Abstract
We investigated whether the yield of the B vitamin folic acid could be elevated in Bacillus subtilis. Strategies for increasing the folic acid yield were investigated by employing computer-aided flux analysis and mutation. Controlling the activity of the enzyme pyruvate kinase by placing it under inducible control was one strategy devised to elevate yield while insuring that a rapid growth rate results. Other single mutation strategies included amplifying the expression of the genes in the folate operon and overexpressing the Escherichia coli aroH gene, which encodes 2-dehydro-3-deoxyphosphoheptonate aldolase. The latter could conceivably elevate the abundance of the folic acid precursor, para-aminobenzoic acid. Strains that combined two or more mutations were also constructed. Overall, a strain possessing inducible pyruvate kinase, overexpressed aroH, and increased transcription and translation of genes from the folic operon exhibited the best yield. The yield was eightfold higher than that displayed by the parent B. subtilis 168 strain.
Metabolic engineering has focused on increasing the yield of high-value, low-quantity products or commodities such as amino acids (4, 18). One product that has received less attention from metabolic engineers is folic acid, which is a member of the B class of vitamins. (Although more chemically precise names exist for the biologically active [e.g., reduced, glutamated] form of the vitamin, the term “folic acid” is commonly used to refer to this B vitamin. Throughout this paper, “folic acid” is used as a generic name for this B vitamin to enable broad referencing and database linking.) Major sources of folic acid for mammals and birds are microbial production in the digestive tract and dietary intake. However, antibiotics (sulfonamides) are now used in animal feeds. To offset the inhibitory effect on folic acid synthesis, feeds are supplemented with folic acid. The supplementation of the human diet with folic acid may also increase because increased folic acid intake has been linked to reduced heart disease and lessened incidence of neural spinal defects in newborns. Indeed, the U.S. Food and Drug Administraion has mandated increased dietary intake and/or that some foods be fortified with folic acid (http://vm.cfsan.fda.gov/∼dms/wh-folic.html).
Folic acid is now primarily produced via chemical synthesis due to the low yields of folic acid provided by current bacterial strains (for example, see U.S. patent 5,968,788, October 1999). Several reasons have, however, prompted new exploration into using microbes for the commercial production of folic acid. First, the glutamated and reduced form of the natural vitamin, which many bacteria produce (10), is what the digestive system is “designed” to process. Prior to absorption, the body initially cleaves all folates to the monoglutamate form via the action of pancreatic and mucosal peptidases. Unlike synthetic folic acid, the “natural” B vitamin also does not require reduction prior to use by the body. Secondly, the U.S. Food and Drug Administration has resisted the use of product labeling language that indicates that the efficacy of synthetic alternatives is equal to or exceeds the natural forms. Thirdly, a single-step fermentation route could prove to be more environmentally benign than a multi-stage chemical process, if an effective production strain can be developed.
Examples of relevant work on improving microbial folic acid production include enhancing the folate production by the dairy starter bacteria Lactococcus lactis (24) and Streptococcus thermophilus (17). A mixed fermentation/chemical synthesis method for the microbial biotransformation of para-aminobenzoic acid (PABA) to folic acid is described in U.S. patent 5,968,788 (October 1999).
The alternative to the biotransformation of PABA or other precursors is the complete microbe-mediated synthesis of folic acid, where inexpensive carbohydrates and salts are the only raw materials used. Production by Bacillus subtilis would also be desirable for numerous reasons. Bacillus-derived products are classified as “generally regarded as safe.” Classical mutagenesis has also been previously used with success to elevate the yield of products that are metabolically related to folic acid such as d-ribose (6), purine nucleosides (12), and riboflavin (19). Looking forward to strain improvement, the gram-positive cell wall of B. subtilis is akin to other microorganisms used in amino acid manufacture such as Corynebacteria. Such strains can be engineered to be leaky by introducing biotin auxotrophy, which could prove to be a desirable trait to later instill into a promising engineered B. subtilis strain. Such a trait could elevate the product yield further by alleviating any end product inhibition effects.
