Abstract
Certain species of marine sponges in the order Dictyoceratida harbor large populations of the cyanobacterial symbiont Oscillatoria spongeliae in the mesohyl (interior) of the sponge. We show that in four of these sponge species (Lamellodysidea herbacea, Lamellodysidea chlorea, Lendenfeldia chondrodes, and Phyllospongia papyracea) from Palau there is a consistent community of α-proteobacteria in addition to O. spongeliae that fall within the Rhodobacter group based on 16S rRNA gene analysis. Some of the α-proteobacteria in Lendenfeldia chondrodes and P. papyracea but not in the Lamellodysidea spp. contained site-specific insertions in the 16S rRNA gene. Reverse transcription-PCR experiments demonstrated that the largest insertion found in this study (63 bp) is present in the mature rRNA. Lendenfeldia chondrodes was the only sponge found to have another cyanobacterium in the tissue, a Synechocystis sp. We found that the Synechocystis sp. was present in both the pinacoderm (surface epithelial tissue) and mesohyl, in contrast to O. spongeliae, which was only found in the mesohyl through the use of specific fluorescence in situ hybridization experiments. Of the four sponge species, only P. papyracea was found to contain a significant number of γ-proteobacteria. These results demonstrate that O. spongeliae-dominated bacterial communities in different sponge species can vary considerably and increase our understanding of the bacterial communities found in marine invertebrates.
Marine sponges (phylum Porifera) are evolutionarily ancient metazoans which have existed relatively unchanged in their general structural organization since the Late Cambrian period (29). Many of these invertebrates have been found to contain diverse bacterial communities which can be intra- or intercellular (24). Investigation of these bacterial communities through phylogenetic analysis and microscopy has shown that the types of bacteria found include cyanobacteria, proteobacteria, firmicutes, bacteroidetes, spirochaetes, actinobacteria, acidobacteria, chloroflexi, and “poribacteria” (18, 19, 25, 47, 51, 53). Although unrelated sponges can harbor similar bacterial groups (25), it is more typical for the bacterial communities to be specific for a given sponge species, irrespective of the collection site in a geographic region (47, 51).
In some cases, certain members of the bacterial community have been shown to benefit their hosts, and therefore are believed to be mutualists. They can serve as sources of nutrition by transferring products of metabolic processes not present in the host. For example, in some cases symbiotic cyanobacteria have been shown to transfer organic carbon obtained through photosynthesis to the host (6). Other benefits of these bacteria include contributing to structural rigidity of the sponge (54) and producing bioactive secondary metabolites which presumably act as chemical defense for the respective sponge (27). In most cases, however, the relationships between the bacteria and the sponges are not well defined. For the purposes of this paper, we define symbiosis as a consistent but not necessarily obligate association between the bacterium and the host.
Recently, the γ-proteobacterial symbiont of the bryozoan Bugula simplex, “Candidatus Endobugula glebosa”, was reported to have a 12-bp stem-loop extension at helix nine (helix numbering used by Brodersen et al. [10]) which is not present in the closely related symbiont of Bugula neritina, “Candidatus Endobugula sertula” (33). The α-proteobacterium Caedibacter caryophila, the parasitic symbiont of Paramecium caudatum, also contained an insertion in the rRNA gene. If it were present in the mature rRNA, there would be a 194-bp insertion in the same helix as “Candidatus E. glebosa.” However, this intervening sequence is removed during processing of the rRNA, which results in fragmentation of the 16S rRNA (45). These bacteria are unusual as none of the alpha- or gammaproteobacteria on the comparative RNA website have insertions at this helix (12). Prior to this study, no insertions in the 16S rRNA gene at helix nine had been identified in sponge symbionts.
We have recently communicated our research (39) on four species of dictyoceratid sponges which harbor large populations of the cyanobacterial symbiont Oscillatoria spongeliae (30 to 50% of the tissue volume). Samples of Lamellodysidea herbacea, Lamellodysidea chlorea, Lendenfeldia chondrodes, and Phyllospongia papyracea (Table 1) were collected from four distant sites in Palau (Fig. 1). Gene sequences were obtained for both host and symbiont from all samples. Each sponge species was found to contain its own strain of cyanobacteria (1% to 2.5% divergence in 16S rRNA gene) regardless of the collection site. Phylogenetic analysis revealed the possibility of at least one host-switching event in the evolution in this symbiosis between O. spongeliae and marine sponges. Of the cyanobacterial strains found in these four Palauan sponge species, only the strain found in Lamellodysidea herbacea contained putative halogenase genes involved in the biosynthesis of peptides containing chlorinated leucine derivatives (39). To date, no phylogenetic studies have been reported which investigate the other bacterial species that are present in sponges containing O. spongeliae.
TABLE 1.
Number and location of collected sponges
| Sponge species | Palau site | Strain designation (SIO IDa) |
|---|---|---|
| Lamellodysidea herbacea | 1 | LaH11 (P1192) |
| 2 | LaH21 (P1193) | |
| 4 | LaH41 (P1194) | |
| Lamellodysidea chlorea | 4 | LaC41 (P1195) |
| Lendenfeldia chondrodes | 1 | LeC11 (P1196) |
| 1 | LeC12 (P1197) | |
| Phyllospongia papyracea | 1 | PhP11 (P1198) |
| 3 | PhP31 (P1199) | |
| 3 | PhP32 (P1200) |
Scripps Institution of Oceanography Benthic Invertebrate identification no.
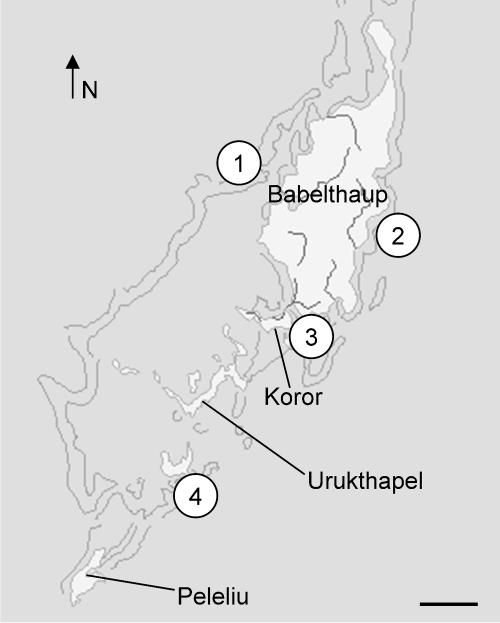
FIG. 1.
Map of the Republic of Palau (excluding Anguar and the Southwest Islands) with the sponge collection sites shown. Scale bar is approximately 7.5 km. The map was provided by ReefBase (http://www.reefbase.org). Reprinted from reference 39 with permission of Elsevier.
