Abstract
Escherichia coli JM101(pSPZ3), containing xylene monooxygenase (XMO) from Pseudomonas putida mt-2, catalyzes specific oxidations and reductions of m-nitrotoluene and derivatives thereof. In addition to reactions catalyzed by XMO, we focused on biotransformations by native enzymes of the E. coli host and their effect on overall biocatalyst performance. While m-nitrotoluene was consecutively oxygenated to m-nitrobenzyl alcohol, m-nitrobenzaldehyde, and m-nitrobenzoic acid by XMO, the oxidation was counteracted by an alcohol dehydrogenase(s) from the E. coli host, which reduced m-nitrobenzaldehyde to m-nitrobenzyl alcohol. Furthermore, the enzymatic background of the host reduced the nitro groups of the reactants resulting in the formation of aromatic amines, which were shown to effectively inhibit XMO in a reversible fashion. Host-intrinsic oxidoreductases and their reaction products had a major effect on the activity of XMO during biocatalysis of m-nitrotoluene. P. putida DOT-T1E and P. putida PpS81 were compared to E. coli JM101 as alternative hosts for XMO. These promising strains contained an additional dehydrogenase that oxidized m-nitrobenzaldehyde to the corresponding acid but catalyzed the formation of XMO-inhibiting aromatic amines at a significantly lower level than E. coli JM101.
Microbial enzymes are useful catalysts for the degradation of organic pollutants in bioremediation but also for the synthesis of added-value products in biocatalytic applications. Various large-scale processes based on bacterial enzymes have been established (17, 26, 30, 47), which illustrate the increasing impact of biocatalysis in the chemical industry (45). Heterologous gene expression has developed to a standard technology for increasing and controlling enzyme activities in bioprocesses. It allows catalysis of a variety of reactions in a limited number of bacterial strains used as recombinant hosts. The focus on a few suitable hosts is emphasized, e.g., by the PlugBug concept (DSM, Heerlen, The Netherlands). Of these strains, Escherichia coli strains are easily accessible for genetic and biochemical engineering and provide high metabolic activity for cofactor regeneration. Their use as recombinant hosts is especially favorable in cofactor-dependent oxygenase-based processes (7) for the conversion of various aromatic hydrocarbons to industrially relevant products (5, 38, 54). E. coli JM101(pSPZ3) (39) contains xylene monooxygenase (XMO) from Pseudomonas putida mt-2 and oxidizes the methyl groups of a variety of toluene and xylene derivatives to the corresponding alcohols, aldehydes, and acids (8, 12, 55).
Nitroaromatics are widely used in large amounts as synthetic intermediates, dyes, pesticides, pharmaceuticals, and explosives (21, 46). Their electrophilic character makes them susceptible to reduction by all kinds of microbial systems (41). Numerous bacterial strains, such as those of Pseudomonas species (1, 43) and E. coli (13), provide enzymes that are able to transform nitro groups of aromatic compounds under aerobic conditions. This indicates that biocatalysis of nitroaromatics might be complicated by influences of the enzymatic background activity of the host strain (52).
In this study, we were interested in the suitability of E. coli JM101(pSPZ3) for the oxidation of m-nitrotoluene by XMO. We investigated the enzymatic background activity of this biocatalyst and parameters determining its efficiency and specificity on a technical scale. A comparison with solvent-tolerant P. putida DOT-T1E (40) and P. putida PpS81 (18), which is deficient for an alcohol dehydrogenase, opened new perspectives for recombinant P. putida strains as efficient biocatalysts in XMO-based processes with nitroaromatic compounds.
MATERIALS AND METHODS
Chemicals.
Chemicals were obtained from Fluka (Buchs, Switzerland) [m-nitrotoluene, ≥99% pure; m-nitrobenzyl alcohol, ≈95%; m-nitrobenzaldehyde, ≈97%; m-nitrobenzoic acid, >98%; m-toluidine, >99.0%; m-aminobenzyl alcohol, ≈97%; m-aminobenzoic acid, ≥97%; and bis(2-ethylhexyl)phthalate, 97%], Riedel-de Haën (Buchs, Switzerland) [p-dimethylaminobenzaldehyde, ≥99%], Aldrich (Buchs, Switzerland) [dicyclopropyl ketone, 95%], and Acros Organics (Geel, Belgium) [n-octane, >98.5%].
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains and plasmids are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) complex medium (Difco, Detroit, Mich.), M9* mineral medium (identical to M9 mineral medium [42] except that it contained three times more phosphate salts in order to increase the buffer capacity and did not contain calcium chloride) (39), and RB mineral medium (6). Both mineral media were supplemented with 1 ml/liter USFe trace element solution (6). Antibiotics (kanamycin, 50 mg/liter; rifampin, 20 mg/liter) and thiamine (10 mg/liter) were added when appropriate. The pH of the M9* mineral medium was adjusted to 7.4 with 10 M NaOH before use in shaking flasks. All cultivations were performed at 30°C, and glucose (0.5% [wt/vol]) was used as a carbon source.
TABLE 1.
Bacterial strains and plasmids used in this study
Whole-cell activity assays.
Whole-cell biotransformations (1-ml scale) with resting cells of E. coli JM101(pSPZ3) were used to determine XMO activity for the sequential oxidation of m-nitrotoluene by using a procedure described elsewhere (8). Bacteria were grown in glucose-containing M9* mineral medium, and XMO synthesis was induced with 0.1% (vol/vol) n-octane. After being harvested, bacteria were resuspended to about 2 g cell dry weight (CDW) per liter in potassium phosphate buffer (pH 7.4) containing glucose. A 1.0-ml portion of cell suspension was incubated with 1.0 mM m-nitrotoluene on a rotary shaker at 250 rpm and 30°C. Product formation was monitored by stopping the reaction at different time points by acidification with 40 μl of 10% (vol/vol) perchloric acid solution. After centrifugation, the supernatant was analyzed for nitroaromatics by reversed-phase high-performance liquid chromatography (RP-HPLC) (see below). Initial activities were calculated from the amount of product formed after 5 min of biotransformation and were expressed in units per gram CDW. One unit is defined as the enzyme activity that forms 1 μmol of product per minute. The experiments were repeated independently at least once. m-Nitrobenzyl alcohol or m-nitrobenzaldehyde at 1.0 mM served as the substrate to determine initial activities of the second or third XMO-catalyzed reaction step, respectively. For studies of inhibition of XMO activity, cell suspensions were incubated with 1.0 mM substrate together with the inhibitor at various concentrations from 0 to 20 mM.
Whole-cell activity assays were also performed using E. coli JM101, P. putida DOT-T1E, and P. putida PpS81 wild-type strains in order to determine enzyme activities converting m-nitrobenzaldehyde to m-nitrobenzyl alcohol or m-nitrobenzoic acid. Analogously, the wild-type strains were incubated with 1.0 mM m-nitrotoluene, m-nitrobenzyl alcohol, m-nitrobenzaldehyde, or m-nitrobenzoic acid to determine the rate of formation of aromatic amines. The incubation time was prolonged to 90 min due to the low formation rates. Aromatic amines were detected by using Ehrlich's reagent (see below).
