was used as an outgroup. Bootstrap values greater than 50% are shown. Accession numbers of previously published sequences are indicated in parentheses. The “T” indicates type strains.
Putative transposase sequences in the genome of Exiguobacterium sp. strain 255-15.
Analysis of the annotated genome of Exiguobacterium sp. strain 255-15 (draft version, http://genome.ornl.gov/microbial/exig/) identified 47 putative transposases, which were grouped into nine homology groups. Seven of these groups contained from 2 to 12 open reading frames (ORFs), which had nucleotide sequence identity higher than 77.3%, whereas two groups were represented by a single transposase sequence (Table 3).
TABLE 3.
Putative transposases in the genome of Exiguobacterium sp. strain 255-15
| IS family | Gene IDa | Homology groupb | G+C content (mol%) | Length
|
Product | |
|---|---|---|---|---|---|---|
| bp | aac | |||||
| IS605/IS200 | 0823*, 0073, 0095, 0413, 0502 | I (77.3-90.5) | 49.19-51.66 | 1,173 | 390 | IS605 orfB |
| 0603 | I (77.3-90.5) | 50.13 | 1,167 | 388 | IS605 orfB | |
| 1900 | I (77.3-90.5) | 49.47 | 750 | 249 | IS605 orfB | |
| 1938*, 0398, 0692, 2634, 0883, 0012 | II (83.8-97.5) | 45.69-48.14 | 1,101 | 366 | IS605 orfB | |
| 1122 | II (83.8-97.5) | 46.18 | 747 | 248 | IS605 orfB | |
| 0987 | II (83.8-97.5) | 44.97 | 696 | 231 | IS605 orfB | |
| 0988 | II (83.8-97.5) | 44.92 | 492 | 163 | IS605 orfB | |
| 1018* | III (NA) | 45.22 | 1,077 | 358 | IS605 orfB | |
| 1017* | IV (NA) | 42.79 | 402 | 133 | IS200 | |
| 0882*, 0011 | V (98.0-100.0) | 44.86, 45.61 | 399 | 132 | IS200 | |
| IS30 | 0411*, 2662, 2492, 2494, 1905, 0995, 1141, 0829, 0075, 2913 | VI (99.0-100.0) | 47.18-47.5 | 939 | 312 | IS30 |
| 0571 | VI (99.0-100.0) | 47.39 | 942 | 313 | IS30 | |
| 2910 | VI (99.0-100.0) | 46.51 | 387 | 128 | IS658 | |
| IS3 | 0173*, 0791, 2226, 1202, 1265 | VII (86.1-99.7) | 43.51-46.76 | 678 | 225 | IS3, orfA |
| 0174*, 0614 | VIII (83.8-100.0) | 45.78-45.87 | 750 | 249 | IS3, orfB | |
| 1203, 1266 | VIII (83.8-100.0) | 45.78-46.2 | 723 | 240 | IS3, orfB | |
| 2227 | VIII (83.8-100.0) | 47.59 | 435 | 144 | IS3, orfB | |
| 2228 | VIII (83.8-100.0) | 44.44 | 342 | 113 | IS3, orfB | |
| ISNCY | 1723* | IX (99.8-100.0) | 46.51 | 516 | 172 | CREd |
| 1032 | IX (99.8-100.0) | 46.35 | 561 | 187 | CRE | |
| 2917 | IX (99.8-100.0) | 46.96 | 939 | 312 | CRE | |
| 0657 | IX (99.8-100.0) | 46.78 | 1,584 | 527 | CRE | |
ORFs used to construct probes are indicated by asterisks. ID, identification number.
The percent identity between transposase coding nucleotide sequences inside the group is shown in parentheses. ORFs chosen for probe design had more than 82% identity to other ORFs inside the group. NA, not applicable (ORFs are unique and have 20.6 to 26.0% [ORF1018], and 20.9 to 25.9% [ORF1017] identity to others).
aa, amino acids.
CRE, cassette chromosome recombinase.