In this study, we report on other members of the bacterial communities discovered in these four Palauan species. We identified members of bacterial community that are present in these four species through microscopy and 16S rRNA gene clone library construction and analysis. Some α-proteobacteria present in the clone libraries had insertions present in the rRNA gene potentially forming a stem-loop extension at helix nine, and reverse transcription (RT)-PCR experiments were performed to evaluate whether the largest insertion observed was present in the mature rRNA. PCR surveys were also carried out to determine which sponges had bacteria that appeared to be unique to certain species based on the clone library results. In addition, we used fluorescence in situ hybridization (FISH) to identify the location of the two cyanobacterial species in the tissue of Lendenfeldia chondrodes. No attempts were made to culture these bacteria.
MATERIALS AND METHODS
Sponge collection.
Specimens of the dictyoceratid marine sponges Lamellodysidea herbacea (Keller, 1889), family Dysideidae; Lamellodysidea chlorea (de Laubenfels, 1954); Lendenfeldia chondrodes (de Laubenfels, 1954), family Thorectidae; and Phyllospongia papyracea (Esper, 1806), family Thorectidae, were collected from four reef sites using self-contained underwater breathing apparatus in the Republic of Palau (Fig. 1). Site 1 was at West Channel Buoy 5 (07°32.33′N, 134°28.30′E) at a depth of 15 to 20 ft; site 2 was an inner reef entrance to Ngatpaet (07°27.94′N, 134°37.65′E) at a depth 45 ft, site 3 was at seamount 2 in the KB channel (07°20.30′N, 134°31.07′E) at a depth of 45 ft, and site 4 was in the Ngerechong channel (07°06.90′N, 134°22.78′E) at a depth of 20 ft. Nine total sponge samples were gathered at these sites (Table 1). While collecting, care was taken to ensure that the entire sample was one piece so that no sample contained more than one species, and any encrusting organisms were removed.
One sample of Lendenfeldia chondrodes was collected in September 2001, while all other sponge samples were collected in September 2002. Voucher samples of these sponges have been deposited in the Scripps Institution of Oceanography Benthic Invertebrate Collection (Table 1). These sponges were previously identified by Patricia Bergquist (University of Aukland) and Mary Kay Harper (University of Utah) (39).
Transmission electron microscopy.
Sponge tissue from each sample was fixed in 2.5% gluteraldehyde in a 0.1 M phosphate buffer (0.3 M sucrose, pH 7.3) and stored at 4°C for 1 year. The samples were incubated twice in water for 15 min each to remove the phosphate buffer, and then stored overnight in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer, pH 7.4. The samples were then dehydrated using a series of ethanol solutions followed by propylene oxide and infiltration with epoxy resin (Scipoxy 812, Energy Beam Sciences). After polymerization at 65°C overnight, thin sections were cut and stained with 4% uranyl acetate in 50% ethanol, followed by bismuth subnitrate. Sections were examined at an accelerating voltage of 60 kV using a Zeiss EM10C electron microscope.
DNA isolation and 16S rRNA gene clone library construction.
DNA was isolated from sponge tissue from each sample stored in RNAlater (Ambion) at −20°C using the animal tissue protocol of a DNeasy kit (QIAGEN). In all cases the optional RNase treatment was carried out. Since PCR inhibitors were present, the isolated DNA was further purified using a Qiaquick PCR purification kit (QIAGEN). The DNA sequences of all PCR primers used in this work are shown in Table 2.
TABLE 2.
Primers and probes used in this study
| Name | Sequence (5′-3′)a | Reference |
|---|---|---|
| Eub27F | AGA GGT TGA TCM TGG CTC AG | 30 |
| Eub1492R | TAC GGC TAC CTT GTT ACG ACT T | 30 |
| Eub338F | ACT CCT ACG GGA GGC AGC | 5 |
| Eub536R | GTA TTA CCG CGG CTG CTG G | 35 |
| alpha303R | CCC AGT GTT GCT GAT CAT | This study |
| ins63bpR | CCA TCA AAT GCC ACC AAA C | This study |
| Eub338 | Bio/GCT GCC TCC CGT AGG AGT | 5 |
| Osc611 | Bio/CCA CAG TTA AGC TGT GGT | This study |
| nonOsc611 | Bio/CCA CAG TTT AGC TGT GGT | This study |
| Syn611 | Bio/CAG TGG TTG AGC CAC TGG | This study |
| nonSyn611 | Bio/CAG TGG TTC AGC CAC TGG | This study |
| H595 | CTT TGA CAG CAG ACT T | This study |
| H629 | CAG TTT CCA CTG CCT TT | This study |
Bio, biotin.
Using the eubacterial primers 27F and 1492R along with the high-fidelity DNA polymerase pfu turbo (Stratagene), 50-μl PCRs were carried out on the isolated genomic DNA from each specimen of Lamellodysidea herbacea, Lamellodysidea chlorea, Lendenfeldia chondrodes, and P. papyracea. The reaction conditions consisted of a 1-min denaturing step at 95°C followed by 25 cycles of 95°C for 1 min, 60°C for 0.5 min, and 72°C for 2 min. In an attempt to minimize heteroduplex formation, reconditioning PCR (50) was then performed on each sample by running another 50-μl PCR using pfu turbo as the polymerase and 5 μl of obtained PCR product as the DNA template. The PCR conditions were three cycles of 95°C for 1 min, 60°C for 0.5 min, and 72°C for 2 min. These reconditioned PCR products were then purified using a Qiaquick PCR cleanup kit (QIAGEN), and adenylated through incubating the PCR product at 72°C for 10 min with Taq DNA polymerase and dATP. This adenylated product was cloned directly into a pCR4-TOPO plasmid vector (Invitrogen) and transformed into chemically competent Escherichia coli TOP10 (Invitrogen) following the instruction manual.
The 16S rRNA gene inserts in the resulting colonies were amplified directly from E. coli cells using the T3 and T7 primers provided in the TOPO TA cloning kit (Invitrogen). The PCR conditions were as follows: 25-μl reactions using Taq polymerase, 1-min 95°C denaturing step followed by 35 cycles of 95°C for 1 min, 60°C for 0.5 min, and 72°C for 2 min. The clone libraries were screened by incubating the PCR products with RsaI (Invitrogen), MspI (New England Biologicals), and EcoRI (New England Biolab) for a period of 3 h. Colonies that contained 16S rRNA gene inserts that displayed a unique restriction fragment digestion pattern on a 1.2% agarose gel were identified, and two or more colonies from each pattern were selected for sequencing. Plasmids were then isolated using a QIAprep miniprep kit (QIAGEN), and sequenced using the T3 and T7 plasmid primers (QIAGEN) as well as internal 338F and 536R primers.