Two-liquid-phase biotransformation.
Biotransformations in the presence of a second, organic liquid phase were performed on a 2-liter scale using a reactor setup and procedure described earlier (6). A 100-ml overnight culture of E. coli JM101(pSPZ3) in RB medium was used as inoculum for a bioreactor containing 900 ml RB medium with 0.7% (wt/vol) glucose as a carbon source. Cells grew in batch mode at 30°C overnight. The pH was maintained constant at 7.4 by regulated addition of phosphoric acid and ammonium hydroxide. The stirrer speed and aeration rate were set to 1,500 rpm and 1 liter air per minute, respectively. Fed-batch cultivation was started by activating the feed of a solution containing 730 g glucose and 19.6 g MgSO4 · 7H2O per liter after complete consumption of the carbon source (corresponds to time zero). The initial feed rate of 5.9 g glucose/h was increased stepwise during fed-batch cultivation (see Fig. 3B and 5B). XMO synthesis was induced by the addition of dicyclopropyl ketone to a concentration of 0.02% (vol/vol) in the aqueous phase and of 60 mmol n-octane in the presence of the second liquid phase. Biotransformation was started by the addition of 1 liter of the second liquid phase, which consisted of bis(2-ethylhexyl)phthalate and 50 mmol m-nitrotoluene as substrate. The stirrer speed and aeration rate were increased to 2,000 rpm and 2 liters per minute, respectively. After separation of the aqueous and organic phase of samples by centrifugation, nitroaromatics were detected by RP-HPLC and gas chromatography, respectively (see below). Glucose and acetic acid concentrations were determined as described elsewhere (6). The cell concentration was determined spectrophotometrically at 450 nm as described elsewhere (53). One absorption unit at 450 nm corresponded to a cell dry weight of 0.29 g/liter.
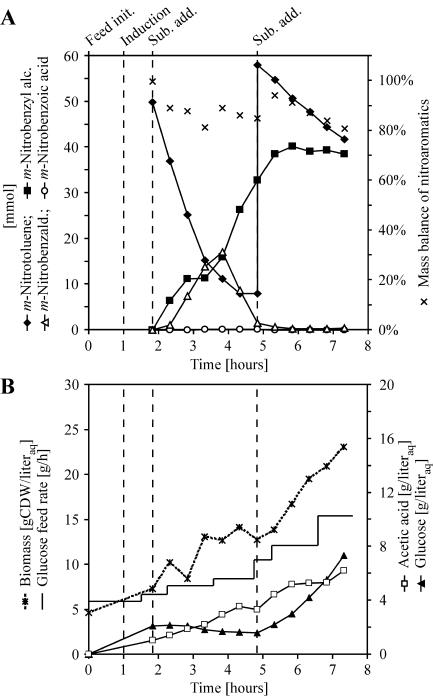
FIG. 3.
Biotransformation of m-nitrotoluene by E. coli JM101(pSPZ3) in a two-liquid-phase system on a 2-liter scale (working volume). (A) XMO synthesis was induced 50 min before substrate addition. Biotransformation was started by the addition of the second organic phase (phase ratio, 0.5) containing 50 mmol m-nitrotoluene. After most of the substrate was converted, another 50 mmol m-nitrotoluene was added. Nitroaromatics are represented as the sum of the respective amounts in the organic and aqueous phase. The mass balance of nitroaromatics describes the overall sum of m-nitrotoluene, m-nitrobenzyl alcohol, m-nitrobenzaldehyde, and m-nitrobenzoic acid given as the percentage (moles/moles) of m-nitrotoluene added as substrate. (B) Cultivation parameters of the biotransformation. The glucose feed was increased stepwise starting at 5.9 g/h. Experimental details are described in Materials and Methods.
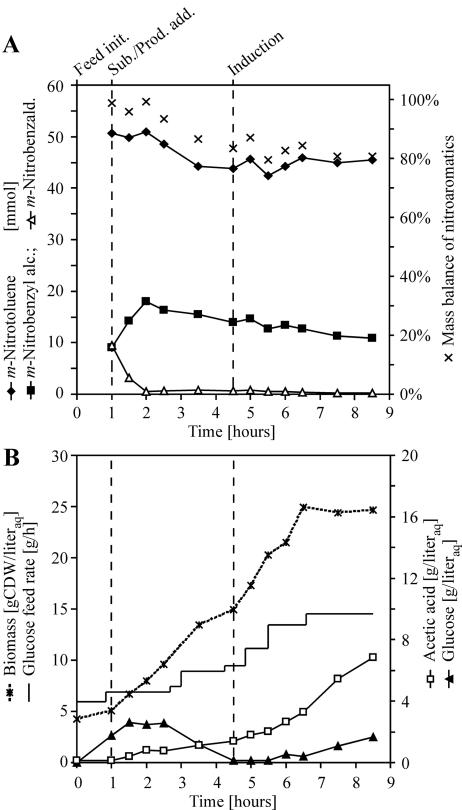
FIG. 5.
Biotransformation by E. coli JM101(pSPZ3) with induction of XMO synthesis 3.5 h after addition of nitroaromatics. (A) Biotransformation was performed using the same initial conditions as in the experiment shown in Fig. 3, except for the addition of m-nitrobenzyl alcohol and m-nitrobenzaldehyde with m-nitrotoluene in the organic phase and the delayed induction of XMO synthesis. During biotransformation, no m-nitrobenzoic acid was detected, and it therefore is not indicated in the figure. (B) Cultivation parameters of the biotransformation.
Analytical procedures.
m-Nitrotoluene, m-nitrobenzyl alcohol, m-nitrobenzaldehyde, and m-nitrobenzoic acid in aqueous samples were separated with an RP-HPLC instrument from Merck Hitachi (interface D-7000, UV detector L-7400, pump L-7100, autosampler L-7200) equipped with a Nucleosil C18 column (pore size, 100 Å; particle size, 5 μm; inner diameter, 4 mm; length, 25 cm) from Macherey-Nagel (Oensingen, Switzerland). The mobile phase consisted of acetonitrile and H2O containing 0.1% perchloric acid. The elution profile was 35 to 43% acetonitrile over 5 min, followed by 43 to 80% over 5 min, and then 80% acetonitrile isocratic for 2 min. The flow rate was 1.0 ml/min. Detection occurred at 210 nm.
Samples from the organic phase of the two-liquid-phase biotransformations were diluted 25-fold in ice-cold diethyl ether containing 0.1 mM n-decane as an internal standard and were dried over sodium sulfate. m-Nitrotoluene, m-nitrobenzyl alcohol, and m-nitrobenzaldehyde were separated using a gas chromatograph instrument from Fisons Instruments equipped with an OPTIMA-5 fused silica capillary column (length, 25 m; inner diameter, 0.32 mm; film thickness, 0.25 μm) from Macherey-Nagel (Oensingen, Switzerland). The temperature profile was as follows: isotherm at 40°C for 2 min, from 40 to 280°C at 18°C/min, and then 280°C isotherm for 5 min. Splitless injection with hydrogen as the carrier gas was used. Detection occurred by flame ionization. Identification and quantification of the substances were done by comparison with commercially available standards.