At the nucleotide sequence level, the putative Exiguobacterium transposases lacked significant identity with other sequences in the database, suggesting that these were novel transposases. Only short nucleotide sequence fragments of ORF1017 (118/142 bp; IS200), of ORF1018 (73/86 bp; IS605), of ORF0411 (66/77 bp; IS30), and of ORF2917 (54/62 bp; cassette chromosome recombinase) had 83 to 87% identity to transposase coding sequences of Bacillus cereus ATCC 14579 and B. halodurans. At the amino acid level, the highest identity of 78% (104/133) and 64% (232/359) was observed for ORF1017 (IS200) and ORF1018 (IS605), respectively, to transposases from B. cereus ATCC 14579.
Putative IS elements in the genome of Exiguobacterium sp. strain 255-15.
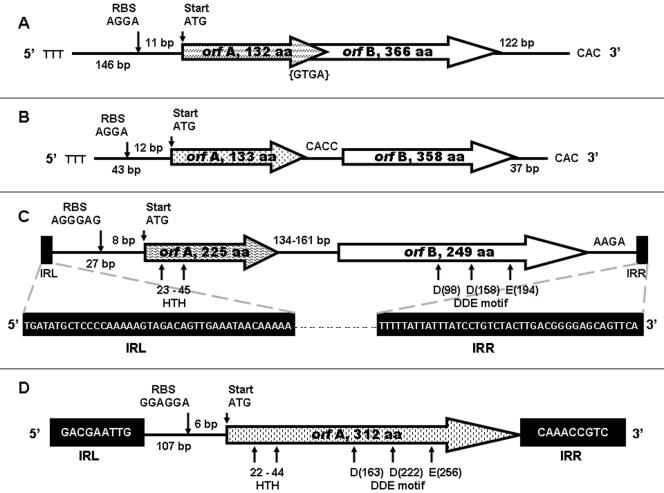
Protein BLAST-based analysis of the deduced polypeptides suggested that the putative transposases were members of insertion sequence (IS) families: IS200/IS605, IS3, IS30, and ISNCY (data not shown). Based on the suggested nomenclature (12), these IS elements were designated ISEsp1 (2 copies) and ISEsp2 (1 copy), both of the IS200/IS605 family; ISEsp3 (5 copies) of the IS3 family; and ISEsp4 (11 copies) of the IS30 family. ISEsp1, ISEsp2, and ISEsp3 contain two consecutive ORFs for two putative unrelated transposases, orfA and orfB, whereas ISEsp4 contains just one ORF, orfA (Fig. 2).
FIG. 2.
Organization of IS elements residing in the genome of Exiguobacterium sp. strain 255-15. The ORFs are represented as cross-hatched (orfA) and open (orfB) boxes; the direction of the transcription is indicated. The black boxes at each end represent the left (IRL) and right (IRR) terminal inverted repeats. (A and B) ISEsp1 (A) and ISEsp2 (B), both are IS605 elements; (C) ISEsp3 from IS3 family; (D) ISEsp4 from IS30 family.
ISEsp1 and ISEsp2 contained two ORFs for the putative transposases of IS200 (orfA) and IS891/IS1136/IS1341 (orfB) and were designed as IS605 elements, as suggested (4). As is typical for IS605 (12), ISEsp1 and ISEsp2 elements lacked terminal repeats, whereas predicted flanking regions terminated with 5′-TTT- - - -CAC-3′. In spite of their similar features (Fig. 2A and B), ISEsp1 and ISEsp2 had less than 26% identity at the nucleotide sequence level.
ISEsp3 contained ORFs for two putative unrelated transposases transcribed in the same direction and separated by an intergenic region of either 161 or 134 bp (Fig. 2C). ISEsp3 was flanked by 41-bp imperfect inverted terminal repeats containing nine mismatches. As is the case with the majority of the elements of the IS3 family (12), ISEsp3 terminated with 5′-TG- - - -CA-3′, and terminal repeats contained an internal block of G/C residues. The predicted primary amino acid sequence of the orfA protein of ISEsp3 exhibited a relatively strong helix-turn-helix motif, whereas orfB harbored a DDE motif. The leucine zipper harbored by many members of the IS3 family at the end of orfA (12) was not detected in ISEsp3.