Reverse transcription-PCR and PCR survey of α-proteobacteria.
RNA was extracted from both samples of Lendenfeldia chondrodes from tissue stored in RNAlater (Ambion) at −20°C using the RNeasy kit (QIAGEN) following the manufacturer's instructions and incorporating the optional DNase I step. The reverse primer alpha303R was designed to bind to an accessible region considering the secondary structure of the α-proteobacterial 16S rRNA (8). The specificity of alpha303R was evaluated using Probe Match on the RDP website (14); 686 hits were reported, of which 685 were in the phylum Proteobacteria and the remaining hit was an unclassified bacterium. In the Proteobacteria, 681 hits were α-proteobacteria, four were γ-proteobacteria, and one was unclassified; 674 of the hits in the α-proteobacteria were in the Rhodobacterales, suggesting that this is a fairly specific probe for members of this order. Additionally, analysis of clone library sequences verified that it would not bind to the other bacteria discovered in the libraries. Another reverse probe, ins63bpR, was designed to bind to the largest insertion sequence found in Lendenfeldia chondrodes.
The RT-PCR conditions were as follows: 20-μl reactions using Superscript II reverse transcriptase (Invitrogen) and alpha303R, and a one-step amplification of 25°C for 10 min, 42°C for 50 min, and inactivation at 70°C for 15 min. Control reactions were performed in parallel where no reverse transcriptase was added but otherwise were treated in the same way. Three μl from each reaction was then used for 25-μl PCRs using the 27F and ins63bpR primers under the following conditions: 25-μl reactions using Taq polymerase, 1-min 96°C denaturing step followed by 35 cycles of 96°C for 1 min, 50°C for 0.5 min, and 72°C for 1.5 min. The cDNA PCR product was purified using a Qiaquick PCR purification kit (QIAGEN) and sequenced using the ins63bpR reverse primer. PCR surveys were conducted on genomic DNA isolated from all the sponges in the study using these PCR conditions with the primer combinations 27F-alpha303R and 27F-ins63bpR.
Sequencing.
All sequencing was performed on an ABI 3100 Analyzer using the ABI Prism BigDye terminator kit. All DNA sequences obtained were assembled and checked by eye using Sequencher 4.0.5 (Gene Codes Corp.). The 16S rRNA gene sequences obtained from the clone libraries were compared with those ofother bacteria using the basic local alignment search tool (BLAST) (4) and theRibosomal Database Project (RDP-II) (14). Four sequences that appeared chimeric by alignment with other obtained sequences and analysis using Check_Chimera (14) were discarded.
Phylogenetic analysis.
Sequences that were closely related to the clone sequences based on the database searches were aligned using Clustal X 1.83 (49), and checked by eye. Bayesian phylograms were generated using Mr. Bayes 3.0 (40). Default priors were used for the Bayesian analysis, and the general time reversible model was used with a gamma distribution of rate variation across sites and a proportion of invariable sites. Four Markov chain Monte Carlo chains were run for three million generations sampling every 100, and the first 2,000 trees were discarded prior to the analysis. Phylogenetic trees were also constructed in PAUP 4.0b10 (46) using the maximum parsimony and neighbor-joining algorithms. For maximum parsimony analysis, transversions were weighted two times transitions (maximum likelihood estimation of ratio was rounded up), and a heuristic search of 10 repetitions with random addition of sequence was completed. For the neighbor joining analysis, a general time reversible nucleotide substitution model with gamma distribution of rate variation across sites was used. Bootstrap analysis for both maximum parsimony and neighbor joining was performed with 1,000 replicates. Figures of the trees were prepared using TreeView (36).
Fluorescence in situ hybridization.
Tissue from Lendenfeldia chondrodes was fixed in 4% buffered paraformaldehyde (0.5 M NaCl, 0.02 M phosphate buffer, pH 7.5) for 4 h at 4°C. The tissue was removed from the paraformaldehyde and placed in 70% ethanol for 18 to 24 h at 4°C, at which point the sample was stored in fresh 70% ethanol at −20°C for 5 months. The tissue was embedded in paraffin, cut into 10 μm sections, mounted on slides, and stored at −80°C.
Osc611 and Syn611 biotinylated oligonucleotide probes were designed to specifically target O. spongeliae and the Synechocystis sp. based on comparison of the respective 16S rRNA gene sequences and evaluation of probe specificity using Probe Match in the RDP (14). All other 16S rRNA gene sequences in the clone libraries were checked to make sure that the probes would not bind to them. The one bp mismatched probes nonOsc611 and nonSyn611 were designed as negative controls, and Eub338 was used as a positive control. Additionally, the need to use unlabeled helper oligonucleotides (21) H595 and H629 was evaluated for successful experiments, as this 16S rRNA target was not readily accessible based on other FISH experiments (8). All probes were purchased from IDT, Inc., and the DNA sequences are given in Table 2.
The sections of tissue were found to contain endogenous biotin and peroxidases, so the use of tyramide signal amplification and biotinylated probes required a prehybridization protocol to remove these causes of false positives. After treating the sectioned slides with histology grade xylenes to remove the paraffin, a 20-min incubation with an immunopure peroxidase suppressor (Pierce) was followed by the steps of an endogenous biotin-blocking kit (Molecular Probes). Afterwards, the slides were incubated for 40 min in 10 mg/ml lysozyme solution in 0.1 M Tris-HCl, 0.01 M EDTA. The hybridization was carried out at 46°C for 4 to 18 h in buffered 35% formamide for Eub338 or buffered 20% formamide (0.9 M NaCl, 20 mM HCl, 0.01% SDS) for all other probes using a probe concentration of 5.6 ng/μl. Hybridizations were attempted for Osc611 and Syn611 with and without H595 and H629 at a 5.6 ng/μl concentration, and it was determined that the helper probes were required for successful detection. Negative controls included one-base mismatch probes with H595 and H629. To remove excess probe(s), the slides were then washed with either buffered 0.07 M NaCl for Eub338 or buffered 0.22 M NaCl for all other probes (buffer: 20 mM Tris-HCl pH 7.0, 0.01% sodium dodecyl sulfate) at 48°C for 15 min.
The posthybridization detection steps were performed using the tyramide signal amplification kit T-20937 (Molecular Probes) following the manufacturer's instructions with two exceptions. The tyramide-conjugated fluorophore-amplification buffer was 1:10 (37), and the working solution of streptavidin-horseradish peroxidase solution was prepared by diluting the stock solution 2:100 in blocking reagent instead of 1:100. The tyramide-conjugated fluorophores used in this study were Cy5 (Perkin-Elmer) and Alexa 350 (Molecular Probes). Additional steps were incorporated for the dual-labeled FISH experiments, taking care to keep slides in the dark once the first probe detection was complete. The peroxidase and endogenous biotin suppressor steps were repeated to prevent any cross talk between the probes prior to repeating the hybridization and detection steps for the second probe. To ensure that cross talk did not occur, a negative control was performed using Syn611 for the first hybridization and nonOsc611 for the second hybridization. Afterwards, the slides were washed with distilled H2O, dried, and mounted with Vectashield (Vector Laboratories).
