Abstract
Variation in genome size and content is common among bacterial strains. Identifying these naturally occurring differences can accelerate our understanding of bacterial attributes, such as ecological specialization and genome evolution. In this study, we used representational difference analysis to identify potentially novel sequences not present in the sequenced laboratory strain Rm1021 of the nitrogen-fixing bacterium Sinorhizobium meliloti. Using strain Rm1021 as the driver and the type strain of S. meliloti ATCC 9930, which has a genome size ∼370 kilobases bigger than that of strain Rm1021, as the tester, we identified several groups of sequences in the ATCC 9930 genome not present in strain Rm1021. Among the 85 novel DNA fragments examined, 55 showed no obvious homologs anywhere in the public databases. Of the remaining 30 sequences, 24 contained homologs to the Rm1021 genome as well as unique segments not found in Rm1021, 3 contained sequences homologous to those published for another S. meliloti strain but absent in Rm1021, 2 contained sequences homologous to other symbiotic nitrogen-fixing bacteria (Rhizobium etli and Bradyrhizobium japonicum), and 1 contained a sequence homologous to a gene in a non-nitrogen-fixing species, Pseudomonas sp. NK87. Using PCR, we assayed the distribution of 12 of the above 85 novel sequences in a collection of 59 natural S. meliloti strains. The distribution varied widely among the 12 novel DNA fragments, from 1.7% to 72.9%. No apparent correlation was found between the distribution of these novel DNA sequences and their genotypes obtained using multilocus enzyme electrophoresis. Our results suggest potentially high rates of gene gain and loss in S. meliloti genomes.
Accumulating evidence reveals that variations in genome size and gene content are common in bacteria (29). For example, the genomes of natural isolates of the common bacterium Escherichia coli can vary by more than 1 million base pairs (6, 7, 17). Among the serotypes of another common bacterium, Salmonella enterica (serotypes Enteritidis, Paratyphi, Typhi, and Typhimurium), chromosome sizes can differ by ∼300 kilobase pairs (21). These genomic differences often contribute to their ecological adaptations, such as host specificity, nutrient utilization, stress tolerance, pathogenicity, and antibiotic resistance (4, 5, 10). Indeed, identifying the naturally occurring differences among bacterial strains has greatly accelerated our understanding of bacterial adaptation and evolution. However, aside from the common human pathogenic bacteria, such as E. coli, S. enterica, and Staphylococcus aureus, relatively little is known about genome size variation and the factors contributing to such variation in other bacterial species, including the symbiotic nitrogen-fixing bacterium Sinorhizobium meliloti.
S. meliloti is a symbiont of legumes belonging to the three genera Medicago, Melilotus, and Trigonella. Aside from its symbiotic lifestyle, S. meliloti also has a free-living phase and is ubiquitous in soils with pH values above 6. It has a global distribution, from tropical arid regions to temperate humid soils. Because of its nonfastidious lifestyle and the ease of genetic manipulations, S. meliloti has become a model species for studying the molecular basis of symbiotic nitrogen fixation. As a result, the genome of a model laboratory strain, Rm1021, became one of the first symbiotic nitrogen-fixing bacteria to be completely sequenced (15). This strain has a tripartite 6.7-Mb genome comprised of a 3.65-Mb chromosome, a 1.35-Mb megaplasmid called pSymA, and a 1.68-Mb megaplasmid called pSymB. Previous laboratory studies have shown that deletions of large portions of the S. meliloti genome often had little or no fitness consequences under certain laboratory conditions (12, 18, 24). These results suggest that genome size among natural strains of S. meliloti might be highly variable and that DNA sequences absent in strain Rm1021 might exist in other natural strains.
To begin investigating this possibility and identify the DNA sequences and potential mechanisms contributing to natural genomic variation in S. meliloti, we first compared the genome sizes of the type strain of S. meliloti, ATCC 9930, and the sequenced model laboratory strain Rm1021 using pulsed-field gel electrophoresis (PFGE). Strain ATCC 9930 was found to have a genome about 370 kb bigger than that of Rm1021 (see below). The potential differences between strains ATCC 9930 and Rm1021 were then examined.
Several subtractive DNA hybridization methods have been developed to reveal natural genomic differences (27). One such method is called representational difference analysis (RDA). The RDA technique was first described in 1993 (20), and the incorporation of PCR to selectively enrich the target sequences has greatly enhanced its effectiveness and attractiveness (2, 4, 5, 9, 10, 11, 23, 27, 31). The PCR steps selectively amplify unique “tester” homoduplexes formed after hybridization against the “driver” DNA, thereby enriching for sequences unique to the tester population. RDA has been used successfully to isolate unique sequences from several bacterial species, including Neisseria meningitidis (4, 5), Neisseria gonorrhoeae (31), Vibrio cholerae (11), Bordetella spp. (23), and E. coli (2, 9, 10).
In this study, we performed RDA using genomic DNA from strain ATCC 9930 as the tester and that from the model laboratory strain Rm1021 as the driver. Two RDA libraries were constructed, and randomly selected clones were sequenced. Abundant novel DNA sequences were found in strain ATCC 9930. PCR primers were then developed from a random subset of these unique DNA sequences and used to screen for their distribution in a diverse collection of natural strains of S. meliloti. This collection of strains has been analyzed previously by Eardly et al. (13) using multilocus enzyme electrophoresis (MLEE). The distribution of these novel DNA fragments among natural strains was analyzed and discussed in the context of genotypes obtained using MLEE analysis. We specifically tested whether strains with the same MLEE type would share more-similar profiles of novel DNA fragment distribution.