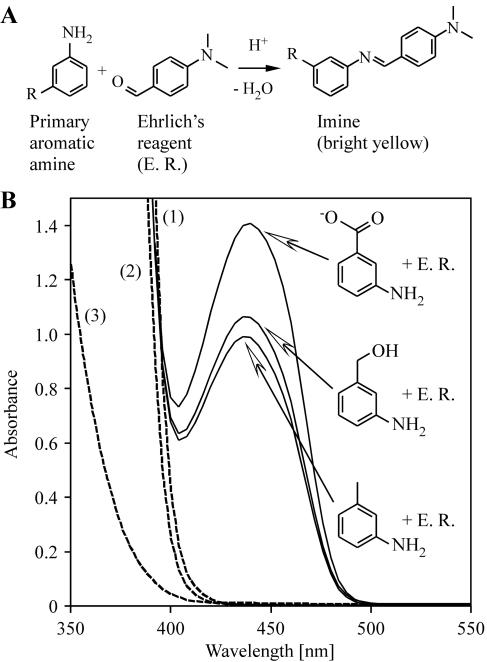
Aromatic amines in samples from whole-cell activity assays were detected by addition of Ehrlich's reagent, consisting of 20 g/liter p-dimethylaminobenzaldehyde in 20% hydrochloric acid (2). p-Dimethylaminobenzaldehyde reacts with primary aromatic amines to form yellow imines in acidic environments (Fig. 1A) (28). Twenty microliters of Ehrlich's reagent was added per ml cell-free supernatant of aqueous samples. Formation of yellow reaction products was monitored by eye or spectrophotometrically at 436 nm, the corresponding absorption maximum (Fig. 1B). m-Toluidine, m-aminobenzyl alcohol, and m-aminobenzoic acid served as standards for the expected amines produced by the host from m-nitrotoluene, m-nitrobenzyl alcohol, and m-nitrobenzoic acid, respectively. m-Aminobenzaldehyde was not available as a standard due to its instability (16). Aromatic amines formed from m-nitrobenzaldehyde were quantified using an equimolar mixture of m-toluidine, m-aminobenzyl alcohol, and m-aminobenzoic acid as a standard, which might have influenced the quantification.
FIG. 1.
Detection of aromatic amines by Ehrlich's reagent. (A) Primary aromatic amines nonspecifically react with p-dimethylaminobenzaldehyde in acidic solution (Ehrlich's reagent) to form yellow imines (28). (B) Spectroscopic detection of imines derived from m-toluidine, m-aminobenzyl alcohol, and m-aminobenzoic acid (1.0 mM each) by Ehrlich's reagent (E. R.) in aqueous solution (with absorption maxima at 436 nm). Controls: 1, mixture of all nitroaromatics (1.0 mM each) with Ehrlich's reagent; 2, Ehrlich's reagent alone; 3, mixture of all nitroaromatics and aromatic amines tested (1.0 mM each) without Ehrlich's reagent.
RESULTS
Biotransformation of m-nitrotoluene by E. coli JM101(pSPZ3).
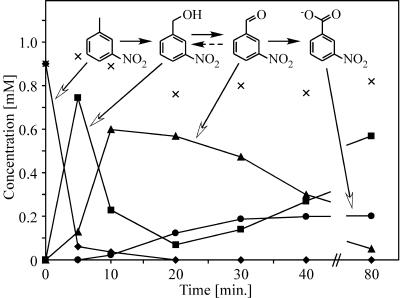
Resting cells of E. coli JM101(pSPZ3) containing XMO were incubated with m-nitrotoluene, which was oxidized sequentially to m-nitrobenzyl alcohol, m-nitrobenzaldehyde, and m-nitrobenzoic acid at maximum formation rates of 105, 60, and 6 U/g CDW, respectively (Fig. 2). When m-nitrobenzyl alcohol was used as a substrate, m-nitrobenzaldehyde was formed at a rate of 90 U/g CDW. With m-nitrobenzaldehyde as a substrate, m-nitrobenzoic acid was formed at 13 U/g CDW. In addition, m-nitrobenzaldehyde was reduced to the corresponding alcohol in E. coli JM101(pSPZ3) (Fig. 2).
FIG. 2.
Multistep oxygenation of m-nitrotoluene by E. coli JM101(pSPZ3). The assay was performed with resting cells (1.7 g CDW/liter) as described in Materials and Methods. m-Nitrotoluene at 1.0 mM (⧫) served as a substrate. m-Nitrobenzyl alcohol (▪), m-nitrobenzaldehyde (▴), and m-nitrobenzoic acid (•) were formed as consecutive products. ×, sum of nitroaromatic reactant concentrations.
The oxidation of m-nitrotoluene by XMO was investigated with higher substrate concentrations and over an extended time period by using growing E. coli JM101(pSPZ3) in a two-liquid-phase biotransformation setup on a 2-liter scale. Partition coefficients between the organic and aqueous phases for m-nitrotoluene, m-nitrobenzyl alcohol, and m-nitrobenzaldehyde were experimentally determined to >1,400, 9.8 ± 0.2, and 73.0 ± 7.4, respectively. XMO synthesis was started by induction 50 min before substrate addition. The course of biotransformation is shown in Fig. 3A. After substrate addition, cells oxidized m-nitrotoluene to the corresponding alcohol and aldehyde at rates of 40 and 23 U/g CDW, respectively. No significant oxidation to the acid was detected (<0.5 mmol). The aldehyde accumulated to a maximum of 17 mmol after about 4 hours from feed initiation. Afterwards, it disappeared at a rate of 21 U/g CDW, whereas at the same time the corresponding alcohol was formed at a rate of 26 U/g CDW. The similar rates for aldehyde disappearance and alcohol formation indicated the predominant formation of m-nitrobenzyl alcohol independently from XMO activity by host intrinsic enzymes of E. coli JM101. The lack of m-nitrobenzyl alcohol formation in the absence of m-nitrobenzaldehyde confirmed the minor XMO activity towards the end of the biotransformation. The process parameters are summarized in Table 2. Acetic acid accumulated to concentrations which, accordingly to Konstantinov et al. (29), do not affect cell growth in a significant way (Fig. 3B).
TABLE 2.
Process parameters of the two-liquid-phase biotransformation of m-nitrotoluene by E. coli JM101(pSPZ3)a
| Parameter | Unit | Value |
|---|---|---|
| Biotransformation timeb | h | 5.5 |
| Final cell concn | g CDW/literaq | 23.1 |
| Total m-nitrotoluene amt added | mmol | 99.8 |
| Final m-nitrotoluene amt | mmol | 41.7 |
| Final m-nitrobenzyl alcohol amt | mmol | 38.5 |
| Final m-nitrobenzaldehyde amt | mmol | 0.4 |
| Final m-nitrobenzoic acid amt | mmol | 0.0 |
| Molar yield | % | 48 |
| Maximal specific XMO activityc | U/g CDW | 40 |
| Maximal specific accumulationalc | U/g CDW | 26 |
| Maximal volumetric activityalc | U/literaq | 344 |
| Avg volumetric activityalc | U/literaq | 117 |
| Avg productivityalc | g/(litertot · h) | 0.54 |
Two-liter scale (working volume); phase ratio of 0.5. aq, aqueous phase; tot, organic and aqueous phases; alc, referring to m-nitrobenzyl alcohol accumulation.