The slides were examined using a Zeiss Axioplan microscope under epifluorescence using either a Cy5 filter (Chroma Technology Corp. set 41008) or a 4′,6′-diamidino-2-phenylindole (DAPI) (Chroma Technology Corp., set 31000) filter. Black-and-white images were taken using a Kodak KA1-2000 camera, and false-color images were generated and stacked using Metamorph 5.0 r4. The same exposure conditions were used for positive and negative controls.
PCR survey of Synechocystis sp.
Using the PCR primers Syn611R (nonbiotinylated Syn611, Table 2) and 27F, PCR surveys were conducted on genomic DNA isolated from all the sponges in the study. The PCR conditions were as follows: 25-μl reactions using Taq polymerase, 1-min 96°C denaturing step followed by 35 cycles of 96°C for 1 min, 50°C for 0.5 min, and 72°C for 1.5 min.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the rRNA gene sequences obtained from the clone libraries are AY615501 to AY615509 for O. spongeliae and AY845227 to AY845245 for all other sequences.
RESULTS AND DISCUSSION
Transmission electron microscopy.
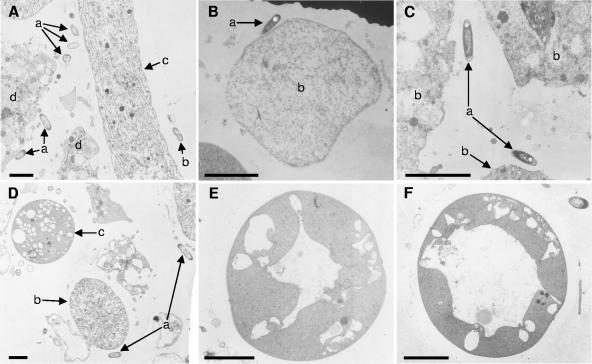
One sample of each of the four species was examined by electron microscopy. The bacterial population in the mesohyl (sponge interior between epithelial tissue) in each species was dominated by filamentous cyanobacteria which matched Oscillatoria spongeliae in appearance and description (9, 31). In all four species, unicellular rod-shaped bacteria were also present, and were occasionally undergoing cell division (Fig. 2A to D). These bacteria are similar in appearance to those found in samples of Lamellodysidea herbacea (=Dysidea herbacea) collected on the Great Barrier Reef in Australia (20), and therefore these unicellular bacteria could be significant members of the bacterial communities in these sponges. Additionally, Lendenfeldia chondrodes was found to contain large spherical cyanobacteria (Fig. 2D) that were not observed in the other three species. These unicellular cyanobacteria resembled the ultrastructure of Synechocystis spp., which has been found in several didemnid species of ascidians (phylum Chordata) as well as three unrelated sponge species (Spirastrella aff. decumbens, Prianus aff. melanus, and an unidentified “brown fleshy sponge”) (17, 26).
FIG. 2.
Transmission electron micrographs of bacteria found in the mesohyl of these sponges. (A) Many unicellular bacteria (a), occasionally dividing (b), are observed in close proximity to O. spongeliae (c) and sponge cells (d) in Lendenfeldia chondrodes (magnification, 5,500×); (B) representative of rod-shaped bacteria (a) close to a sponge cell (b) in Lamellodysidea herbacea (magnification, 5,500×); (C) unicellular bacteria (a) in vicinity of sponge cells (b) in Lamellodysidea chlorea (magnification, 17,325×); (D) unicellular bacteria (a) with O. spongeliae filament (b) and Synechocystis sp. (c) found in Lendenfeldia chondrodes (magnification, 4,400×); (E) Synechocystis cell with an intermediate-size distended thylakoid (magnification, 17,325×); (F) Synechocystis cell with a large distended thylakoid (magnification, 17,325×). All scale bars, 3 μm.
These cells contained various numbers and sizes of vacuole-like structures (Fig. 2D-F), which are in fact distended thylakoids. In some cases, a distended thylakoid occupied most of the cellular volume in the cyanobacterium (Fig. 2F). The presence of these “vacuoles” is a distinctive characteristic of Synechocystis (17) and closely related Prochloron spp. (16) which are symbionts of didemnid ascidians (32).
Community composition.
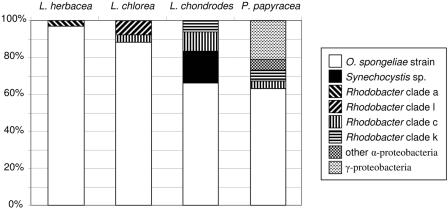
From the three samples of Lamellodysidea herbacea, 17 to 27 clones were screened, for a total of 66. Only two restriction patterns were observed by gel electrophoresis. From Lamellodysidea chlorea, 25 clones were screened and three restriction patterns were observed; 19 and 34 clones, respectively, were screened from the two samples of Lendenfeldia chondrodes for a total of 53. Five restriction patterns were observed for this species. From the three collections of P. papyracea, 31 to 40 clones were screened from each for a total of 107, and six restriction digest patterns were observed. For those species where more than one sample was collected, the relative proportion of the restriction pattern types varied slightly but the types and resulting 16S rRNA gene sequences were consistent between samples. The bacterial communities identified in these sponge species are given in Fig. 3. The dominant bacterial species in all of these sponges was identified as O. spongeliae (39) based on sequence identity to the previously published O. spongeliae sequence (48).
FIG. 3.
Summary of clone library results. Each pattern represents different bacteria that were detected in these sponges. The identification of the 16S rRNA gene sequences based on BLAST and RDP-2 database searches is given in the legend. For the Rhodobacter group sequences, the clades where the taxa are located in Fig. 4B are indicated.
We found that 17% of the clone library of Lendenfeldia chondrodes was comprised of another cyanobacterial sequence which was not closely related to O. spongeliae cyanobacterium present in the same sponge (Fig. 4A). Consistent with the TEM results, the rRNA gene sequence had 97% 16S rRNA sequence identity to Synechocystis trididemni, a symbiont of the ascidian Trididemnum solidum (43). Four clones of the Synechocystis sp. restriction pattern were sequenced, two which were identical to each other (29P18). The other two clones (29P35 and 36P1) had 2 of 1,407 bp divergence relative to 29P18, possibly indicating that closely related strains inhabit these sponges. Alternatively, this could reflect divergence of rRNA genes between operons in the genome of this cyanobacterium (1). Both specimens of Lendenfeldia chondrodes collected a year apart contained the symbiont, indicating that this is likely a stable association.