Another IS element, ISEsp4 (1,056 bp), was present in 12 copies in the genome of 255-15 and belonged to the IS30 family. ISEsp4 contained a single ORF of 939 bp, which spanned almost the entire length. ISEsp4 was flanked by 9-bp imperfect inverted terminal repeats containing two mismatches (Fig. 2D). The predicted amino acid sequence of the orfA protein of ISEsp4 exhibited a relatively strong helix-turn-helix motif and a conserved DDE motif.
Southern blots with group-specific transposase probes confirm the number of putative transposases of each group in the genome of Exiguobacterium sp. strain 255-15.
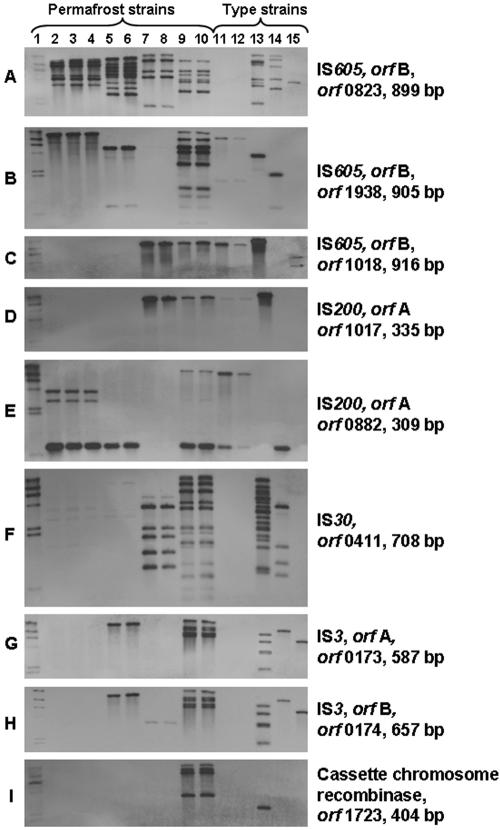
Southern blots were performed by using as probes PCR-amplified internal fragments of selected transposase coding sequences, chosen so as to include one sequence from each of the nine groups identified on the basis of DNA homology (Table 3). In the genome of Exiguobacterium sp. strain 255-15, none of the 47 putative transposase coding sequences harbored internal EcoRI sites, and the number of EcoRI fragments expected to hybridize with each probe would therefore correspond to the number of transposase genes within each homology group. Exceptions may be due to fragments of similar sizes, or fragments harboring more than one transposase from the same group. The Southern blot results with Exiguobacterium sp. strain 255-15 were indeed in close agreement with those expected on the basis of the number of homologous genes identified in each group, and only minor deviations were observed. In one case, the probe derived from ORF0823 (IS605, homology group I) resulted in six bands (Fig. 3A) when seven were expected. Analysis of the genomic data indeed revealed that two homologous ORFs were located on EcoRI fragments of similar size (data not shown). In the second case, the probe derived from ORF0411 (IS30, homology group VI) resulted in 13 bands (Fig. 3F), 1 more than predicted. Further screening of the genomic sequences, however, identified one truncated fragment (580 bp) with 100% identity to an internal fragment of ORF0411 that had not been listed in the draft annotation, thus accounting for the additional hybridizing fragment observed in the Southern blot with this probe. In a third case, the probe derived from ORF0174 (IS3, homology group VIII) resulted in five hybridizing bands (Fig. 3H), whereas six were expected, due to the fact that two of the transposases in this group (ORF2227 and ORF2228) were localized on a single EcoRI fragment.
FIG. 3.
Southern hybridization analysis of genomic DNAs from Exiguobacterium strains. EcoRI-digested genomic DNA was resolved on 0.8% agarose gel, transferred to nylon membrane, and hybridized with the corresponding probe as described in Materials and Methods. Unless otherwise indicated, bacteria were grown at 24°C. The ORF designation and size of probe are indicated on the right-hand side. Lane 1, DNA molecular weight marker II digoxigenin-labeled, 0.12 to 23.1 kbp (Roche). Lanes 2 to 15 are Exiguobacterium strains, as follows: 2, strain 7-3; 3, strain 7-3 grown at 4°C; 4, strain 7-3 after eight cycles of freeze-thawing and subsequent growth at 24°C; 5, strain 5138; 6, strain 5138 grown at 4°C; 7, strain 190-11; 8, strain 190-11 grown at 4°C; 9, strain 255-15; 10, strain 255-15 grown at 4°C; 11, E. aurantiacum DSM 6208; 12, E. aurantiacum DSM 6208 grown at 4°C; 13, E. antarcticum DSM 14480; 14, E. undae DSM 14481; 15, E. acetylicum DSM 20416.