MATERIALS AND METHODS
Strains.
Two strains were used for RDA: one was the sequenced model laboratory strain Rm1021, and the other was the type strain of S. meliloti ATCC 9930. Rm1021 is a streptomycin-resistant derivative of strain SU47, originally isolated from alfalfa in Australia. Strain ATCC 9930 was isolated in the United States, and its genome size, structure, and genomic sequence are unknown. Both Rm1021 and ATCC 9930 have the same MLEE type, ET1, as determined by Eardly et al. (13). In RDA, genomic DNA from strain Rm1021 was used as the “driver,” while that of strain ATCC 9930 was used as the “tester.” Fifty-seven additional natural strains were included in the analysis of distributions of the novel DNA sequences discovered from RDA. These strains were generously provided by Bert Eardly of Pennsylvania State University, Berks-Lehigh Valley College, Reading, Pennsylvania. The strain names, MLEE types, isolation hosts, and geographic origins for all 59 strains are presented in Table S1 in the supplemental material. Among the 59 strains, 27 shared the same MLEE type, ET1. The remaining 32 strains each had a different MLEE type (13). These strains are available from the senior author at jpxu@mcmaster.ca.
Genome size determination for strain ATCC 9930.
The genome size of strain ATCC 9930 was estimated by PFGE using a contour-clamped homogeneous electric field apparatus (CHEF-DR II; Bio-Rad Laboratories). We followed the protocol given in the instrument manual for bacterial cultures, with the following minor modifications. Briefly, bacteria were first grown in liquid LBmc broth (per liter, 10 g of pancreatic digest of casein, 5 g of NaCl, 5 g of yeast extract, 2.5 mM MgSO4, and 2.5 mM CaCl2, pH 7). Cells were incubated at 30°C with constant agitation at 120 rpm and harvested through centrifugation when the population density reached an optical density at 600 nm (OD600) between 0.8 and 1.0. The harvested cells were resuspended in the suspension buffer (10 mM Tris-Cl, 20 mM NaCl, 50 mM EDTA). Agarose plugs were prepared by mixing equal volumes of the bacterial suspension with 2% CleanCut agarose (Bio-Rad Laboratories). Cells in the agarose plugs were lysed by the lysozyme buffer (10 mM Tris-Cl, 50 mM NaCl, 0.2% sodium deoxycholate, 0.5% N-sodium lauroyl sarcosine, 1 mg/ml lysozyme) and then treated with proteinase K buffer (100 mM EDTA, 0.2% sodium deoxycholate, 1% N-sodium lauroyl sarcosine, 1 mg/ml proteinase). Plugs were then washed three times using Tris-EDTA buffer (20 mM Tris, 50 mM EDTA; pH 8.0) and stored at 4°C. The plugs were run in a 1% agarose gel in 0.5× Tris-borate-EDTA buffer (45 mM Tris, 45 mM borate, 1.0 mM EDTA, pH 8.3) at a voltage gradient of 4.5 V/cm and a switch time of 60 to 200 seconds for 48 h at 14°C. In addition to using strain Rm1021, chromosomal size standards of two yeast species, Saccharomyces cerevisiae and Schizosaccharomyces pombe, from Bio-Rad Laboratories were also used to help estimate replicon sizes of strain ATCC 9930.
Genome DNA extraction.
To obtain genomic DNA for analysis, strains were first grown in LBmc broth. For strains Rm1021 and ATCC 9930, cells were grown into 150 ml of LBmc broth. For the other 57 strains, cells were inoculated into 5 ml LBmc broth. Cells were incubated at 30°C with constant agitation at 120 rpm and harvested when the population density reached an OD600 reading between 0.8 and 1.0. Genomic DNA was extracted according to a protocol described previously for S. meliloti (12). The quantity and quality of DNA were assessed using the UltraSpec 2000 pro spectrophotometer.
RDA.
Genomic DNA RDA was performed according to a protocol described by Allen et al. (3) with minor modifications (see Fig. S1 in the supplemental material). Briefly, 500 μl of the driver (strain Rm1021) genomic DNA (1.0 μg/μl) was sheared through sonication (30 s; output, 30; VibraCell) to generate fragments between 1 kb and 10 kb in length. Five microliters (1.0 μg/μl) of the tester genomic DNA (strain ATCC 9930) was digested using restriction endonuclease Sau3AI (New England Biolabs). After enzyme inactivation, the digested tester DNA was ligated to two partially complementary oligonucleotides, ASau12 (5′GATCTGTTCATG 3′) and ASau24 (5′ACCGACGTCGACTATCCATGAACA 3′), in a ratio of 1 μl tester DNA to 0.5 nM of each of the two oligonucleotides. The tester/driver DNA ratio of 1:200 was used, and the remaining subtraction and PCR enrichment steps followed those described in Allen et al. (3). The second round of hybridization used two different hybridization ligators, TSau12 (5′GATCTTCCCTCG 3′) and TSau24 (5′AGGCAACTGTGCTATCCGAGGGAA3′). All cleanup steps used QIAquick spin columns (QIAGEN) followed by phenol extraction/ethanol precipitation. Two rounds of subtraction and PCR amplification were performed. After each round, the enriched RDA products were visualized on a 1% agarose gel and used for the construction of subtractive libraries.
Construction of subtractive libraries.