Organic phase addition is defined as the start point.
Based on the XMO-catalyzed formation of m-nitrobenzyl alcohol, aldehyde, and acid.
Stability of m-nitrobenzaldehyde in different bacterial strains.
Resting E. coli JM101 was incubated with m-nitrobenzaldehyde in whole-cell activity assays in order to investigate the stability of this product towards host intrinsic enzyme activities. m-Nitrobenzaldehyde was reduced to m-nitrobenzyl alcohol at an initial rate of 24 U/g CDW (Fig. 4A).
FIG. 4.
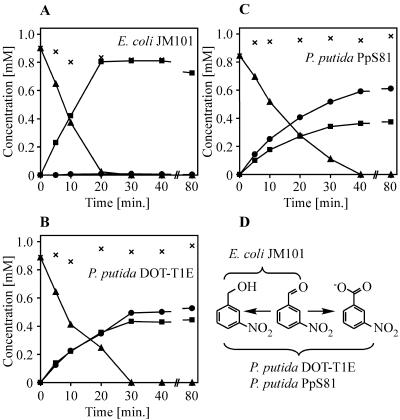
Stabilities of m-nitrobenzaldehyde in different bacterial strains. (A) E. coli JM101 (1.9 g CDW/liter), (B) P. putida DOT-T1E (2.3 g CDW/liter), and (C) P. putida PpS81 (1.7 g CDW/liter) were incubated with 1.0 mM m-nitrobenzaldehyde (▴). m-Nitrobenzyl alcohol (▪) and m-nitrobenzoic acid (•) were formed as products. ×, sum of nitroaromatic reactant concentrations. (D) Scheme of m-nitrobenzaldehyde-degrading reactions catalyzed by host-intrinsic enzymes of E. coli JM101, P. putida DOT-T1E, and P. putida PpS81.
In addition, the enzymatic background activities of other promising host strains were determined. We incubated m-nitrobenzaldehyde with solvent-tolerant P. putida DOT-T1E (40) and P. putida PpS81, a strain which is deficient for an alcohol dehydrogenase and thus unable to oxidize alkanols (3, 18, 33). Reductive activities for m-nitrobenzaldehyde were present in both Pseudomonas strains and formed m-nitrobenzyl alcohol at rates of 12 U/g CDW (Fig. 4B and C). Oxidative activities formed m-nitrobenzoic acid from m-nitrobenzaldehyde at initial rates of 11 and 17 U/g CDW in P. putida DOT-T1E and P. putida PpS81, respectively. In E. coli JM101, m-nitrobenzaldehyde was oxidized to m-nitrobenzoic acid at a negligible rate of below 1 U/g CDW (Fig. 4A). All strains under investigation showed oxidoreductase activities for the carbonyl group of m-nitrobenzaldehyde (Fig. 4D), but no oxidation of the methyl group of m-nitrotoluene was detected.
Inhibition of xylene monooxygenase by aromatic amines, side products formed from nitroaromatics in the E. coli host.
Two-liquid-phase biotransformation was repeated using the identical biocatalyst and protocol except for the time point of induction in order to verify whether the relatively fast decrease of XMO activity during biotransformation correlates with the presence of nitroaromatics. Instead of substrate addition 50 min after induction, cells were incubated with nitroaromatics for 3.5 h before induction of XMO synthesis. In addition to 50 mmol m-nitrotoluene as a substrate, the second liquid phase also contained 10 mmol each of m-nitrobenzyl alcohol and m-nitrobenzaldehyde.
The enzymatic background activity of E. coli JM101(pSPZ3) reduced all m-nitrobenzaldehyde to the corresponding alcohol at an initial rate of 29 U/g CDW, immediately after the nitroaromatics were added (Fig. 5A). No formation of m-nitrobenzyl alcohol, m-nitrobenzaldehyde, or m-nitrobenzoic acid was detected after the delayed induction, which indicated a complete absence of XMO activity, while biomass, glucose, and acetic acid concentrations (Fig. 5B) were similar to those seen in the previously performed biotransformation (Fig. 3B). During the experiment, bacteria were withdrawn from the bioreactor and diluted 80-fold in phosphate buffer for whole-cell activity assays with m-nitrotoluene as a substrate. No m-nitrobenzyl alcohol formation was detected in samples withdrawn before induction at 4.5 h. In samples withdrawn after induction at 6.5 and 8.5 h, the respective m-nitrobenzyl alcohol formation rates were recovered to 75% and 49%, compared to rates obtained in whole-cell activity assays using bacteria from the same inoculum. This pointed to a reversible inhibition of XMO.
The sum of nitroaromatics steadily decreased in concentration throughout both two-liquid-phase biotransformations, independent of the time point of induction (Fig. 3A and 5A). This indicated the formation of side products from nitroaromatics by native enzymes of the E. coli host. Many microorganisms are able to reduce nitro groups to amine groups (24, 37, 56). In order to investigate whether E. coli JM101 forms aromatic amines, resting cells were incubated with m-nitrotoluene, m-nitrobenzyl alcohol, m-nitrobenzaldehyde, or m-nitrobenzoic acid. Primary aromatic amines were detected as condensation products of Ehrlich's reagent for all nitroaromatics tested in E. coli JM101 and, with lower formation rates, also in P. putida DOT-T1E and P. putida PpS81 (Table 3). The formation of m-aminobenzaldehyde from m-nitrobenzaldehyde remained unclear, since a prior reduction or oxidation to m-nitrobenzyl alcohol or m-nitrobenzoic acid, followed by the conversion to m-aminobenzyl alcohol or m-aminobenzoic acid, respectively, in the tested bacteria might have occurred.
TABLE 3.
Formation of aromatic amines from corresponding nitroaromatics
| Strain | Aromatic amine formation rate (U/g CDW) ona:
|
|||
|---|---|---|---|---|
| m-Nitrotoluene | m-Nitrobenzyl alcohol | m-Nitrobenzaldehyde | m-Nitrobenzoic acid | |
| E. coli JM101 | 1.18 ± 0.04 | 0.70 ± 0.11 | 2.63 ± 0.24 | 3.39 ± 0.32 |
| P. putida DOT-T1E | 0.14 ± 0.03 | 0.10 ± 0.03 | 0.55 ± 0.06 | 0.55 ± 0.07 |
| P. putida PpS81 | 0.09 ± 0.07 | 0.02 ± 0.02 | 0.15 ± 0.06 | 0.09 ± 0.05 |
Formation rates (means ± standard deviations) are based on aromatic amine concentrations detected by Ehrlich's reagent in whole-cell activity assays using resting cells (≈2 g CDW/liter). See Materials and Methods for details. One unit is the enzyme activity that forms 1 μmol of product per min.