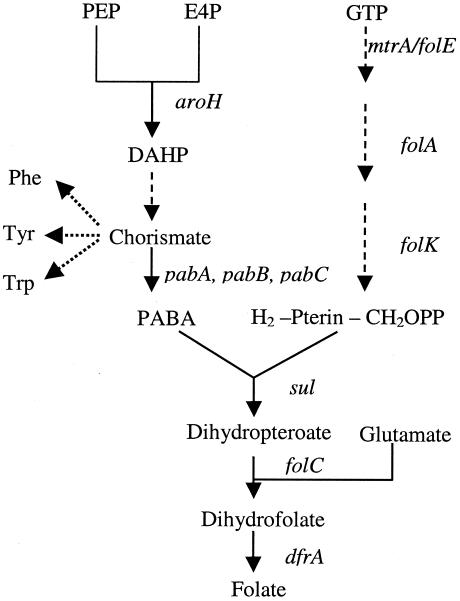
Figure 1, which shows the pathway for folic acid synthesis, provides a starting point for conceiving metabolic engineering strategies. The precursors of folic acid include phosphoenolpyruvate (PEP) from glycolysis, d-erythrose-4-phosphate (E4P) from the hexose monophosphate (HMP) pathway, GTP from purine synthesis, and glutamate from the tricarboxylic acid (TCA) cycle. E4P and PEP combine to form 3-deoxy-d-arabinoheptulosonate-7-phosphate (DAHP) via the aroH-encoded activity of 2-dehydro-3-deoxyphosphoheptonate aldolase. DAHP is the precursor of the intermediate chorismate, which is also an intermediate in aromatic amino acid metabolism. The genes that encode the utilization of chorismate and other folic acid intermediates are located at four different sites on the B. subtilis chromosome, but the major genes are located at 7 degrees, (Fig. 2). The first three genes in this operon are pabB, pabA, and pabC; they are involved in converting chorismate to PABA as shown in Fig. 1. The next three genes, sul, folA, and folK, encode dihydropteroate synthase, dihydroneopterin aldolase, and pyrophosphokinase, respectively. The first enzyme that commits GTP to folic acid production, GTP cyclohydrolase I, is encoded by mtrA, which is located in a two-gene operon with mtrB (Fig. 1), which codes the tryptophan RNA-binding attenuation protein (TRAP) (27). TRAP has been investigated extensively, and it has been found to terminate the transcription and translation of the folate operon. By analyzing B. subtilis strains that overexpress each of the five genes (sul, folA, folK, mtrA, and folC), Eichler et al. (7) reported that folK might encode the rate-limiting enzyme (pyrophosphokinase). However, it is conceivable that amplifying all five genes could elevate folic acid production.
FIG. 1.
Folic pathway schematic showing the flow of metabolites and the genes that encode the enzymes involved. Metabolite abbreviations not found in the text are Phe (phenylalanine), Tyr (tyrosine), Trp (tryptophan), and GTP. The transketolase-catalyzed reaction that forms E4P is omitted on the figure for brevity.
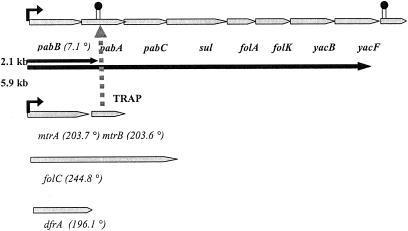
FIG. 2.