To determine the sequences of enriched RDA products, we first constructed two DNA libraries. The first library was constructed using product from the first round of subtraction, and the second library was constructed using product from the second round of subtraction. To construct these libraries, the RDA products were first purified using the StrataPrep PCR purification kit (Stratagene, La Jolla, CA), and these purified products were then ligated onto the pPCR-script Amp SK (+) cloning vector and transformed into Epicurian Coli XL 10 Gold Kan ultracompetent cells (PCR-script Amp cloning kit; Stratagene, La Jolla, CA) according to the manufacturer's instructions. The library clones were screened by direct PCR from bacterial colonies using the T7 forward and T3 reverse primers. These PCR products were first confirmed by agarose gel electrophoresis and then cleaned using the MicroCLEAN PCR purifying reagents according to the manufacturer's instructions (Microzone Limited, West Sussex, United Kingdom) before being sent for sequencing.
DNA sequencing and BLAST analysis.
The purified PCR products from the subtractive library clones were sent for sequencing to the MOBIX laboratory of McMaster University. Sequencing was performed using a multicolor-fluorescence-based DNA analysis system, the ABI PRISM 3100 genetic analyzer, according to the manufacturer's instructions. These sequences were then used to search for homologs in the S. meliloti database and other public databases using a BLAST score with an E value of <10−5 as a threshold value. Only the best match for each clone is presented in Results.
PCR assay for the distribution of novel sequences among natural strains.
After identifying DNA fragments present in strain ATCC 9930 but absent in strain Rm1021, we proceeded to determine the distributions of some of these sequences in other natural strains. To achieve this goal, specific PCR primers were designed, and these primers were then used, through PCR, to screen 57 natural strains. In these screens, strains ATCC 9930 and Rm1021 were used as positive and negative controls, respectively. A typical PCR contained 1× PCR buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl [pH 8.3]), 7.5 pmol of each primer, 0.1 μg of template DNA, 60 μM deoxynucleoside triphosphate mixture (15 μM [each] dATP, dCTP, dGTP, and dTTP), and 0.75 U of Taq DNA polymerase (Invitrogen Life Technologies) in a total volume of 15 μl. Typical running conditions were 3 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at the appropriate annealing temperature specific to individual primer pairs, and 30s at 72°C, and finally, 10 min of extension at 72°C. PCR products were run on 1% agarose gels, stained with ethidium bromide, revealed using UV light, and scored for the presence/absence of expected DNA fragments.
Analyses of the distribution of novel DNA sequences among natural strains.
To analyze the distribution patterns of these novel DNA fragments, each DNA fragment was treated as a locus with two alternative alleles in the population: the presence of the fragment and the absence of the fragment. Because the S. meliloti genome is haploid, the allelic assignment for each strain at each locus is therefore unambiguous. A multilocus genotype based on these assayed DNA fragments was derived for each of the 59 strains.
To determine whether strains with the same MLEE type would have more-similar profiles of novel DNA fragment distribution, we used the topology-dependent permutation tail probability (T-PTP) function implemented in the phylogenetic program PAUP* 4.0 (30). In the T-PTP test, the length of the maximum parsimony tree for which all strains of the same MLEE type were considered to belong to a monophyletic group was compared to the length of the tree without this monophyletic-group constraint. If the constrained tree was significantly longer than the unconstrained tree, this result would suggest that MLEE type is not a good indicator of novel DNA fragment distribution (14). The statistical significance of this test is derived from data permutation under the assumption of nonmonophyly to generate a null distribution of tree lengths. Statistical support for nonmonophyly is achieved when over 95% of all permuted data sets have tree lengths shorter than that of the maximum parsimony tree generated with the constraint of monophyly (14). A thousand permuted data sets were generated and analyzed for this test. Potential patterns of association with host species and geographic origins as constraints were also analyzed using the T-PTP test.
The multilocus genotypes inferred from the distribution of these novel DNA fragments were also used to analyze the allelic associations among these markers. Such an analysis can be used to infer whether this population of strains has a predominantly clonal or recombining population structure. To analyze the association among alleles, we calculated the overall index of association (IA) and the proportion of pairwise loci that are phylogenetically incompatible. A phylogenetic incompatibility occurs between two loci with two alleles each when all four possible genotypes are found in the population. A low IA value and a high proportion of pairwise loci that are phylogenetically incompatible suggest evidence of recombination in the population. These tests are implemented in the program Multilocus version 1.3 (1). The underlying assumptions, formula, and inferences of statistical significance of these tests can be found on the program homepage.
RESULTS
Genome size of strain ATCC 9930.
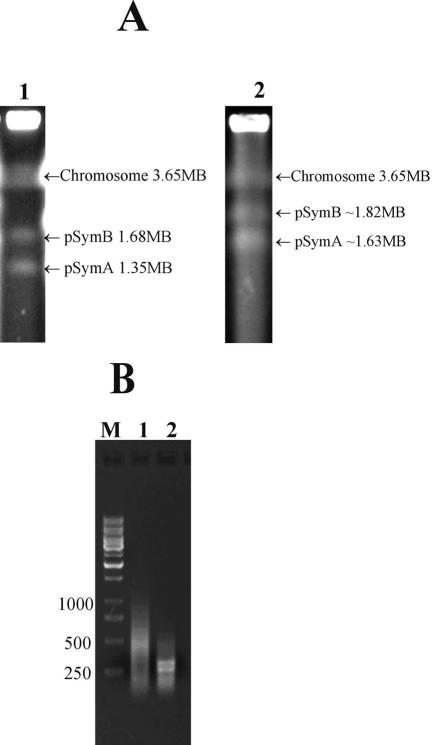
Like the model laboratory strain Rm1021, strain ATCC 9930 was found to have a tripartite genome containing three replicons (Fig. 1A). Based on comparisons with replicon sizes of strain Rm1021, the estimated sizes of the three replicons for strain ATCC 9930 were 3.65 Mb, 1.82 Mb, and 1.63 Mb. Therefore, the total genome size of strain ATCC 9930 is approximately 7.1 Mb, which is about 370 kb larger than that of strain Rm1021 (Fig. 1A).