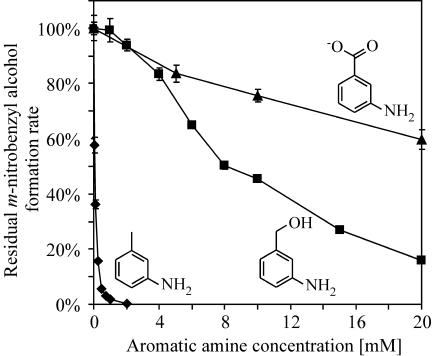
We investigated a possible relationship between the inhibition of XMO observed during the two-liquid-phase biotransformation and the formation of aromatic amines. Therefore, whole-cell activity assays were performed using E. coli JM101(pSPZ3) as a biocatalyst with m-nitrotoluene as the substrate in the presence of various concentrations of the expected side products m-toluidine, m-aminobenzyl alcohol, and m-aminobenzoic acid. A lowered cell density (0.8 to 0.9 g CDW/liter) used in this experiment allowed focusing on the first XMO-catalyzed reaction step. All three aromatic amines inhibited the XMO-catalyzed oxidation of m-nitrotoluene (Fig. 6). m-Toluidine was found to be the most effective inhibitor, completely repressing m-nitrobenzyl alcohol formation at concentrations of above 2.0 mM.
FIG. 6.
Effect of aromatic amines on the XMO-catalyzed m-nitrotoluene oxidation to the corresponding alcohol. The remaining activities of the first XMO-catalyzed reaction step in the presence of different concentrations of m-toluidine (⧫), m-aminobenzyl alcohol (▪), and m-aminobenzoic acid (▴) in resting E. coli JM101(pSPZ3) (0.8 to 0.9 g CDW/liter) are represented relative to the noninhibited XMO activity.
Incubation with m-nitrotoluene, m-nitrobenzyl alcohol, or m-nitrobenzaldehyde as a substrate in combination with 200 μM m-toluidine, 8 mM m-aminobenzyl alcohol, or 20 mM m-aminobenzoic acid resulted in inhibition of each of the XMO-catalyzed reaction steps in E. coli JM101(pSPZ3) (Table 4). The applied m-toluidine and m-aminobenzyl alcohol concentrations completely inhibited the formation of m-nitrobenzaldehyde from the corresponding alcohol. The determined residual XMO activities, however, might have been influenced by host-intrinsic enzymes of the E. coli host which reduced m-nitrobenzaldehyde to m-nitrobenzyl alcohol (Fig. 4A).
TABLE 4.
Effects of aromatic amines on the XMO-catalyzed oxidation of m-nitrotoluene, m-nitrobenzyl alcohol, and m-nitrobenzaldehyde
| Inhibitor | Residual activity (%) fora:
|
||
|---|---|---|---|
| m-Nitrotoluene | m-Nitrobenzyl alcohol | m-Nitrobenzaldehdye | |
| m-Toluidine, 200 μM | 29 ± 6 | 0 | 6 ± 0 |
| m-Aminobenzyl alcohol, 8 mM | 33 ± 6 | 0 | 20 ± 1 |
| m-Aminobenzoic acid, 20 mM | 83 ± 14 | 39 ± 3 | 17 ± 1 |
Residual activities (means ± standard deviations) of E. coli JM101(pSPZ3) as a biocatalyst (1.7 g CDW/liter) are given as percentages of the activities in the absence of aromatic amines.
DISCUSSION
The increasing knowledge about microbial metabolic pathways, the enzymes involved, and influences of environmental conditions on biodegradation (15) might lead to the characterization of preferred groups of microorganisms as hosts for the biocatalysis of specific reaction types. This development might simplify the selection and design of bacterial host strains for future applications (27) and motivated our investigation of the biotransformation of m-nitrotoluene by recombinant E. coli containing XMO.
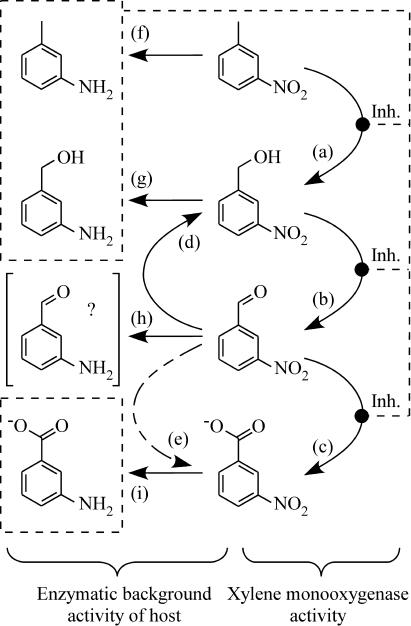
The successive oxidation of m-nitrotoluene to the corresponding alcohol, aldehyde, and acid (Fig. 2) revealed an XMO-catalyzed multistep oxygenation, as reported for toluene and pseudocumene (8). The results of the two-liquid-phase biotransformation of m-nitrotoluene by E. coli JM101(pSPZ3) were promising and illustrate the usefulness of this in situ extraction system for the accumulation of m-nitrobenzyl alcohol in technically relevant concentrations (Table 2). The further oxidation to m-nitrobenzaldehyde and m-nitrobenzoic acid, however, was hindered by the activity of host-intrinsic enzymes, which turned out to play a key role in reaction selectivity (Fig. 7).
FIG. 7.
Proposed pattern of enzyme-catalyzed reactions during biotransformation of m-nitrotoluene by E. coli JM101(pSPZ3). Vertical direction: XMO catalyzed the multistep oxygenation of m-nitrotoluene to the corresponding alcohol (a), aldehyde (b), and acid (c). m-Nitrobenzaldehyde formation was counteracted by alcohol dehydrogenase activities of the enzymatic background of the E. coli JM101 host, which reduced it to the corresponding alcohol (d). Additionally, in P. putida DOT-T1E and P. putida PpS81, m-nitrobenzaldehyde was oxidized to the corresponding acid (e). Horizontal direction: all tested bacterial strains reduced nitroaromatics to aromatic amines (f to i), which strongly inhibited XMO-catalyzed oxidation. The formation of unstable m-aminobenzaldehyde by nitroreductases (h) remains unclear.
Effects of aromatic amine formation on xylene monooxygenase activity.
Degradation of nitroaromatics can be initialized by the enzymatic reduction of the nitro group to the amine group via a nitroso group and a hydroxylamino group in various microorganisms (10, 32, 43). E. coli also contains a set of oxygen-insensitive nitroreductases (4, 51) which aerobically reduce nitro aromatic compounds to the corresponding amines (13). Although the catalyzed reduction of aromatic hydroxylamines to the corresponding amines in E. coli is still unclear (25, 37), we observed aromatic amine formation from all nitroaromatics tested (Table 3). However, it remains unclear whether the assay based on Ehrlich's reagent can distinguish between aromatic amines and hydroxylamines. The simple detection of aromatic amines as yellow condensation products of Ehrlich's reagent makes this method a useful tool for future investigations on the biodegradation of nitroaromatics or for the screening for microorganisms providing nitroreductase activities.