Folic acid biosynthetic and regulatory genes in B. subtilis from the National Center for Biotechnology Information website (http://www.ncbi.nih.gov). The major folic operon located at 7° encodes 6 genes (pabB, pabA, pabC, sul, folA, and folK) required for folate synthesis. GTP cyclohydrolase I (mtrA) and trp RNA-binding attenuation protein (mtrB) are located on an operon at 203.7°. The last two enzymes for folate synthesis, folC and dfrA, are located at 244.8° and 196.1°. The dashed arrow highlights the transcriptional and translational control of TRAP on the folic operon. Binding of TRAP to folic operon-derived mRNA terminates the translation of the DNA (2.1-kb mRNA would be the major product) and translation of the mRNA as well. Disruption of mtrB increases the expression level of 5.9-kb mRNA.
Elevating the production of precursors such as PEP and E4P is among the general metabolic engineering strategies that emerge. Both metabolites can fuel folic acid synthesis, where PEP is a direct folic acid precursor and glucose-6-phosphate reacts to form the precursor E4P. One means to accomplish this strategy is to create a B. subtilis mutant deficient in the enzyme pyruvate kinase (PYK) by interrupting the pyk gene. A prior publication reported that such a mutant exhibited significantly increased PEP and glucose-6-phosphate pools (8). Moreover, acetate production was significantly reduced, which could ultimately enable high density cell culture and thus improved volumetric yield. Amplifying aroH expression is another strategy that could also enhance folic acid production by elevating PABA production.
For many conceivable strategies, however, the interconnectivity of central metabolism suggests that benefits and losses may result when a given strategy is employed. For example, elevating PEP by restricting PEP uptake into the TCA cycle could positively influence folic acid synthesis due to the elevation of a precursor's mass action potential. However, a potential downside is that the formation of other folic acid precursors such as glutamate by the TCA cycle could be impaired. Thus, no net increase in folic acid productivity may result. Alternately, poor cell viability could result. Amplifying transketolase (e.g., tktA from Escherichia coli) is another example of a strategy that could have a beneficial or detrimental effect on folic acid synthesis. Overexpression could increase the formation of E4P. But amplification could also increase the rate of conversion of E4P and xylulose 5-phosphate (X5P) to fructose 6-phosphate (F6P) and glyceraldehyde 3-phosphate. Table 1 summarizes some potential positive and negative effects of single mutations that could conceivably be used for improving the folic acid productivity of bacteria.
TABLE 1.
Potential effects of single mutations on folic acid productiona
| Mutation | Potential positive effect | Potential negative effect |
|---|---|---|
| Disruption of pyk gene or put pyk under inducible control to provide low flux | Increase PEP pool and the flux to DAHP | Decrease the TCA flux and formation of the precursor glutamate |
| Amplify (E. coli) tktA on plasmid in B. subtilis | Increase the synthesis of the precursor DAHP | Decrease the flux to guanosine |
| Amplify (E. coli) aroH on a plasmid in B. subtilis | Increase the flux to DAHP | Plasmid maintenance and expression reduce the growth rate |
| Disrupt mtrB in B. subtilis | Increase the expression of full-length folate operon | Metabolic disruption or toxicity |
In this paper, we present the results of implementing single and combined mutation strategies for enhancing folic acid synthesis in B. subtilis. The strategies are summarized in Table 1, and they target both upstream and downstream steps in folic acid synthesis. The strategies were first investigated theoretically by using the convex analysis-based software Metabologica (31). The analysis provides bounds on yield, illuminates potential tradeoffs, and provides flux distributions that can serve as targets for metabolic engineering. For brevity, the flux analysis results will emphasize the potential effects of attenuating pyruvate kinase activity (elevates precursor PEP) and tktA expression (may elevate precursor E4P). The folic acid production capabilities B. subtilis with single and multiple mutations will then be summarized. Overall, the simulation trends and yield data concur. Moreover, most of the single mutations increased the folic acid yield, and one combination of mutations resulted in a strain that yielded eightfold more folic acid than B. subtilis 168 while retaining a doubling time under 100 min.
MATERIALS AND METHODS
Strains and culture conditions.
Bacillus subtilis 168 was used as the host strain for folic acid production; it was obtained from the Bacillus Genomic Stock Center (BGSC, Columbus, OH). Escherichia coli strain DH5a was used for the construction of plasmids. All the strains and plasmids used in this study are listed in Table 2.