FIG. 1.
Agarose gel electrophoresis. (A) PFGE of replicon DNA. Lane 1, the three replicons of strain Rm1021; lane 2, the three replicons of strain ATCC 9930. (B) Gel electrophoresis of RDA subtractive products. Lane M, 1-kb size standard; lane 1, subtractive products from the first round of RDA; lane 2, subtractive products from the second round of RDA.
RDA and subtractive library construction.
After each round of RDA, the presence of enriched, potentially unique DNA sequences in strain ATCC 9930 was checked using agarose gel electrophoresis (Fig. 1B). The first round of differential products showed a smear of enriched DNA ranging from 250 to 750 bp (Fig. 1B, lane 1). The second round of differential products showed two clearly visible bands about 250 bp and 300 bp in size (Fig. 1B, lane 2). The two purified differential products were ligated onto the cloning vector pPCR-script, and two subtractive libraries were constructed.
Sequencing and BLAST searches.
Clones from the two subtractive libraries were randomly selected and screened by PCR using vector-specific primers T7 and T3. A total of 161 clones were sequenced. Two clones were found redundant, yielding a total of 159 nonredundant clones, with each containing a unique sequence. Based on BLAST search results and for discussion purposes, we divided these 159 clones into six groups. The distribution of these clones in the six groups is summarized in Table 1. There was no significant difference in distribution pattern among the six groups between the two subtractive libraries (data not shown). Below are brief descriptions for each of the six groups of clones.
TABLE 1.
Summary information of BLAST analysis of 159 RDA clones
| Group | Group characteristic | Repliconsa (no. of clones) | Total no. of clones | % Identity | Length (bp) |
|---|---|---|---|---|---|
| I | Homologous to DNA sequences in the genome of Rm1021 | C (24), A (32), B (18) | 74 | 67-100 | 113-475 |
| II | Part of the sequence homologous to Rm1021 | C (7), A (12), B (5) | 24 | 75-100 | 20-319/88-468b |
| III | Homologous to published sequences in other S. meliloti strains | 3 | 98-99 | 282-469 | |
| IV | Homologous to published sequences in other nitrogen-fixing bacterial species | 2 | 85-100 | 137-211 | |
| V | Homologous to published genes in non-nitrogen-fixing bacterium | 1 | 87 | 332 | |
| VI | Sequences with no homolog in public databases | 55 | 135-671 |
C, chromosome; A, pSymA; B, pSym.
Homologous portions/novel portions.
The first group (I) consisted of clones with entire DNA sequences homologous to those in the genome of strain Rm1021. Among the total of 74 clones in this group, 24 were homologous to sequences on the chromosome of Rm1021, 32 were homologous to those on pSymA of Rm1021, and 18 were homologous to those on pSymB of Rm1021 (Table 1). Among the 24 clones with homologs on the chromosome of Rm1021, 15 (62.5%) showed 100% sequence identity to DNA in Rm1021 and the remaining 9 had a range (90 to 99%) of sequence identities. Among the 32 DNA fragments with homologs on pSymA of strain Rm1021, 5 (16.7%) showed 100% sequence identity to strain Rm1021 and the remaining 27 had a range (70 to 99%) of sequence identities. Of the 18 clones with homologs on pSymB of Rm1021, 3 were completely identical, 5 showed 99% sequence identity, and 10 exhibited a range (67 to 97%) of sequence identities to sequences in strain Rm1021.
The second group (II) contained DNA fragments in which at least one section was homologous to a sequence in strain Rm1021 and other sections contained novel sequences not found in strain Rm1021. Among the 24 sequenced fragments found in this group, BLAST searches identified that 7 contained segments with significant homology to chromosomal regions of strain Rm1021, 12 contained segments with significant homology to regions of pSymA in strain Rm1021, and 5 contained segments with significant homology to regions of pSymB in strain Rm1021 (Table 1). The lengths of the homologous portions varied between 20 and 319 bp, with sequence identities ranging from 75 to 100%. The novel portions of these sequences ranged in length between 88 bp and 468 bp (Table 1 and see Table S2 in the supplemental material).
The third group (III) contained three clones. They ranged between 282 and 469 bp in length. While these three clones had no homolog in the Rm1021 genome, they were homologous to DNA sequences found on plasmid pRmeGR4b of another strain, GR4 of S. meliloti (22, 36). Specifically, two clones had high sequence identities to a hypothetical protein with unknown functions, while the third was homologous to a tyrosinase gene involved in melanin production in strain GR4 (22). Sequence identities among the three clones and those on pRmeGR4b varied between 98 and 99% (see Table S2 in the supplemental material).
The fourth group (IV) contained sequences with homologs in other nitrogen-fixing bacterial species (Table 1 and see Table S2 in the supplemental material). This group had two representative clones. One clone was 137 bp long and had 85% sequence identity to a probable peptide synthetase gene in the symbiotic gene region reported for strain USDA110 of Bradyrhizobium japonicum (16). The second clone was 211 bp long and contained a 54-bp fragment with 94% sequence identity to the virB8 gene in Rhizobium etli CE3 (8).
The fifth group (V) contained one representative clone that was 332 bp long and had 87% sequence identity to the 6-aminohexanoate-dimer hydrolase gene on the plasmid of Pseudomonas spp. strain NK87 (see Table S2 in the supplemental material). This protein was found capable of degrading nylon oligomers (19, 34). Nylon oligomers are among the compounds not present in natural environments until they were synthesized and released by humans very recently.