Aromatic amine formation took place at relatively low rates compared to the detected oxidoreductase activities and therefore had only a minor impact on product degradation. Nevertheless, we elucidated the effective inhibition of XMO by aromatic amines in E. coli JM101(pSPZ3) (Fig. 6; Table 4). This inhibition apparently takes place on the protein level and in a reversible fashion. The high polarity of aromatic amines suggests a predominant partitioning into the aqueous phase during two-liquid-phase biotransformation. Considering the determined amine formation rates (Table 3) and the resulting inhibiting effects (Fig. 6; Table 4), XMO activity is thought to be completely inhibited about 2 to 3 hours after substrate addition during two-liquid-phase biotransformation, which correlates well with our observations (Fig. 3A). XMO was active at least fourfold longer when the same experimental setup was used but with pseudocumene instead of m-nitrotoluene as the substrate (6, 9). The prevention of nitroreduction therefore seems to be crucial for efficient XMO-based biotransformation of m-nitrotoluene. Although mutants of E. coli that are deficient in oxygen-insensitive nitroreductases have been identified (34), the suitability of such strains for process applications remains to be investigated.
Significantly lower rates of aromatic amine formation were detected with P. putida DOT-T1E and P. putida PpS81 than with E. coli JM101 (Table 3). This was surprising since P. putida strains are known as effective degraders of many aromatic compounds (23, 49). However, the slower accumulation of aromatic amines might be explained by the subsequent degradation in P. putida, which may be absent in E. coli (32, 36). The generally broad substrate spectra of nitroreductases suggest a lower aromatic amine accumulation rate not only for the tested compounds but also for other nitroaromatics for the P. putida strains under investigation (44). An increased efficiency of XMO-based biotransformations on various nitroaromatics might therefore be achieved by using P. putida DOT-T1E or P. putida PpS81 instead of E. coli JM101 as the recombinant host strain. Furthermore, the solvent tolerance of P. putida DOT-T1E might be exploited to enlarge the spectrum of solvents applicable for in situ product extraction and for the production of a broader range of organic compounds in two-liquid-phase bioreactor setups (11). m-Nitrotoluene was not oxidized in P. putida strains DOT-T1E and PpS81, yet the synthesis of oxidative enzymes involved in the mineralization of m-nitrotoluene via m-nitrobenzyl alcohol, m-nitrobenzaldehyde, and m-nitrobenzoic acid has been reported for P. putida strain OU83 upon induction with m-nitrotoluene (1, 50). Interestingly, m-toluidine accumulated as the main product, apparently without significant inhibition of the m-nitrotoluene hydroxylation.
Effects of dehydrogenases on product formation patterns.
The multistep oxygenation of m-nitrotoluene by XMO (Fig. 7a to c) was counteracted by host-intrinsic enzyme activities from the E. coli host, which reduced m-nitrobenzaldehyde to m-nitrobenzyl alcohol (Fig. 4A and 7d). This reduction has been proposed to be catalyzed by an unspecific alcohol dehydrogenase activity (8, 31). All of the m-nitrobenzaldehyde previously formed by XMO was reduced to the corresponding alcohol at the end of the two-liquid-phase biotransformation (Fig. 3A). In contrast to XMO, alcohol dehydrogenase(s) was apparently not inhibited by aromatic amines formed as side products. A biocatalyst which provides a lower alcohol dehydrogenase activity and a prolonged XMO presence might further increase the efficiency of m-nitrobenzyl alcohol accumulation and allow the production of m-nitrobenzaldehyde or m-nitrobenzoic acid.
P. putida PpS81 and P. putida DOT-T1E both reduced m- nitrobenzaldehyde to m-nitrobenzyl alcohol with identical conversion rates (Fig. 4B and C and 7d). This suggests that alcohol formation was catalyzed by enzymes different from the alcohol dehydrogenase lacking in P. putida PpS81 (18). The m-nitrobenzaldehyde reduction rates detected in the P. putida strains were only half as high as those in E. coli JM101 and would therefore counteract the XMO-catalyzed oxidation of m-nitrobenzyl alcohol at a significantly lower level in P. putida DOT-T1E and P. putida PpS81 used as hosts.
However, both P. putida strains also oxidized m-nitrobenzaldehyde to the corresponding acid (Fig. 4B and C and Fig. 7e), presumably with an aldehyde dehydrogenase such as NtnC, a p-nitrobenzaldehyde dehydrogenase from Pseudomonas sp. strain TW3 (22). On the other hand, the similar rates of alcohol and acid formation from m-nitrobenzaldehyde in P. putida DOT-T1E (Fig. 4B) point to the activity of a single enzyme that catalyzed both reactions. Such an alcohol dehydrogenase with aldehyde dismutase activity was reported to catalyze the stoichiometric disproportion of acetaldehyde into one equivalent of ethanol and acetic acid in yeast (48). Taking reductive and oxidative activities into account, the net conversion rates on the carbonyl group of m-nitrobenzaldehyde were similar in P. putida DOT-T1E, P. putida PpS81, and E. coli JM101, making all of them equally useful as hosts for the production of m-nitrobenzaldehyde. Nevertheless, the additional aldehyde dehydrogenase supports XMO-catalyzed oxidation of m-nitrobenzaldehyde and makes P. putida DOT-T1E and P. putida PpS81 interesting as hosts in an XMO-based process for the production of m-nitrobenzoic acid from m-nitrotoluene. Further studies will be necessary to clarify whether the substrate range is limited to m-nitrobenzaldehyde and whether these oxidoreductases are also present in other interesting P. putida strains.
Effect of enzymatic background activity on cofactor regeneration.
Both the XMO-catalyzed oxidation of m-nitrobenzyl alcohol (Fig. 7b) (19) and the alcohol dehydrogenase-catalyzed reduction of the resulting m-nitrobenzaldehyde in the reverse direction (Fig. 7d), require NADH as a cofactor. This “futile cycle” might decrease process efficiency not only by the counteracting product formation but also by limiting NADH availability. No significant cofactor shortage was expected in our setup, since detected m-nitrobenzaldehyde reduction rates were similar in the two-liquid-phase biotransformations using E. coli JM101(pSPZ3) (Fig. 3A and 5A) and in whole-cell activity assays with E. coli JM101 (Fig. 4A). However, this issue may become important for biotransformations with higher and more enduring oxygenase activities (14). Closer identification of the enzymes involved might simplify the generation of knockout mutants unable to reduce m-nitrobenzaldehyde to the alcohol. Such an engineered strain might be more efficient as a host for the biocatalytic production of m-nitrobenzaldehyde or m-nitrobenzoic acid from m-nitrotoluene.
Reductions catalyzed by NAD+-dependent dehydrogenases are associated with NADH consumption, whereas oxidations generate NADH. The detected conversion of m-nitrobenzaldehyde to m-nitrobenzoic acid in P. putida DOT-T1E and P. putida PpS81 should therefore support cofactor regeneration in an XMO-based process using these strains as hosts. Such an uncoupling from metabolic cofactor regeneration was shown to be useful in synthetic applications with the selective oxidation of 2-methylquinoxaline by recombinant E. coli (G. Ionidis, D. Meyer, and A. Schmid, unpublished data) and might also be relevant for environmental applications such as the biodegradation of o-chlorotoluene by an engineered Pseudomonas strain (20).