FIG. 4.
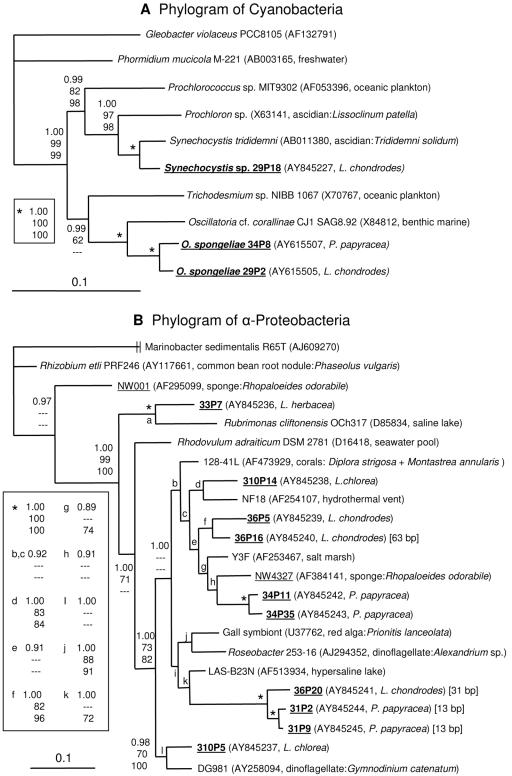
Phylogenetic trees generated with Mr. Bayes based on 16S rRNA gene sequences of cyanobacteria (A) and α-proteobacteria (B). Gleobacter violaceus is the outgroup in tree A, while the γ-proteobacterium Marinobacter sedimentalis is the outgroup in tree B. Sequences from sponges are underlined, and those from the Palauan sponges are in bold type. Next to the clone or culture designation in parentheses is given the GenBank accession number and the source of the sequence or culture. In the case of symbiotic bacteria, the host species is indicated. For the α-proteobacteria which contain insertions in the 16S rRNA gene, the size is indicated in brackets. Bootstrap values are shown only for branches which have a value over 60%, while Bayesian posterior probability is given for all branches (top, Bayesian posterior probability; middle, maximum parsimony bootstrap; bottom, neighbor joining bootstrap). A dash indicates a bootstrap value of less than 60% or clade not observed with that method. The scale bars indicate the number of substitutions per nucleotide position.
All four species of sponges also contained α-proteobacteria which best fit in the Rhodobacter group compared to sequences in the BLAST and RDP-2 databases. A phylogram of these clone sequences and other selected α-proteobacteria is shown in Fig. 4B. This group of bacteria is commonly found in unrelated marine sponges. In addition to NW4327 (52) shown in Fig. 4B, clones isolated from Halichondria panicea (3), Homaxinella balfourensis, Kirkpatrickia varialosa, and Latrunculia apicalis (51) also fell within this group of bacteria when included in the phylogenetic analysis. Specifically, two clones from sponge samples of Halichondria panicea collected from the Adriatic, Baltic, and North seas fell within clade e (GenBank accession numbers Z88581 and Z88582) in Fig. 4B. Twelve clones from the Antarctic sponges Homaxinella balfourensis (AY321379 to AY321381, AY321428 to AY321430, AY321432, AY321384, and AY321385), Kirkpatrickia varialosa (AY321387), and Latrunculia apicalis (AY321396 and AY321420) were interspersed in clade c. In order to keep the tree at a manageable size and in consideration of the short sequences (<1,000 bp) available for these clones, they are not presented in Fig. 4B. Also closely related are bacteria associated with other marine invertebrates such as red algae, dinoflagellates, and coral (Fig. 4B). Interestingly, environmental bacteria from salt marshes, hydrothermal vents, and saline lakes are also present.
Exactly what environmental role(s) these sponge symbionts have is not presently known. Several of the bacteria in this clade appear to be pathogenic to their hosts. NW4327, related to 34P11 and 24P35 from P. papyracea, degrades spongin fibers of Rhopaloeides odorabile, which causes necrosis of the sponge tissue (52). Clone 128-41L (clade b, Fig. 4B) is found on the corals Diploria strigosa and Montastrea annularis that have black band disease but not on their healthy counterparts (15). In addition, the symbiont of the red alga Prionitis lanceolata (clade j, Fig. 4B) is associated with the formation of galls on the host (7). However, the sponges in this study showed no sign of infection and no tissue necrosis was observed when the tissue was examined by microscopy. Although this does not mean that the α-proteobacteria found in the Palauan sponges cannot act as pathogens under certain circumstances, it does suggest they have different roles. Other metabolic possibilities for these sponge symbionts that have been attributed to Rhodobacter group bacteria include degradation of aromatic compounds (11), aerobic and anaerobic anoxygenic photosynthesis (2, 23), nitrogen fixation (13), nitrate respiration, and denitrification (42).
The clone libraries of P. papyracea contained the most diverse 16S rRNA gene sequences. Three α-proteobacterial sequences (31P4, 31P10, and 31P16) were found that did not fall in the Rhodobacter clade. They were not closely related to other α-proteobacteria, with the closest sequences ranging from 88 to 90% sequence identities with one exception. Sequence 31P4 was only found in one sponge and had 96% sequence identity to a clone recovered from seawater in Monterey Bay (AY627379), and therefore may be a seawater contaminant and not truly associated with the sponge. P. papyracea was also the only species in this study where γ-proteobacteria were found in the clone libraries. Database searches revealed that the γ-proteobacteria are not closely related to other reported bacteria, with the closest 16S sequences ranging from 88 to 91% identity based on the BLAST results. Clones 31P6, 34P16, and 34P38 comprised 29%, 12%, and 59%, respectively, of the γ-proteobacteria in the clone library based on their restriction digest patterns.
Coverage of the clone libraries was estimated with using the formula C = 1 - (N/n), where C is coverage, N is the number of unique restriction patterns, and n is the total number of clones screened (22, 44), which suggested that relatively high coverage of the bacterial communities was achieved. For Lamellodysidea herbacea, coverage estimates was 91, 92, and 94% for the individual sponges and if combined it was 97%. For Lamellodysidea chlorea, the estimated coverage was 88%. For Lendenfeldia chondrodes, coverage was estimated at 84 and 91% individually and if combined it was 94%. Similarly, the estimates for P. papyracea were 81, 83, and 85% for the individual samples and 94% for all three samples combined.