The last group (VI) had 55 clones. Clones in this group contained no obvious homologs in the public databases.
More-detailed information about clones in groups II, III, IV, and V is presented in Table S2 in the supplemental material. For each clone, all information on the clone's identification, sequence length, closest homolog in the public database with assigned or putative function, sequence identity, and GenBank accession number is included.
Distribution of novel DNA sequences among natural strains.
From the above-described novel DNA sequences, we selected 12 random clones and designed PCR primers to screen for their distributions among natural strains. The primer information and predicted fragment size based on information from novel DNA sequences in strain ATCC 9930 are presented in Table S3 in the supplemental material. As expected, strain Rm1021 was found to contain none of the 12 fragments, while strain ATCC 9930 was found to contain all 12 fragments of predicted sizes (see Table S4 in the supplemental material). A representative profile is shown in Fig. S2 in the supplemental material. Interestingly, for each primer pair, the PCR-amplified products from all strains containing the sequence were all identical in size (see Fig. S2 in the supplemental material).
The multilocus genotypes based on PCR screening of the 12 novel DNA fragments are presented in Table S4 in the supplemental material. The frequency distribution among the 12 DNA fragments varied widely, from 1.7% for clone RDA-35 (found only in the tester strain ATCC 9930) to 72.9% (found in 43 of the 59 natural strains) for clone RDA-25.
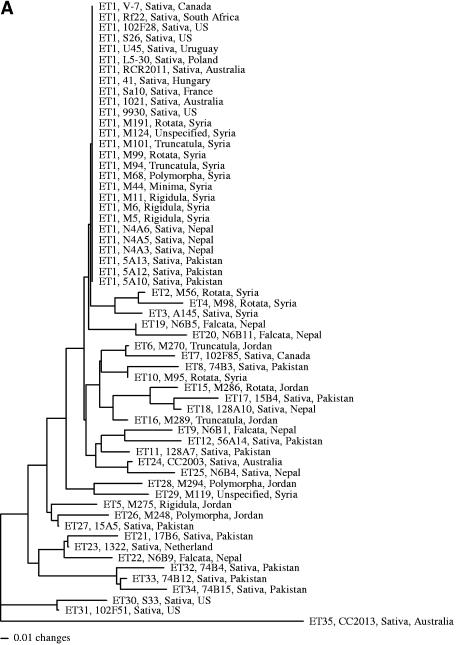
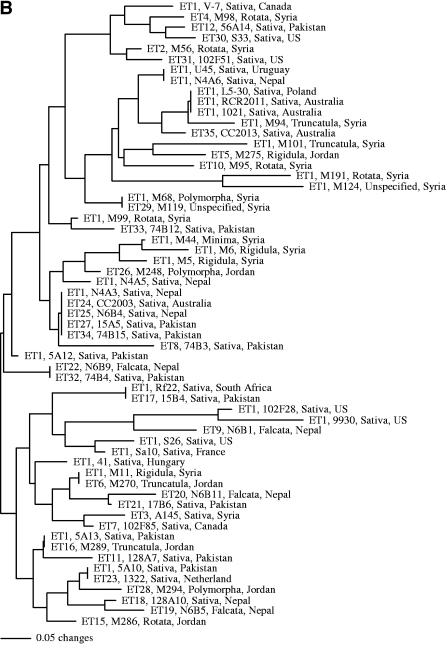
This genotyping method identified a total of 46 multilocus genotypes among the 59 strains. To facilitate the comparison of MLEE data and the novel DNA sequence distribution data, the relationships among strains inferred from these two data sets are presented side by side in Fig. 2, with Fig. 2A and B based on MLEE data and novel DNA sequence distribution data, respectively. The 27 strains with the same MLEE type, ET1, were found to have 24 different novel DNA sequence distribution profiles. Only two genotypes were shared among strains with ET1. One included three strains, RCR2011, Rm1021, and L5-30. This genotype was found to contain none of the 12 assayed fragments. Strains RCR2011 and Rm1021 were different derivatives of the same isolate, SU47, and their identical patterns confirmed the robustness of this genotyping method. However, strain L5-30 was found in a location (Poland) far from that of SU47 (Australia). Their identical patterns in both novel sequence distribution and MLEE suggest a recent common origin for the three strains.
FIG. 2.
Strain relationships inferred from MLEE data (A) and the novel DNA sequence distribution data (B). The MLEE data were from Eardly et al. (13) and are used here for comparison. In both analyses, the neighbor-joining algorithm was used. For each strain, the following four pieces of information were presented: MLEE type, strain name, host species name (all belong to the genus Medicago), and geographic origin.
Of the remaining 32 strains belonging to different MLEE types, a total of 27 profiles were found, with one profile shared by two strains and another profile shared by four strains. Interestingly, six profiles were shared between strains from ET1 and those from other MLEE types. The most frequent profile contained five strains, and all five strains had different MLEE types. These results suggest that MLEE genotype data are not reliable predictors of novel sequence distributions and vice versa. The T-PTP test supports this conclusion. Without monophyletic constraints, all randomized data generated trees with lengths significantly shorter than those with monophyletic constraints for strains belonging to MLEE ET1 (P < 0.001). Similarly, we found no evidence of monophyly for strains that either belong to the same geographic region or have the same host species (P < 0.001).
Association among alleles at different loci.