This work confirms that the enzymatic background activity of microbial biocatalysts can interfere with biotransformation by substrate and product degradation. Side product formation often can be avoided by using recombinant biocatalysts based on host strains such as E. coli, which provide a “neutral” enzymatic background due to a narrow catabolic substrate spectrum. We identified host-specific factors limiting the catabolic performance of m-nitrotoluene biotransformation by XMO in recombinant E. coli JM101. Our results suggest a significantly higher efficiency for the successive oxidation of m-nitrotoluene to m-nitrobenzyl alcohol, m-nitrobenzaldehyde, and m-nitrobenzoic acid with P. putida DOT-T1E or P. putida PpS81 as the host than with E. coli JM101. Future research activities are therefore directed towards the evaluation and development of recombinant P. putida strains for biocatalytic applications.
Acknowledgments
This work was supported by grants from the European Commission (QLK3-2001-00435 [BARTOLO]).
We are indebted to Bruno Bühler for fruitful discussions, to Andreas Schenzle for helpful advice concerning the detection of aromatic amines, and to Juan L. Ramos for providing P. putida DOT-T1E.
REFERENCES
- 1.Ali-Sadat, S., K. S. Mohan, and S. K. Walia. 1995. A novel pathway for the biodegradation of 3-nitrotoluene in Pseudomonas putida. FEMS Microbiol. Ecol. 17:169-176. [Google Scholar]
- 2.Bogensberger, S., N. Boss, M. Büttner, R. Jäckle, S. Jäckle-Kirchhoff, G. Meier, P. Nawrocki, S. Parzhuber, R. Pilsinger, W. Rempe-Baldin, B. Scheele, C. Striebeck, and G. Wangerin. 1998. Roche-Lexikon Medizin, 4th ed. Hoffmann-La Roche AG and Urban & Fischer, Munich, Germany.
- 3.Bosetti, A., J. B. van Beilen, H. Preusting, R. G. Lageveen, and B. Witholt. 1992. Production of primary aliphatic alcohols with a recombinant Pseudomonas strain, encoding the alkane hydroxylase enzyme system. Enzyme Microb. Technol. 14:702-708. [Google Scholar]
- 4.Bryant, D. W., D. R. McCalla, M. Leeksma, and P. Laneuville. 1981. Type I nitroreductases of Escherichia coli. Can. J. Microbiol. 27:81-86. [DOI] [PubMed] [Google Scholar]
- 5.Bühler, B., I. Bollhalder, B. Hauer, B. Witholt, and A. Schmid. 2003. Chemical biotechnology for the specific oxyfunctionalization of hydrocarbons on a technical scale. Biotechnol. Bioeng. 82:833-842. [DOI] [PubMed] [Google Scholar]
- 6.Bühler, B., I. Bollhalder, B. Hauer, B. Witholt, and A. Schmid. 2003. Use of the two-liquid phase concept to exploit kinetically controlled multistep biocatalysis. Biotechnol. Bioeng. 81:683-694. [DOI] [PubMed] [Google Scholar]
- 7.Bühler, B., and A. Schmid. 2004. Process implementation aspects for biocatalytic hydrocarbon oxyfunctionalization. J. Biotechnol. 113:183-210. [DOI] [PubMed] [Google Scholar]
- 8.Bühler, B., A. Schmid, B. Hauer, and B. Witholt. 2000. Xylene monooxygenase catalyzes the multistep oxygenation of toluene and pseudocumene to corresponding alcohols, aldehydes, and acids in Escherichia coli JM101. J. Biol. Chem. 275:10085-10092. [DOI] [PubMed] [Google Scholar]
- 9.Bühler, B., B. Witholt, B. Hauer, and A. Schmid. 2002. Characterization and application of xylene monooxygenase for multistep biocatalysis. Appl. Environ. Microbiol. 68:560-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerniglia, C. E., and C. C. Somerville. 1995. Reductive metabolism of nitroaromatic and nitropolycyclic aromatic hydrocarbons, p. 99-115. In J. C. Spain (ed.), Biodegradation of nitroaromatic compounds. Plenum Press, New York, N.Y.
- 11.de Bont, J. A. M. 1998. Solvent-tolerant bacteria in biocatalysis. Trends Biotechnol. 16:493-499. [Google Scholar]
- 12.Delgado, A., M. G. Wubbolts, M.-A. Abril, and J. L. Ramos. 1992. Nitroaromatics are substrates for the TOL plasmid upper-pathway enzymes. Appl. Environ. Microbiol. 58:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz, E., A. Ferrandez, M. A. Prieto, and J. L. Garcia. 2001. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 65:523-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duetz, W. A., J. B. van Beilen, and B. Witholt. 2001. Using proteins in their natural environment: potential and limitations of microbial whole-cell hydroxylations in applied biocatalysis. Curr. Opin. Biotechnol. 12:419-425. [DOI] [PubMed] [Google Scholar]
- 15.Ellis, L. B. M., C. D. Hershberger, and L. P. Wackett. 1999. The University of Minnesota Biocatalysis/Biodegradation Database: specialized metabolism for functional genomics. Nucleic Acids Res. 27:373-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng, K., M. Tsushima, T. Matsumoto, and T. Kurosaki. 1998. Synthesis and properties of novel photosensitive polyimides containing chalcone moiety in the main chain. J. Polym. Sci. Polym. Chem. 36:685-693. [Google Scholar]
- 17.Glöckler, R., and J. P. Roduit. 1996. Industrial bioprocesses for the production of substituted aromatic heterocycles. Chimia 50:413-415. [Google Scholar]
- 18.Grund, A., J. Shapiro, M. Fennewald, P. Bacha, J. Leahy, K. Markbreiter, M. Nieder, and M. Toepfer. 1975. Regulation of alkane oxidation in Pseudomonas putida. J. Bacteriol. 123:546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harayama, S., M. Kok, and E. L. Neidle. 1992. Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 46:565-601. [DOI] [PubMed] [Google Scholar]
- 20.Haro, M.-A., and V. de Lorenzo. 2001. Metabolic engineering of bacteria for environmental applications: construction of Pseudomonas strains for biodegradation of 2-chlorotoluene. J. Biotechnol. 85:103-113. [DOI] [PubMed] [Google Scholar]
- 21.Hartter, D. R. 1985. The use and importance of nitroaromatic chemicals in the chemical industry, p. 1-13. In D. E. Rickert (ed.), Toxicity of nitroaromatic compounds. Chemical Industry Institute of Toxicology Series. Hemisphere, Washington, D.C.