Putative transposase genes are conserved in the genomes of other Exiguobacterium spp. from permafrost and other sources.
Southern blots with the probes representing each homology group were used to determine whether the putative transposases identified in the sequenced genome of Exiguobacterium sp. strain 255-15 had homologs in the genomes of the other three Exiguobacterium sp. isolates from permafrost. With the exception of the probe derived from ORF1723 (cassette chromosome recombinase ISNCY, group IX) which hybridized only with Exiguobacterium sp. strain 255-15, each of the eight remaining probes hybridized with one or more of the other permafrost isolates. Three probes, derived from ORF0823 (IS605, group I), ORF1938 (IS605, group II), and ORF0411 (IS30, group VI), hybridized with all four permafrost genomes, with the ORF0823 probe yielding multiple, strongly hybridizing bands with each of the isolates (Fig. 3A, B, and F and Table 4). Of the remaining five probes, the probe derived from ORF0882 (IS200, group V) hybridized with two of the three genomes, as did the probe derived from ORF0174 (IS3, group VIII), and three probes hybridized with only one of the additional genomes (Fig. 3 and Table 4). The patterns of reactivity of each isolate with the panel of nine probes could clearly differentiate among each of the four isolates (Table 4).
TABLE 4.
Observed distribution of the transposase coding sequences among Exiguobacterium strains
| Transposase | Representative ORF | Probe size (bp) | No. of bandsa for strain:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 255-15 | 7-3 | 5138 | 190-11 | DSM 6208 | DSM 14480 | DSM 14481 | DSM 20416 | |||
| IS605, orfB | ORF0823 | 899 | 6 | 6 | 10 | 5 | 0 | 8 | 5 | 1 |
| IS605, orfB | ORF1938 | 905 | 6 (3) | 1 (4) | 1 (2) | ((1)) | (2) | 1 (1) | 1 (1) | 0 |
| IS605, orfB | ORF1018 | 916 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 2 |
| IS200 | ORF1017 | 335 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| IS200 | ORF0882 | 309 | 1 (1) | 3 | 1 | 0 | 2 | 0 | 1 | 0 |
| IS30 | ORF0411 | 708 | 7 (6) | (4) | (2) | 5 (1) | 0 | 16 | 4 (2) | 0 |
| IS3, orfA | ORF0173 | 587 | 5 | 0 | 1 | 0 | 0 | 5 | 1 | 1 |
| IS3, orfB | ORF0174 | 657 | 5 | 0 | 1 | 1 | 0 | 5 | 1 | 1 |
| CREb | ORF1723 | 404 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Total | 46 | 18 | 18 | 15 | 6 | 39 | 16 | 5 | ||
Numbers indicate bands detected by Southern hybridization. Hybridizing bands with low hybridization signal are given in parentheses. Double parentheses indicate a low hybridization signal that could be detected over long-time exposure.
CRE, cassette chromosome recombinase.
As mentioned above, on the basis of 16S rRNA sequence analysis Exiguobacterium sp. strain 255-15 was closest to Exiguobacterium sp. strain 7-3, having sequence identity of 99.7% (Fig. 1). However, of the five probes that failed to hybridize with all three genomes only the probe derived from ORF0882 (IS200, group V) hybridized with the genome of Exiguobacterium sp. strain 7-3. In contrast, three of these probes hybridized with the genome of the phylogenetically more distant Exiguobacterium sp. strain 190-11 (the 16S rRNA gene sequence of which was 97.9% identical to that of Exiguobacterium sp. strain 255-15). Such data suggest that the distribution of these transposases and the corresponding IS elements is not congruent with the phylogeny of the organisms, possibly as a result of IS transfer between Exiguobacterium spp.