The allelic data for all 12 loci were analyzed for their potential association with each other at the population level. For both the IA analysis and the phylogenetic incompatibility test, three samples were analyzed. The first included the 27 strains belonging to MLEE ET1, the second included the 32 strains of other MLEE types, and the third included all 59 strains. Results of these analyses are presented in Table 2. Briefly, the IA analysis suggested that there was an overall statistically significant association among alleles at different loci in all three samples (P ≤ 0.02). Similarly, phylogenetic incompatibility tests suggested that the majority of pairwise loci comparisons were phylogenetically compatible (P ≥ 0.97). These results are consistent with a predominantly clonal population structure. However, all three samples showed various proportions of loci that were clearly phylogenetically incompatible, suggesting a low-level genetic recombination in natural populations of this species (Table 2).
TABLE 2.
Analyses of the association among novel DNA fragments in samples of Sinorhizobium meliloti
| Sample types | Index of association
|
Phylogenetic incompatibility
|
||
|---|---|---|---|---|
| IA | P | % Incompatibility | P | |
| All 59 strains | 0.554 | <0.01 | 3.03 | 1.00 |
| 27 Strains of ET1 | 1.204 | <0.01 | 6.06 | 0.97 |
| Strains of non-ET1 | 0.234 | 0.02 | 33.33 | 0.99 |
| All strains using MLEE data (13) | 0.309 | 0.03 | 83.52 | <0.01 |
DISCUSSION
This study compared the genome size differences of the model laboratory strain Rm1021 and the type strain of S. meliloti, ATCC 9930. Strain ATCC 9930 was found to have a genome size about 370 kb bigger than that of strain Rm1021. This difference is comparable to those found among strains in other species, such as E. coli and S. enterica (6, 7, 21). Using the RDA method, we identified a large number of DNA sequences not present in the genome of Rm1021. These novel DNA sequences were classified based on their relationships to sequences in public databases. Surprisingly, 55 of the 161 sequenced RDA clones had no obvious homolog anywhere in the public databases. The distribution of 12 novel DNA fragments among a collection of 59 natural strains was surveyed, and our analyses suggested that the relationships among these strains were very different from those inferred from MLEE genotype data.
While the exact physical locations of the novel sequences isolated in this study are unknown at present, a preliminary analysis of our data suggests that these novel sequences likely exist on all three replicons and are more likely to be found on the megaplasmids than on the chromosome. This conclusion is based on sequence information from the 24 clones in strain ATCC 9930 that contained both novel fragments and sequences homologous to strain Rm1021 (group II) (Table 1). Among the homologous portions of the 24 clones, 7 were found on the chromosome and 17 were found on the megaplasmids (12 on pSymA and 5 on pSymB) of strain Rm1021. Based on the relative sizes of the three replicons and assuming a random novel sequence distribution among the three replicons, of the 24 clones, the expected counts are 13 and 11 for those on the chromosome and the megaplasmids, respectively. The significantly biased distribution of these 24 clones towards megaplasmids (especially pSymA) (chi-square value = 6.04; df = 1; P < 0.05) is consistent with the differences in replicon sizes observed here between strains ATCC 9930 and Rm1021 (Fig. 1).
Several factors and processes can contribute to genome size variation in bacteria. These include the following: (i) independent gains of DNA from external sources through conjugation, transformation, and transduction, especially of those DNAs involved in niche-specific ecological adaptations, such as host-specific pathogenicity/symbiosis, substrate utilization, and antibiotic resistance; (ii) gene duplication; and (iii) independent random losses of genes (6, 7). Because the exact evolutionary relationship between strains Rm1021 and ATCC 9930 is not known, the precise contribution of each of these processes to the genome size variation between these two strains cannot be unambiguously inferred using the existing data. However, several lines of evidence suggest that multiple independent gene losses and/or gains have probably played a significant role.
First, the homologous portions of the 24 group II clones with partially novel and partially homologous sequences to Rm1021 are distributed broadly across all three replicons of strain Rm1021 (see Table S2 in the supplemental material). Therefore, a single gain or loss cannot explain the observed differences between the two strains, and the simplest explanation for such a dispersed distribution of novel DNA fragments is that many of those novel DNA sequences in strain ATCC 9930 were gained independently or that the lack of such sequences in strain Rm1021 was the result of multiple independent losses.
Second, we found no shared features among the homologous portions of the 24 sequences mentioned above (data not shown). These sequences are highly divergent from each other. This result suggested that there was probably no common functional attribute to these novel DNA sequences. Both strains Rm1021 and ATCC 9930 were isolated from the same host plant species, Medicago sativa, and have the same MLEE genotype ET1. Therefore, it is possible that these two strains shared a recent common ancestry but had recently diverged from each other through independent gains or losses of DNA fragments. If so, the genomic differences between the two strains observed are remarkable.
The suggestion of frequent gains or losses of DNA fragments in natural strains of S. meliloti is consistent with an earlier phylogenetic analysis of the Rm1021 pSymB replicon sequence (33). Using a whole-replicon nearest-neighbor analysis, Wong and Golding identified that pSymB in strain Rm1021 had a complex evolutionary history, with closest sequence matches coming from diverse groups of organisms (33). However, their study was a theoretical investigation of one replicon in one strain and did not examine the issue of genome size variation among S. meliloti strains (33).
The suggestion of frequent gains and losses of DNA fragments in S. meliloti is also consistent with laboratory studies that showed that the deletions of large fragments of DNA in the S. meliloti genome often had very little effect on fitness under laboratory conditions (12, 24). A similar phenomenon could exist in nature for the ancestor of strain Rm1021. While it is highly possible that these novel DNA fragments do contribute to fitness differences in highly variable natural environments, these results are consistent with the hypothesis of a high plasticity of the S. meliloti genomes.