These calculations may not accurately reflect the coverage of the bacterial diversity in these sponges due to several factors. The observed dominance of O. spongeliae in the clone libraries (63 to 97% of the clones) and in the sponge tissue may obscure the presence of bacteria present in small concentrations. The use of restriction digest patterns as the operational taxonomic unit might provide an additional source of error, as some microvariation in the clone library might be missed (34). Also, the 27F and 1492R eubacterial primers have demonstrated primer bias, where sequences with higher GC content in the primer site amplify preferentially (38). Therefore, the results in this study should not be considered a quantitative analysis of the bacterial communities in these sponges, but rather as a qualitative survey of bacteria other than O. spongeliae that are present.
16S rRNA gene insertions.
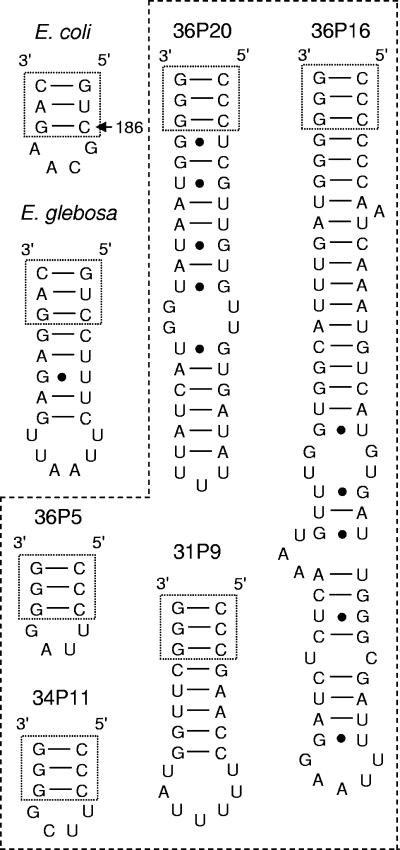
While examining the secondary structure, it became apparent that some of the α-proteobacterial clones from the Lendenfeldia chondrodes and P. papyracea 16S rRNA gene libraries had sizeable sequence insertions that would form extensions at helix nine (helix numbering used by Brodersen et al. [10]) if present in the mature rRNA. Insertions of 13 bp (31P2 and 31P9), 31 bp (36P20), and 63 bp (36P16) were observed, but all other related sequences had no insertion. The insertions all had a predicted stem extension and a terminal loop when analyzed using the Vienna RNA secondary-structure server (28) as shown in Fig. 5. Since the 13-bp and 63-bp insertions were found in clones from multiple sponge samples, they are not PCR artifacts. The RT-PCR experiments indicated that the 63-bp insertion is present in the mature rRNA like the 12-bp insertion in “Candidatus E. glebosa,” and not excised like the 194-bp insertion in Caedibacter caryophila.
FIG. 5.
16S rRNA structure of E. coli and the predicted stem-loop structure of “Candidatus Endobugula glebosa” and representative α-proteobacterial clones. The stem-loops found in this study are indicated with a dashed line. Clones 31P9 and 34P11 are from P. papyracea, while 36P5, 36P16, and 36P20 are from Lendenfeldia chondrodes.
PCR surveys.
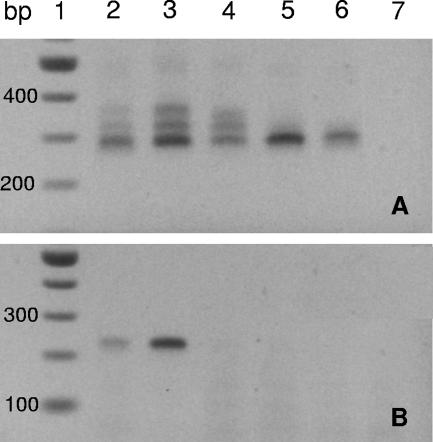
All the sponge samples in this study were investigated for the presence of insertions at helix nine by PCR amplification with the primers 27F-alpha303R (Fig. 6A). Only Lendenfeldia chondrodes and P. papyracea appear to have α-proteobacteria which contain insertions in this range based on the pattern of the PCR products. The pattern from all three P. papyracea samples from site one and three were identical (not shown) suggesting a consistent association of the rRNA insertion-containing bacteria. Although there appeared to be some variability in the intensity of the banding pattern in the two samples of Lendenfeldia chondrodes, the pattern was the same indicating that the same may be true of this species (Fig. 6A). Since both Lamellodysidea herbacea and Lamellodysidea chlorea do not appear to contain bacteria with insertions in this range (within the detection limits of PCR and the size resolution of the agarose gels), these bacteria may be specifically associated with Lendenfeldia chondrodes and P. papyracea.
FIG. 6.
PCR products from genomic DNA samples amplified using the primer combination 27F and alpha303R (A) or 27F and ins63bpR (B) on 2% agarose gels. Lane 1 contains a 100-bp DNA ladder (New England Biologicals), while the remaining lanes contain the PCR products from the following sponges: lanes 2 and 3, Lendenfeldia chondrodes; lane 4, P. papyracea; lane 5, Lamellodysidea herbacea; lane 6, Lamellodysidea chlorea; lane 7, no-DNA negative controls. All PCR products shown were amplified from samples collected at site 1 except for Lamellodysidea chlorea, which was from site 4. All samples collected in this study were surveyed, and the patterns of Lamellodysidea herbacea and P. papyracea from the other sites were identical to the representative sample for the species (not shown).
As many 16S rRNA gene studies of bacterial communities in sponges to date have used primer combinations that would miss insertions at this helix, it is not currently known how widespread these insertion-containing bacteria are in the Porifera. Although both Lendenfeldia chondrodes and P. papyracea contain bacteria with insertions, the communities appear different in these species, as PCR amplifications with a primer pair specific to the 63 bp insertion (27F-insH9R) only yielded product from both samples of Lendenfeldia chondrodes and not from P. papyracea (Fig. 6B). The PCR surveys with the 27F-Syn611R only yielded product from both Lendenfeldia chondrodes specimens (data not shown), which provides further evidence that the Synechocystis sp. is only present in this species.
Fluorescence in situ hybridization.