The type strains included in the present study represented four different species, isolated from different environments (Table 1). Interestingly, several of the transposase genes in the genome of Exiguobacterium sp. strain 255-15 appeared to have closely related homologs in the genomes of one or more of the type strains. The ORF0823-based probe, which hybridized with all four permafrost isolates, hybridized strongly with the genomes of the type strains as well, except for E. aurantiacum DSM 6208 (Fig. 3A and Table 4). E. antarcticum DSM 14480 was noteworthy among the type strains in hybridizing with all but one of the nine probes, yielding multiple hybridization bands with several of the probes. A strong hybridization signal was obtained with the genome of this strain even with the probe derived from ORF1723 (cassette chromosome recombinase), which had not hybridized with any of the isolates from permafrost (other than Exiguobacterium sp. strain 255-15) (Fig. 3I and Table 4).
The data summarized in Table 4 indicate that Exiguobacterium spp. from permafrost sediments, microbial mat from Lake Fryxell in the Antarctic, or surface water had variable but typically multiple (15-19) transposase hybridization bands with transposase probes derived from Exiguobacterium sp. strain 255-15 (Table 4). Food processing plant-derived strains E. aurantiacum DSM 6208 and E. acetylicum DSM 20416 appeared to have a significantly smaller copy number of sequences homologous to these probes, yielding a total of only six and five hybridizing bands, respectively.
Transposase-derived probes as molecular subtyping tools for Exiguobacterium.
Each of the eight Exiguobacterium spp. isolates could be distinguished from each other not only on the basis of its reactivity with the panel of the nine probes but also by its hybridization pattern (number and size of hybridizing fragments) with specific probes. The probes derived from ORF0823, ORF1938, or ORF0411 appeared most suitable for such differentiation, since they yielded multiple, strongly hybridizing fragments with several isolates (Fig. 3A, B, and F and Table 4).
The hybridization patterns following growth at 24 and 4°C or repeated freeze-thaw cycles were stable in all strains (Fig. 3 and data not shown). Selected strains were also examined after prolonged (4 months) storage at −20°C, and no changes in hybridization patterns were detected (data not shown).
Tandem transposases in the genomes of Exiguobacterium spp. correspond to bipartite elements IS3 and IS200/IS605.
Probes derived from ORF0173 (orfA of IS3 family) and its immediately adjacent ORF0174 (orfB of IS3 family) hybridized with the same EcoRI fragment in the genome of Exiguobacterium sp. strain 255-15, as well as in several other genomes (permafrost isolate Exiguobacterium sp. strain 5138, as well as E. antarcticum DSM 14480, E. undae DSM 14481, and E. acetylicum DSM 20416). In the case of E. antarcticum DSM 14480 the same five hybridizing bands were obtained with both probes, suggesting five copies of the element, each with an orfA and orfB transposase on the same EcoRI fragment (Fig. 3G and H and Table 4). Isolates that harbored orfA also harbored orfB, with the exception of Exiguobacterium sp. strain 190-11, derived from permafrost, which hybridized with ORF0174 (orfB) but not ORF0173 (orfA) (Fig. 3G and H and Table 4).
Identical hybridizing bands were also obtained with probes derived from ORF1017 (orfA of IS200 family) and its immediately adjacent ORF1018 (orfB of IS605 family) in Exiguobacterium spp. 255-15 and 190-11, as well as E. aurantiacum DSM 6208 and E. antarcticum DSM 14480, with a single band hybridizing in each case (Fig. 3C and D). Each IS200 element was immediately adjacent to transposase IS605 orfB in the genome of strain 255-15 (Table 3), and the Southern blot data suggest that the tandem gene arrangement of IS200/IS605 transposases also exists in Exiguobacterium sp. strain 190-11, E. aurantiacum DSM 6208 and E. antarcticum DSM 14480. There was only one possible exception, E. acetylicum DSM 20416, which hybridized with ORF1018 (orfB), but not with the ORF1017 (orfA)-derived probe (Fig. 3C and D and Table 4). It is not clear whether the lack of hybridization signals reflected the absence of the second ORF sequences or the presence of a divergent ORF that lacked sufficient homology with the probes.