- 22.James, K. D., and P. A. Williams. 1998. ntn genes determining the early steps in the divergent catabolism of 4-nitrotoluene and toluene in Pseudomonas sp. strain TW3. J. Bacteriol. 180:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimenez, J. I., B. Minambres, J. L. Garcia, and E. Diaz. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4:824-841. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, G. R., and J. C. Spain. 2003. Evolution of catabolic pathways for synthetic compounds: bacterial pathways for degradation of 2,4-dinitrotoluene and nitrobenzene. Appl. Microbiol. Biotechnol. 62:110-123. [DOI] [PubMed] [Google Scholar]
- 25.Kadiyala, V., L. J. Nadeau, and J. C. Spain. 2003. Construction of Escherichia coli strains for conversion of nitroacetophenones to ortho-aminophenols. Appl. Environ. Microbiol. 69:6520-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiener, A. 1992. Enzymatic oxidation of methyl groups on aromatic heterocycles: a versatile method for the preparation of heteroaromatic carboxylic acids. Angew. Chem. Int. Ed. Engl. 31:774-775. [Google Scholar]
- 27.Kieslich, K. 1993. How to select a useful biocatalyst. Chimia 47:99-101. [Google Scholar]
- 28.Kolsek, J., N. Novak, and M. Perpar. 1957. p-Dimethylaminobenzaldehyd als Reagens zur Charakterisierung primärer aromatischer Amine. Z. Anal. Chem. 159:113-117. [Google Scholar]
- 29.Konstantinov, K., M. Kishimoto, T. Seki, and T. Yoshida. 1990. A balanced DO-stat and its application to the control of acetic acid excretion by recombinant Escherichia coli. Biotechnol. Bioeng. 36:750-758. [DOI] [PubMed] [Google Scholar]
- 30.Liese, A., K. Seelbach, and C. Wandrey. 2000. Industrial biotransformations, 1st ed. Wiley-VCH, Weinheim, Germany.
- 31.Maruyama, T., H. Iida, and H. Kakidani. 2003. Oxidation of both termini of p- and m-xylene by Escherichia coli transformed with xylene monooxygenase gene. J. Mol. Catal. B Enzym. 21:211-219. [Google Scholar]
- 32.Marvin-Sikkema, F. D., and J. A. M. de Bont. 1994. Degradation of nitroaromatic compounds by microorganisms. Appl. Microbiol. Biotechnol. 42:499-507. [DOI] [PubMed] [Google Scholar]
- 33.Mathys, R. G., A. Schmid, and B. Witholt. 1999. Integrated two-liquid phase bioconversion and product-recovery processes for the oxidation of alkanes: process design and economic evaluation. Biotechnol. Bioeng. 64:459-477. [DOI] [PubMed] [Google Scholar]
- 34.McCalla, D. R., C. Kaiser, and M. H. L. Green. 1978. Genetics of nitrofurazone resistance in Escherichia coli. J. Bacteriol. 133:10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Messing, J. 1979. A multipurpose cloning system based on single-stranded DNA bacteriophage M13. Recomb. DNA Tech. Bull. 2:43-49. [Google Scholar]
- 36.Nadeau, L. J., and J. C. Spain. 1995. Bacterial degradation of m-nitrobenzoic acid. Appl. Environ. Microbiol. 61:840-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishino, S. F., J. C. Spain, and Z. He. 2000. Strategies for aerobic degradation of nitroaromatic compounds by bacteria: process discovery to field application, p. 7-61. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. Lewis Publishers, Boca Raton, Fla.
- 38.Panke, S., M. Held, M. G. Wubbolts, B. Witholt, and A. Schmid. 2002. Pilot-scale production of (S)-styrene oxide from styrene by recombinant Escherichia coli synthesizing styrene monooxygenase. Biotechnol. Bioeng. 80:33-41. [DOI] [PubMed] [Google Scholar]
- 39.Panke, S., A. Meyer, C. M. Huber, B. Witholt, and M. G. Wubbolts. 1999. An alkane-responsive expression system for the production of fine chemicals. Appl. Environ. Microbiol. 65:2324-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramos, J. L., E. Duque, M.-J. Huertas, and A. Haidour. 1995. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J. Bacteriol. 177:3911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rieger, P.-G., and H.-J. Knackmuss. 1995. Basic knowledge and perspectives on biodegradation of 2,4,6-trinitrotoluene and related nitroaromatic compounds in contaminated soil, p. 1-18. In J. C. Spain (ed.), Biodegradation of nitroaromatic compounds. Plenum Press, New York, N.Y.
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schackmann, A., and R. Müller. 1991. Reduction of nitroaromatic compounds by different Pseudomonas species under aerobic conditions. Appl. Microbiol. Biotechnol. 34:809-813. [Google Scholar]
- 44.Schenzle, A., H. Lenke, J. C. Spain, and H.-J. Knackmuss. 1999. Chemoselective nitro group reduction and reductive dechlorination initiate degradation of 2-chloro-5-nitrophenol by Ralstonia eutropha JMP134. Appl. Environ. Microbiol. 65:2317-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmid, A., J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts, and B. Witholt. 2001. Industrial biocatalysis today and tomorrow. Nature 409:258-268. [DOI] [PubMed] [Google Scholar]
- 46.Spain, J. C. 1995. Biodegradation of nitroaromatic compounds. Annu. Rev. Microbiol. 49:523-555. [DOI] [PubMed] [Google Scholar]
- 47.Straathof, A. J. J., S. Panke, and A. Schmid. 2002. The production of fine chemicals by biotransformations. Curr. Opin. Biotechnol. 13:548-556. [DOI] [PubMed] [Google Scholar]
- 48.Trivic, S., V. Leskova, and G. W. Winston. 1999. Aldehyde dismutase activity of yeast alcohol dehydrogenase. Biotechnol. Lett. 21:231-234. [Google Scholar]
- 49.Wackett, L. P. 2003. Pseudomonas putida—a versatile biocatalyst. Nat. Biotechnol. 21:136-138. [DOI] [PubMed] [Google Scholar]
- 50.Walia, S. K., S. Ali-Sadat, and G. R. Chaudhry. 2003. Influence of nitro group on biotransformation of nitrotoluenes in Pseudomonas putida strain OU83. Pestic. Biochem. Physiol. 76:73-81. [Google Scholar]
- 51.Whiteway, J., P. Koziarz, J. Veall, N. Sandhu, P. Kumar, B. Hoecher, and I. B. Lambert. 1998. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J. Bacteriol. 180:5529-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson, D., J. M. Ward, and J. M. Woodley. 1996. Choice of microbial host for the naphthalene dioxygenase bioconversion. J. Ind. Microbiol. 16:274-279. [DOI] [PubMed] [Google Scholar]
- 53.Witholt, B. 1972. Method for isolating mutants overproducing nicotinamide adenine dinucleotide and its precursors. J. Bacteriol. 109:350-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wubbolts, M. G., J. Hoven, B. Melgert, and B. Witholt. 1994. Efficient production of optically active styrene epoxides in two-liquid phase cultures. Enzyme Microb. Technol. 16:887-894. [Google Scholar]
- 55.Wubbolts, M. G., P. Reuvekamp, and B. Witholt. 1994. TOL plasmid-specified xylene oxygenase is a wide substrate range monooxygenase capable of olefin epoxidation. Enzyme Microb. Technol. 16:608-615. [DOI] [PubMed] [Google Scholar]
- 56.Ye, J., A. Singh, and O. P. Ward. 2004. Biodegradation of nitroaromatics and other nitrogen-containing xenobiotics. World J. Microbiol. Biotechnol. 20:117-135. [Google Scholar]