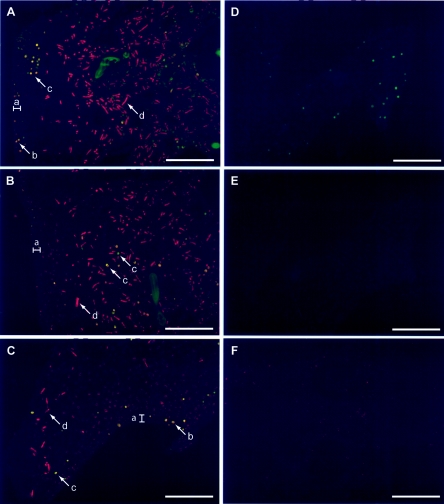
Cyanobacteria are difficult targets for FISH, as they possess photosynthetic pigments which are autofluorescent. Synechocystis and O. spongeliae both possess chlorophyll a and phycoerythrin (17, 31), and therefore required signal amplification steps to detect successful binding of probes. We chose to use tyramide signal amplification or catalyzed reporter deposition FISH, which had been shown to differentiate between an artificial mixture of cyanobacteria (41). Here we show that this FISH technique can be adapted to study cyanobacteria in sponge tissue sections (Fig. 7). Interestingly, there was considerable variability in the population density of the cyanobacteria throughout the tissue section. In general, O. spongeliae dominated the mesohyl (Fig. 7A and B) with the Synechocystis sp. present in the minority, but in some areas there were fewer Oscillatoria filaments (Fig. 7C), and in other areas there were no cyanobacteria present (not shown). In one area, only the Synechocystis sp. was present (Fig. 7D). The Synechocystis cells were also found in the pinacoderm (epithelial surface tissue), where the Oscillatoria filaments were not observed (Fig. 7A and 7C).
FIG. 7.
Tyramide signal amplification FISH epifluorescence micrographs of tissue sections of Lendenfeldia chondrodes. Sections A to C are areas in one tissue section (top to bottom) subjected to labeling with Synechocystis sp. (Syn611, Alexa 350-tyramide detection, green) followed by O. spongeliae (Osc611, Cy5-tyramide detection, red)-specific probes. In A to C, Synechocystis cells appear green or yellow due to various amounts of autofluorescence of these cells in the Cy5 channel. The epithelial tissue range is indicated (a), and representative Synechocystis cells present in this surface tissue are given (b). Representative Synechocystis (c) and O. spongeliae (d) in the mesohyl are also shown. (D) Area of the same tissue section as in A to C which did not contain any O. spongeliae filaments shown in the Alexa 350 channel (green). (E) nonSyn611 probe (Alexa350-tyramide detection, green) negative control. (F) Dual-labeled negative control Syn611 (Alexa 350-tyramide detection) followed by nonOsc611 (tyramide-Cy5 detection, red) shown in the Cy5 channel. All scale bars, 100 μm.
In summary, we investigated the bacterial communities of the Palauan sponges Lamellodysidea herbacea, Lamellodysidea chlorea, Lendenfeldia chondrodes, and P. papyracea known to be dominated by O. spongeliae with the focus on the other bacterial constituents. Only two species (Lendenfeldia chondrodes and P. papyracea) were found to contain unusual α-proteobacteria with insertions present in a stem-loop of their rRNA. Lendenfeldia chondrodes was the only species that contained another cyanobacterium, a Synechocystis sp., while P. papyracea was the only sponge found to contain γ-proteobacteria in the clone libraries. This indicates that despite the common presence of O. spongeliae strains, the bacterial communities of these sponges are quite distinct. This work on these unique sponges enhances our knowledge about sponge/bacterial communities and further demonstrates the microbial diversity that can exist in these sponges.
Acknowledgments
We thank the Coral Reef Research Foundation (CRRF) for their invaluable assistance in collecting these sponges, the Republic of Palau for the research permits, and Bill Fenical for his support of this project. We also thank R. Bencheikh for assistance with RNA isolation and RT-PCR, G. Lim for help with phylogenetic analysis, and K. Sharp for assistance with FISH. The paraffin tissue sections of Lendenfeldia chondrodes were prepared by the Cancer Department Histology Shared Resource at UCSD, and the transmission electron microscopy was completed with Pat Reid at the VA San Diego Hospital (La Jolla).
The DNA sequencing was performed by the DNA Sequencing Shared Resource, UCSD Cancer Center, which is funded in part by NCI Cancer Center Support Grant 2 P30 CA23100-18. This research was supported by a grant from the National Science Foundation (CHE 98-16169).
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allgaier, M., H. Uphoff, A. Felske, and I. Wagner-Döbler. 2003. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl. Environ. Microbiol. 69:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Althoff, K., C. Schütt, R. Steffen, R. Batel, and W. E. G. Müller. 1998. Evidence for a symbiosis between bacteria of the genus Rhodobacter and the marine sponge Halichondria panicea: harbor also for putatively toxic bacteria? Mar. Biol. 130:529-536. [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann, R., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arillo, A., G. Bavestrello, B. Burlando, and M. Sarà. 1993. Metabolic integration between symbiotic cyanobacteria and sponges: a possible mechanism. Mar. Biol. 117:159-162. [Google Scholar]
- 7.Ashen, J. B., and L. J. Goff. 1996. Molecular identification of a bacterium associated with gall formation in the marine red alga Prionitis lanceolata. J.Phycol. 32:286-297. [Google Scholar]
- 8.Behrens, S., C. Rühland, J. Inácio, H. Huber, Á. Fonseca, I. Spencer- Martins, B. M. Fuchs, and R. Amann. 2003. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borowitzka, M. A., R. Hinde, and F. Pironet. 1988. Carbon fixation by the sponge Dysidea herbacea and its endosymbiont Oscillatoria spongeliae, p. 151-156. In J. H. Choat, D. J. Barnes, M. A. Borowitzka, J. C. Coll, P. J. Davies, P. Flood, B. G. Hatcher, D. Hopley, P. A. Hutchings, D. Kinsey, G. R. Orme, M. Pichon, P. F. Sale, P. W. Sammarco, C. C. Wallace, C. R. Wilkinson, E. Wolanski, and O. Bellwood (ed.), Proceedings of the Sixth International Coral Reef Symposium, Australia, vol. 3. Sixth International Coral Reef Symposium Executive Committee, Townsville, Australia. [Google Scholar]
- 10.Brodersen, D. E., W. M. Clemons, A. P. Carter, B. T. Wimberly, and V. Ramakrishnan. 2002. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 316:725-768. [DOI] [PubMed] [Google Scholar]
- 11.Buchan, A., L. S. Collier, E. L. Neidle, and M. A. Moran. 2000. Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacter lineage. Appl. Environ. Microbiol. 66:4662-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structural information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantera, J. J. L., H. Hawasaki, and T. Seki. 2004. The nitrogen-fixing gene (nifH) of Rhodopseudomonas palustris: a case of lateral gene transfer? Microbiology 150:2237-2246. [DOI] [PubMed] [Google Scholar]
- 14.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kalum, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooney, R. P., O. Pantos, M. D. A. Le Tissier, M. R. Barer, A. G. O'Donnell, and J. C. Bythell. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4:401-413. [DOI] [PubMed] [Google Scholar]