The distribution patterns of these novel DNA sequences among natural strains are also consistent with frequent gains or losses of DNA fragments in natural strains of S. meliloti. Specifically, we found no correlation between the relationship inferred from MLEE data and that inferred from novel DNA distribution data (Fig. 2). Based on genotypes inferred from novel DNA distributions, the 27 strains of MLEE type ET1 had genotype diversity similar to that of the other 32 strains with different MLEE types.
Alternatively, the dispersed and relatively random distribution of novel DNA sequences could be the result of multiple, unequal crossovers when ancestors of these two strains exchanged genetic materials. Evidence for genetic exchange among strains has been found in several nitrogen-fixing bacterial species (28), including Rhizobium leguminosarum biovar trifolii (35) and Rhizobium leguminosarum biovar phaseoli (25). Indeed, several studies of natural populations of S. meliloti (13, 26) identified evidence for recombination. Our analysis of novel DNA sequence distribution is consistent with these studies and suggests that natural populations of S. meliloti have a predominantly clonal population structure with low-level genetic recombination (Table 2). The lack of geographic clustering in novel DNA sequence distribution is also consistent with significant gene flow among geographic regions, as previously suggested by Eardly et al. (13) based on MLEE data.
The discovery of a large number of novel DNA sequences in a natural strain of S. meliloti and their highly varied distribution among natural strains pose many unanswered questions. For example, where did these and potentially other novel sequences come from? Were these sequences present in the first symbiotic nitrogen-fixing S. meliloti strain, but during subsequent evolution, did different progeny lose different parts of their genomes? As identified previously and mentioned above, large portions of the S. meliloti genome in strains Rm1021 (12) and Rm2011 (24) could be deleted without any obvious fitness effect under laboratory conditions (both on artificial media and in association with plants); therefore, the Rm1021 genome size is unlikely to be the smallest among natural strains. It is highly possible that there are unique sequences present only in strain Rm1021 and absent in strain ATCC 9930 or other natural strains. Large-scale genome sequencing of additional natural strains, such as ATCC 9930, could help address these and other issues. Such studies could significantly enhance our understanding of genome evolution in S. meliloti.
The existence of a large number of novel DNA sequences among natural strains also invites questions with regard to their functional significance. The proposed functions of the six clones in groups 3, 4, and 5 not found in strain Rm1021 but with homologs in the public databases were very diverse. Specifically, clone RDA-47 was homologous to the virB8 gene found in the type IV secretion system involved in transporting DNA and DNA-protein complexes (8). Clone RDA-81 was homologous to a gene in Pseudomonas sp. strain NK87 that degrades nylon oligomers, a type of xenobiotic compound (19, 34). Clone RDA-104 was homologous to a probable peptide synthetase gene in the symbiotic island of B. japonicum that may function in host specialization (16). Clone RDA-34 was homologous to a copper-binding tyrosinase involved in melanin synthesis in strain GR4 of S. meliloti (22). Melanin is an extracellular component capable of protecting cells from physical, chemical, and biological stresses and exists in many soil microorganisms. The last two of the six clones in this category were homologous to a hypothetic protein with an unknown function (32, 36). This protein exists on the same plasmid, pRmeGR4b, as the copper-binding tyrosinase (22). The existence of a group of functionally diverse genes in one strain but absent in another or others indicates potential niche specificity of natural strains. The recently completed genome sequencing of strain Rm1021 has generated a large number of functional genomics projects and exciting research findings. Inclusion of these and other novel DNA sequences in genome-wide expression studies could help reveal their functional properties.
Supplementary Material
Acknowledgments
We thank Bert Eardly for providing strains and Brian Golding for comments.
Financial support for this study is provided by Genome Canada, the Canadian Foundation for Innovation, the Ontario Innovation Trust, and the Ontario Research and Challenge Development Fund.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Agapow, P. M., and A. Burt. 2001. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1:101-102. [Google Scholar]
- 2.Allen, N. L., A. C. Hilton, R. Betts, and C. W. Penn. 2001. Use of representational difference analysis to identify Escherichia coli O157-specific DNA sequences. FEMS Microbiol. Lett. 197:195-201. [DOI] [PubMed] [Google Scholar]
- 3.Allen, N. L., C. W. Penn, and A. C. Hilton. 2003. Representational difference analysis: critical appraisal and method development for the identification of unique DNA sequences from prokaryotes. J. Microbiol. Methods 55:73-81. [DOI] [PubMed] [Google Scholar]
- 4.Bart, A., J. Dankert, and A. van der Ende. 2000. Representational difference analysis of Neisseria meningitidis identifies sequences that are specific for the hyper-virulent lineage III clone. FEMS Microbiol. Lett. 188:111-114. [DOI] [PubMed] [Google Scholar]
- 5.Bart, A., Y. Pannekoek, J. Dankert, and A. van der Ende. 2001. NmeSI restriction-modification system identified by representational difference analysis of a hypervirulent Neisseria meningitides strain. Infect. Immun. 69:1816-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergthorsson, U., and H. Ochman. 1995. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J. Bacteriol. 177:5784-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergthorsson, U., and H. Ochman. 1998. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 15:6-16. [DOI] [PubMed] [Google Scholar]