TABLE 2.
Origin of strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| 168 | trpC2 | BGSC |
| IA96 | trpC2 pheA | BGSC |
| BG4233 | argC4 mtrB::cm (deletion from position 1001 to 1176) | 27 |
| BSZT0402 | trpC2 pyk(Ind) | This work |
| BSZT0408 | trpC2 mtrB::erm | This work |
| BSZT0410 | trpC2 aroH(Over) | This work |
| BSZT0412 | trpC2 aroH(Over) tktA(Over) | This work |
| BSZT0419 | trpC2 mtrB::cm pyk(Ind) | This work |
| BSZT0425 | trpC2 aroH(Over) pyk(Ind) | This work |
| BSZT0437 | trpC2 aroH(Over) mtrB::cm pyk(Ind) | This work |
| Plasmids | ||
| pMutin4 | Ampr Ermr | BGSC |
| pRB374 | Ampr Kanr | This work |
| pMTPYK | Ampr Ermr; contains 740 bp of pyk gene | This work |
| pRA1 | Ampr Kanr aroH | This work |
| pRAT2 | Ampr KanraroH tktA | This work |
| pVp2 | Ampr Ermr; contains Ermr cassette between 5′ and 3′ fragments of mtrB | 27 |
Strains were cultured in Luria-Bertani (LB) or glucose minimum medium (1). When appropriate, the medium contained chloramphenicol (5 mg/liter), kanamycin (10 mg/liter), ampicillin (100 mg/liter), or erythromycin (0.3 mg/liter). Baffled 300-ml shake flasks were used to grow the different strains. Two seed cultures were prepared prior to culture. The first seed culture consisted of 25 ml of LB-rich medium with suitable antibiotics, and the second culture consisted of 25 ml of glucose minimum medium with suitable antibiotics. The folic acid production culture consisted of 25 ml of glucose minimum medium without antibiotics. The culture conditions were 37°C and shaking at 200 rpm. Optical density was measured offline using a Lambda 6 spectrophotometer (Perkin-Elmer, Norwalk, CT) using our calibrations (1 optical density unit at 660 nm [OD660] = 0.35 g of cell dry weight/liter). The glucose concentration was measured enzymatically (kit 16-UV; Sigma Chemicals, St. Louis, MO).
PCR amplification and DNA manipulation.
Several E. coli or B. subtilis genes were amplified from chromosomal DNA by PCR with 50 ng of DNA in a final volume of 50 μl containing deoxyribonucleoside triphosphates (0.5 mM each), oligonucleotides (50 pM) (Table 2), and 1 U of Pfx polymerase (Invitrogen, Paisley, Great Britain). Amplification was performed in a Mastercycler with 35 cycles of denaturation at 94°C for 30 s, annealing at 50 to 60°C for 30 s, and elongation at 68°C for 1 to 3 min.
Isolation of E. coli plasmid DNA was performed using the Promega Wizard SV gel and PCR Clean-Up system. Restriction enzymes were purchased from New England Biolabs and other DNA modification enzymes were obtained from Invitrogen.
Construction of plasmids and transformation of strains.
Plasmid pMTPYK was created by amplifying the 5′ end of the B. subtilis 168 pyk gene by PCR, using primers PYKF (5′-CCGGATCCGAAGATTTCAGAAGGAAGTGAACC-3′) and PYKR (5′-CCGGATCCGGAATTTCCACACCTAAGTCTCC-3′). These primers were designed to include BamHI restriction sites (shown in boldface) to facilitate cloning. The PCR fragment was inserted into plasmid pMutin4 (25). The recombinant plasmids were transformed into competent B. subtilis cells, and transformants were selected on antibiotic medium plates containing 0.3 mg/liter erythromycin. The mutants were analyzed by PCR to confirm the integration of a single copy of the plasmid into the target gene on the chromosome, using a strategy similar to that described previously (22). The mutants were also confirmed by the fact that they cannot grow in two days on a glycerol minimal plate unless IPTG (isopropyl-β-d-thiogalactopyranoside) is added.