- 16.Cox, G. C. 1986. Comparison of Prochloron from different hosts. I. Structural and ultrastructural characteristics. New Phytol. 104:429-445. [Google Scholar]
- 17.Cox, G. C., R. G. Hiller, and A. W. D. Larkum. 1985. An unusual cyanophyte, containing phycourobilin and symbiotic with ascidians and sponges. Mar. Biol. 89:149-163. [Google Scholar]
- 18.Erpenbeck, D., J. A. J. Breeuwer, H. C. van der Velde, and R. W. M. van Soest. 2002. Unravelling host and symbiont phylogenies of halichondrid sponges (Demospongiae, Porifera) using a mitochondrial marker. Mar. Biol. 141:377-386. [Google Scholar]
- 19.Fieseler, L., M. Horn, M. Wagner, and U. Hentschel. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flowers, A. E., M. J. Garson, R. I. Webb, E. J. Dumdei, and R. D. Charan. 1998. Cellular origin of chlorinated diketopiperazines in the dictyoceratid sponge Dysidea herbacea (Keller). Cell Tissue Res. 292:597-607. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs, B. M., F. O. Glöckner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labelled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-240. [Google Scholar]
- 23.Gregor, J., and G. Klug. 1999. Regulation of bacterial photosynthesis by oxygen and light. FEMS Microbiol. Lett. 179:1-9. [DOI] [PubMed] [Google Scholar]
- 24.Hentschel, U., L. Fieseler, M. Wehrl, C. Gernert, M. Steinert, J. Hacker, and M. Horn. 2003. Microbial diversity of marine sponges, p. 60-88. In W. E. G. Müller (ed.), Molecular marine biology of sponges. Springer-Verlag, Heidelburg, Germany. [DOI] [PubMed]
- 25.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68: 4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernádez-Mariné, M., X. Turon, and J. Catalan. 1990. A marine Synechocystis (Chroococcales, Cyanophyta) epizoic on didemnid ascidians from the Mediterranean Sea. Phycologia 29:275-284. [Google Scholar]
- 27.Hildebrand, M., L. E. Waggoner, G. E. Lim, K. H. Sharp, C. P. Ridley, and M. G. Haygood. 2004. Approaches to identify, clone, and express symbiont bioactive metabolite genes. Nat. Prod. Rep. 21:122-142. [DOI] [PubMed] [Google Scholar]
- 28.Hofacker, I. L. 2003. Vienna RNA secondary structure server. Nucleic Acids Res. 31:3429-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooper, J. N. A., and R. W. M. van Soest. 2002. Introduction, p. 1-3. In J. N. A. Hooper and R. W. M. van Soest (ed.), Systema Porifera: a guide to the classification of sponges, vol. 1. Kluwer Academic/Plenum Publishers, New York, N.Y. [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellows (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 31.Larkum, A. W. D., G. C. Cox, R. G. Hiller, D. L. Parry, and T. P. Dibbayawan. 1987. Filamentous cyanophytes containing phycourobilin and in symbiosis with sponges and an ascidian of coral reefs. Mar. Biol. 95:1-13. [Google Scholar]
- 32.Lewin, R. A., and L. Cheng. 1993. Discovery, early history and phylogeny of Prochloron. Korean J. Phycol. 8:91-97. [Google Scholar]
- 33.Lim, G. E., and M. G. Haygood. 2004. “Candidatus Endobugula glebosa,” a specific bacterial symbiont of the marine bryozoan Bugula simplex. Appl. Environ. Microbiol. 70:4921-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moyer, C. L., J. M. Tiedje, F. C. Dobbs, and D. M. Karl. 1996. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl. Environ. Microbiol. 62:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otoguro, M., M. Hayakawa, T. Yamazaki, and Y. Iimura. 2001. An integrated method for the enrichment and selective isolation of Actinokineospora spp. in soil and plant litter. J. Appl. Microbiol. 91:118-130. [DOI] [PubMed] [Google Scholar]
- 36.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 37.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridley, C. P., P. R. Bergquist, M. K. Harper, D. J. Faulkner, J. N. A. Hooper, and M. G. Haygood. 2005. Speciation and biosynthetic variation in four dictyoceratid sponges and their cyanobacterial symbiont, Oscillatoria spongeliae. Chem. Biol. 12:397-406. [DOI] [PubMed] [Google Scholar]
- 40.Ronquist, F., and J. P. Huelsenbeck. 2003. Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 41.Schönhuber, W., B. Zarda, S. Eix, R. Rippka, M. Herdman, W. Ludwig, and R. Amann. 1999. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 65:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwintner, C., M. Sabaty, B. Berna, S. Cahors, and P. Richaud. 1998. Plasmid content and localization of the genes encoding the denitrification enzyme in two strains of Rhodobacter sphaeroides. FEMS Microbiol. Lett. 165:313-321. [DOI] [PubMed] [Google Scholar]
- 43.Shimada, A., N. Yano, S. Kanai, R. A. Lewin, and T. Maruyama. 2003. Molecular phylogenetic relationship between two symbiotic photo-oxygenic prokaryotes, Prochloron sp. and Synechocystis trididemni. Phycologia 42: 193-197. [Google Scholar]
- 44.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Springer, N., W. Ludwig, R. Amann, H. J. Schmidt, H. D. Görtz, and K. H. Schleifer. 1993. Occurrence of fragmented 16S rRNA in an obligate bacterial endosymbiont of Paramecium caudatum. Proc. Natl. Acad. Sci. USA 90:9892-9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 47.Taylor, M. W., P. J. Schupp, I. Dahllöf, S. Kjelleberg, and P. D. Steinberg. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6:121-130. [DOI] [PubMed] [Google Scholar]
- 48.Thacker, R. W., and S. Starnes. 2003. Host specificity of the symbiotic cyanobacterium Oscillatoria spongeliae in marine sponges, Dysidea spp. Mar. Biol. 142:643-648. [Google Scholar]
- 49.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX-Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, J. R., L. A. Marcelino, and M. F. Polz. 2002. Heteroduplexes in mixed-template amplifications: formation, consequences, and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 30:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webster, N. S., A. P. Negri, M. H. G. Munro, and C. N. Battershill. 2004. Diverse microbial communities inhabit Antarctic sponges. Environ. Microbiol. 6:288-300. [DOI] [PubMed] [Google Scholar]
- 52.Webster, N. S., A. P. Negri, R. I. Webb, and R. T. Hill. 2002. A spongin-boring α-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 232: 305-309. [Google Scholar]
- 53.Webster, N. S., K. J. Wilson, L. L. Blackall, and R. T. Hill. 2001. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 67:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkinson, C. R., M. Nowak, B. Austin, and R. R. Colwell. 1981. Specificity of bacterial symbionts in the Mediterranean and Great Barrier Reef sponges. Microb. Ecol. 7:13-21. [DOI] [PubMed] [Google Scholar]