- 8.Bittinger, M. A., J. A. Gross, J. Widom, J. Clardy, and J. Handelsman. 2000. Rhizobium etli CE3 carries vir gene homologs on a self-transmissible plasmid. Mol. Plant-Microbe Interact. 13:1019-1021. [DOI] [PubMed] [Google Scholar]
- 9.Blanc-Potard, A.-B., C. Tinsley, I. Scaletsky, C. Le Bouguenec, J. Guignot, A. L. Servin, X. Nassif, and M.-F. Bernet-Camard. 2002. Representational difference analysis between Afa/Dr diffusely adhering Escherichia coli and nonpathogenic E. coli K-12. Infect. Immun. 70:5503-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonacorsi, S. P. P., O. Clermont, C. Tinsley, I. Le Gall, J.-C. Beaudoin, J. Elion, X. Nassif, and E. Bingen. 2000. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect. Immun. 68:2096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calia, K. E., M. K. Waldor, and S. B. Calderwood. 1998. Use of representational difference analysis to identify genomic differences between pathogenic strains of Vibrio cholerae. Infect. Immun. 66:849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles, T., and T. M. Finan. 1991. Analysis of a 1600-kilobase Rhizobium meliloti megaplasmid using defined deletions generated in vivo. Genetics 127:5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eardly, B. D., L. A. Materon, N. H. Smith, D. A. Johnson, M. D. Rumbaugh, and R. K. Selander. 1990. Genetic structure of natural populations of the nitrogen-fixing bacterium Rhizobium meliloti. Appl. Environ. Microbiol. 56:187-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faith, D. P. 1991. Cladistic permutation tests for monophyly and nonmonophyly. Syst. Zool. 40:366-375. [Google Scholar]
- 15.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 16.Göttfert, M., S. Röthlisberger, C. Kundig, C. Beck, R. Marty, and H. Hennecke. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harsono, K. D., C. W. Kaspar, and J. B. Luchansky. 1993. Comparison and genomic sizing of Escherichia coli O157:H7 isolates by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 59:3141-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.House, B. L., M. W. Mortimer, and M. L. Kahn. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70:2806-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanagawa, K., M. Oishi, S. Negoro, I. Urable, and H. Okada. 1993. Characterization of the 6-aminohexanoate-dimer hydrolase from Pseudomonas sp. NK87. J. Gen. Microbiol. 139:787-795. [DOI] [PubMed] [Google Scholar]
- 20.Lisitsyn, N., N. Lisitsyn, and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 21.Liu, S.-L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-CeuI, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercado-Blanco, J., F. García, M. Fernández-López, and J. Olivares. 1993. Melanin production by Rhizobium meliloti GR4 is linked to nonsymbiotic plasmid pRmeGR4b: cloning, sequencing, and expression of the tyrosinase gene mepA. J. Bacteriol. 175:5403-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Middendorf, B., and R. Gross. 1999. Representational difference analysis identifies a strain-specific LPS biosynthesis locus in Bordetella species. Mol. Gen. Genet. 262:189-198. [DOI] [PubMed] [Google Scholar]
- 24.Oresnik, I. J., S.-L. Liu, C. K. Yost, and M. F. Hynes. 2000. Megaplasmid pRme2011a of Sinorhizobium meliloti is not required for viability. J. Bacteriol. 182:3582-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinero, D., E. Martinez, and R. K. Selander. 1988. Genetic diversity and relationships among isolates of Rhizobium leguminosarum biovar phaseoli. Appl. Environ. Microbiol. 54:2825-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roumiantseva, M. L., E. E. Andronov, L. A. Sharypova, T. Dammann-Kalinowski, M. Keller, J. P. W. Young, and B. V. Simarov. 2002. Diversity of Sinorhizobium meliloti from the Central Asian alfalfa gene center. Appl. Environ. Microbiol. 68:4694-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sagerstrom, C. G., B. I. Sun, and H. L. Sive. 1997. Subtractive cloning: past, present, and future. Annu. Rev. Biochem. 66:751-783. [DOI] [PubMed] [Google Scholar]
- 28.Schofield, P. R., A. H. Gibson, W. F. Dudman, and J. M. Watson. 1987. Evidence for genetic exchange and recombination of Rhizobium symbiotic plasmids in a soil population. Appl. Environ. Microbiol. 53:2942-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stêpkowski, T., and A. B. Legocki. 2001. Reduction of bacterial genome size and expansion resulting from obligate intracellular life style and adaptation to soil habitat. Acta Biochim. Pol. 48:367-381. [PubMed] [Google Scholar]
- 30.Swofford, D. L. 2004. PAUP*. Phylogenetic Analysis Using Parsimony (* and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts.
- 31.Tinsley, C. R., and X. Nassif. 1996. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc. Natl. Acad. Sci. USA 93:11109-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toro, N., F. Martinez-Abarca, M. Fernandez-Lopez, and E. Munoz-Adelantado. 2003. Diversity of group II introns in the genome of Sinorhizobium meliloti strain 1021: splicing and mobility of RmInt1. Mol. Genet. Genomics 268:628-636. [DOI] [PubMed] [Google Scholar]
- 33.Wong, K., and G. B. Golding. 2003. A phylogenetic analysis of the pSymB replicon from Sinorhizobium meliloti genome reveals a complex evolutionary history. Can. J. Microbiol. 49:269-280. [DOI] [PubMed] [Google Scholar]
- 34.Yomo, T., I. Urable, and H. Okada. 1992. No stop codons in the antisense strands of the genes for nylon oligomer degradation. Proc. Natl. Acad. Sci. USA 89:3780-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young, J. P. W., and M. Wexler. 1988. Sym plasmid and chromosomal genotypes are correlated in field populations of Rhizobium leguminosarum. J. Gen. Microbiol. 134:2731-2739. [Google Scholar]
- 36.Zekri, S., M. J. Soto, and N. Toro. 1998. ISRm4-1 and ISRm9, two novel insertion sequences from Sinorhizobium meliloti. Gene 207:93-96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.