B. subtilis plasmid pRA1 was created by amplifying the E. coli K-12 aroH gene by PCR, using primers AroHF (5′-CCGAAGCTTCTGCCGTAGAAGCAAC AAAT-3′) and AroHR (5′-CCTCTAGAGCATTCAGAAGCGGGTATC-3′) and E. coli K-12 genomic DNA as the template. These primers were designed to include HindIII and XbaI restriction sites (shown in boldface) to facilitate cloning. The PCR fragment was inserted into plasmid pRB374. The recombinant plasmids were transformed into competent B. subtilis cells, and transformants were selected on antibiotic medium plates containing 10 mg/liter kanamycin. The mutants were confirmed by restriction digestion of plasmid prepared from the culture.
B. subtilis plasmid pRAT2 was created by amplification of the E. coli K-12 tktA gene by PCR, using primers tktAF (5′-CCTCTAGAGCGCTGTCGTCAAGTC GTTAA-3′) and tktAR (5′-CCGGATCCATAAAAAAGGTCGCCGAAGCAC-3′). These primers were designed to include XbaI and BamHI restriction sites (shown in boldface) to facilitate cloning. The PCR fragment was inserted into plasmid pRA1. The recombinant plasmids were transformed into competent B. subtilis cells (28), and transformants were selected on antibiotic medium plates containing 10 mg/liter kanamycin. The mutants were confirmed by restriction digestion of plasmid prepared from the culture. The resultant plasmid possessed both the aroH and tktA genes.
B. subtilis plasmid pVp2 was generously donated by P. Gollinick (27). Plasmid pVp2 was linearized with EcoRI prior to transforming B. subtilis. pVp2 and pMTPYK both use erythromycin as a selective marker. When these two mutations were introduced into the same strain, genomic DNA from strain BG4423 (27) was used to transform competent B. subtilis because BG4423 also has a chloramphenicol-disrupted mtrB gene. To increase the efficiency of transformation, a PCR fragment of disrupted mtrB from strain BG4423 was used. The forward and reverse primers used in the PCR amplification are mtrBF (5′-CAAAGATGTATGCCGAAGTAT-3′) and mtrBR (5′-TTTTTGTTTTCGTCTCGTTTT-3′).
Highly competent E. coli cells were prepared and stored at −80°C for later transformation. The transformation of competent E. coli was performed chemically (11). B. subtilis transformation was also performed chemically using the method from BGSC (28).
Assay of folic acid.
We found that the majority of folic acid is not secreted into the medium; hence, intracellular folic acid was extracted. The intracellular folic acid concentration was also found to be time dependent (data not shown). The intracellular folic acid concentration was consistently found to be at its highest plateau level about 1 h after the maximum cell density was reached. Depending on the mutation, however, some strains exhibited the plateau level earlier, but the measurement point used accurately captured the intracellular level of folic acid. The difference in plateau times may reflect variations in competition for intracellular resources or product feedback inhibition effects. Additionally, we have found that the level of pyruvate kinase expression towards the end of exponential growth differs in wild-type and engineered B. subtilis (data not shown), which could be attributed to the properties of the pSpac promoter from plasmid pMTPYK, which controls the expression of the pyk gene. These variations are under further investigation, and further work with leaky mutants may provide more mechanistic insights as well as even higher folic acid yields. Extracted folic acid was measured in triplicate samples, and the average value and deviation of folic acid concentration are reported. Values are reported on a per cell mass (e.g., μmol folic acid/g cell) and total culture volume basis.