DISCUSSION
Exiguobacterium spp. have been isolated from or molecularly detected in an impressive diversity of habitats, including Arctic permafrost, mats of Lake Fryxell in the Antarctic, surface water, several types of food processing plants, and a range of saline or alkaline environments. In the present study, we have shown that putative transposase genes identified in the genome of Exiguobacterium sp. strain 255-15, isolated from ancient (2 to 3 million years old) Siberian permafrost, had highly conserved homologs in the genomes of other Exiguobacterium spp. not only from permafrost but also from other, diverse habitats.
Transposase probes (such as ORF0823) hybridized strongly with most screened Exiguobacterium spp. genomes, and each of the nine probes had homologs in one or more of the other Exiguobacterium spp. isolates, including representatives of four different species. The findings suggest that the transposases (and associated IS elements) in the genome of the ancient permafrost isolates entered the Exiguobacterium spp. gene pool prior to the time that selected lineages became entrapped in the permafrost, and that modern Exiguobacterium spp. lineages, in spite of their extensive geographical range and their ecologically markedly diverse habitats, still harbor transposase genes highly conserved with those of strain 255-15. This may indicate that Exiguobacterium isolates from diverse sources comprise a common gene pool permissive of IS element transfer. Alternatively, currently unidentified selective pressures have driven conservation and maintenance of the transposase genes (and associated IS elements) in these isolates.
Of the different isolates investigated here, including those from permafrost, it was E. antarcticum DSM 14480 that was found to have the greatest similarity to the ancient permafrost isolate (Exiguobacterium sp. strain 255-15) in terms of the reactivity pattern with the nine transposase probes. Exiguobacterium sp. strain 255-15 was also phylogenetically closer to E. antarcticum DSM 14480 than to any of the other type strains, suggesting that their shared transposase repertoire preceded their geographic separation and, in the case of the permafrost isolate, the entrapment into the frozen sediment. Further studies are needed to characterize the genomic differentiation between these two isolates and to identify possible transposases and IS elements that may be harbored by E. antarcticum DSM 14480 but not by Exiguobacterium sp. strain 255-15. In addition, the actual nucleotide sequences of the homologous transposases in E. antarcticum DSM 14480 (and other Exiguobacterium strains) will need to be determined in order to more accurately evaluate their evolutionary relationship with their counterparts in the genome of the ancient permafrost isolate. Such studies are currently under way in our laboratory.
Multiple, strongly hybridizing bands suggested multiple copies of the corresponding transposase gene in several of the genomes (especially IS605 orfB ORF0823 and IS30 ORF0411 in the permafrost isolates, E. antarcticum DSM 14480 and E. undae DSM 14481). Multiple copies with high genetic similarity to each other would be expected to result from transposition events within the genome, suggesting that the corresponding IS elements are active in these organisms. Hybridization data did not yield evidence for activity under selected laboratory conditions and signals that may induce activity of the transposases remain to be identified. Functional transposases hold promise as tools for transposon mutagenesis of Exiguobacterium spp. Although a mercury resistance transposon has been described in Exiguobacterium (2), genetic tools for these bacteria have not been reported.
In conclusion, we describe the detection of highly conserved putative transposases in the genomes of Exiguobacterium spp. from ancient permafrost and from widely diverse environments. These sequences hold potential as molecular subtyping tools for tracing the place of origin or the genetic ancestry of these hitherto poorly characterized organisms that exhibit impressive diversity in ecological habitats and geographic distribution.
Acknowledgments
This study was supported by NASA Astrobiology Institute and DOE Joint Genome Institute, U.S. Department of Energy.
We thank James Tiedje, Debora Rodrigues, Monica Ponder, and Vera S. Soina for some of the Exiguobacterium isolates used in this study. We are grateful to David A. Gilichinsky for support and feedback throughout this work, Eric Altermann for assistance with the phylogenetic tree constructions, and Edward Lanwermeyer for help with editing the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bogdanova, E. S., I. A. Bass, L. S. Minakhin, M. A. Petrova, S. Z. Mindlin, A. A. Volodin, E. S. Kalyaeva, J. M. Tiedje, J. L. Hobman, N. L. Brown, and V. G. Nikiforov. 1998. Horizontal spread of mer operons among gram-positive bacteria in natural environments. Microbiology 144:609-620. [DOI] [PubMed] [Google Scholar]
- 3.Brambilla, E., H. Hippe, A. Hagelstein, B. J. Tindall, and E. Stackebrandt. 2001. 16S rDNA diversity of cultured and uncultured prokaryotes of a mat sample from Lake Fryxell, McMurdo Dry Valleys, Antarctica. Extremophiles 5:23-33. [DOI] [PubMed] [Google Scholar]
- 4.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Graig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II, vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 5.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, M. D., B. M. Lund, J. A. E. Farrow, and K. H. Schleifer. 1983. Chemotaxonomic study of an alkalophilic bacterium, Exiguobacterium aurantiacum gen. nov., sp. nov. J. Gen. Microbiol. 129:2037-2042. [Google Scholar]
- 7.Farrow, J. A. E., S. Wallbanks, and M. D. Collins. 1994. Phylogenetic interrelationships of round-spore-forming bacilli containing cell-walls based on lysine and the non-spore-forming genera Caryophanon, Exiguobacterium, Kurthia, and Planococcus. Int. J. Syst. Bacteriol. 44:74-82. [DOI] [PubMed] [Google Scholar]
- 8.Fruhling, A., P. Schumann, H. Hippe, B. Straubler, and E. Stackebrandt. 2002. Exiguobacterium undae sp. nov. and Exiguobacterium antarcticum sp. nov. Int. J. Syst. Evol. Microbiol. 52:1171-1176. [DOI] [PubMed] [Google Scholar]
- 9.Gilichinsky, D. 2002. Permafrost as a microbial habitat, p. 932-956. In G. Briton (ed.), Encyclopedia of environmental microbiology. John Wiley & Sons, Inc., New York, N.Y.
- 10.Knudston, K. E., E. J. Haas, P. C. Iwen, W. C. Ramaley, and R. F. Ramaley. 2001. Characterization of a gram-positive, non-spore-forming Exiguobacterium-like organism isolated from a Western Colorado (USA) hot spring. Abstr. Annu. Meet. Am. Soc. Microbiol. 2001, I-92, p. 30.
- 11.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 12.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miteva, V. I., P. P. Sheridan, and J. E. Brenchley. 2004. Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl. Environ. Microbiol. 70:202-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pewe, T. 1995. Permafrost and frozen soils, p. 172-759. In Encyclopedia Britannica, vol. 20. Encyclopedia Britannica, Inc., New York, N.Y. [Google Scholar]
- 15.Reiter, B., U. Pfeifer, H. Schwab, and A. Sessitsch. 2002. Response of endophytic bacterial communities in potato plants to infection with Erwinia carotovora subsp. atroseptica. Appl. Environ. Microbiol. 68:2261-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivkina, E., D. Gilichinsky, S. Wagener, J. Tiedje, and J. McGrath. 1998. Biogeochemical activity of anaerobic microorganisms from buried permafrost sediments. Geomicrobiol. J. 15:187-193. [Google Scholar]
- 17.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 18.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. ARTEMIS: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 19.Saitou, N., and M. Nei. 1987. The maximum-likelihood method for molecular phylogeny. Jpn. J. Genet. 62:547-548. [Google Scholar]
- 20.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 21.Shi, T., R. H. Reeves, D. A. Gilichinsky, and E. I. Friedmann. 1997. Characterization of viable bacteria from Siberian permafrost by 16S rDNA sequencing. Microb. Ecol. 33:169-179. [DOI] [PubMed] [Google Scholar]
- 22.Soina, V. 2002. Personal communication.
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vishnivetskaya, T., S. Kathariou, J. McGrath, D. Gilichinsky, and J. M. Tiedje. 2000. Low-temperature recovery strategies for the isolation of bacteria from ancient permafrost sediments. Extremophiles 4:165-173. [DOI] [PubMed] [Google Scholar]
- 25.Vorobyova, E., V. Soina, M. Gorlenko, N. Minkovskaya, N. Zalinova, A. Mamukelashvili, D. Gilichinsky, E. Rivkina, and T. Vishnivetskaya. 1997. The deep cold biosphere: facts and hypothesis. FEMS Microbiol. Rev. 20:277-290. [Google Scholar]