To prepare a sample for the measurement of intracellular folic acid content, collected cells were lysed with a French press and then passed through a 0.22-μm filter. Filtered samples were diluted 40 to 200 times based on the estimated folic acid concentration. Thereafter, the following microbial bioassay (Difco Manual, 11th edition, [Difco, Detroit, Mich.]) was used to measure the folic acid concentration. A stock culture of the test strain ATCC 7469 was prepared in a 15-ml tube with 10 ml Micro Inoculum Broth. These inocula were incubated at 37°C for 18 h. Under aseptic conditions, the cultures were centrifuged to sediment the cells and then washed three times with the folic acid assay medium. The cells were diluted 100-fold, and 10 μl of this suspension was used to inoculate each of the assay tubes (in a 2-ml microcentrifuge tube). To each tube, 1 ml double-strength folic acid assay medium and 1 ml of sample were added. The optical density of the assay tubes was measured after 18 to 24 h of incubation at 37°C. It was essential that a standard curve be constructed for each separate assay. The standard curve was generated by using 0.0, 0.05, 0.1, 0.15, and 0.2 ng of folic acid per assay tube (2 ml). Optical density was measured using a Lambda 6 Perkin-Elmer spectrophotometer (Perkin-Elmer, Norwalk, CT) at 600 nm.
RESULTS
Network model and flux predictions.
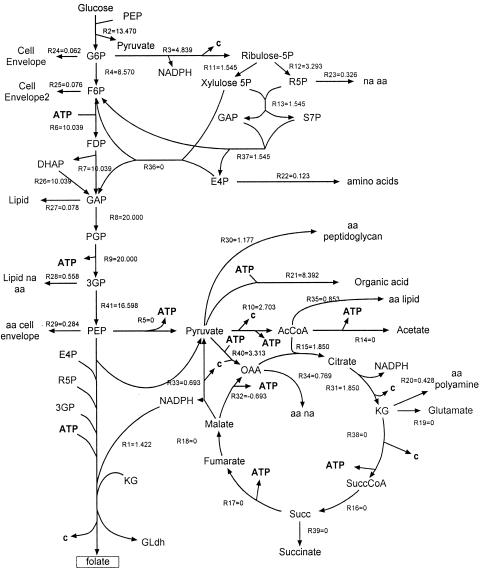
A stoichiometric model of the central carbon metabolism with a lumped reaction leading to folic acid was built using the software Metabologica (30; http://www.metabologica.com/). This software permits the in silico construction of a network model. First, the metabolic network “picture” of interest is drawn on a computer screen. Thereafter, the “model picture” is automatically transformed to mathematical metabolite balance equations, and a number of computations can then be performed. The central metabolic reactions and the composition of B. subtilis used in the computation have been previously described (23). The composition information assigns values for a particular reaction's “load” such as the amount of acetyl coenzyme A that must be produced for membrane biosynthesis. Additionally, system-wide constraints such as net NADPH formation for reductive biosynthesis and minimum ATP requirements, which link groups of reactions, have also been described (14-16, 23). Such “linking constraints” limit the metabolite trafficking alternatives as well as insure that a particular solution is physiologically feasible. A reference specific growth rate equal to 0.4 h−1 was used to generate flux units (e.g., mmol g cell−1 h−1); the results can be scaled when other growth rates are considered. The overall stoichiometry put into Metabologica for the lumped reaction that leads to folic acid (FA) production is 2PEP + E4P + 2 3GP + KG + 11ATP + R5P + 4NADPH → FA + PYR + CO2, where PEP (phosphoenolpyruvate), E4P (erythrose-4-phosphate), 3GP (3-phosphoglyceric acid), KG (oxoglutarate), R5P (ribose-5-phosphate), and PYR (pyruvate) are both FA precursors and intermediate metabolites present in central carbon metabolism.
We found that 32 different feasible solutions exist for the growth of folic-acid producing B. subtilis in glucose minimal medium. Specific mutations can also be explored by, for example, constraining a flux to a zero value to simulate a knockout mutation. For brevity, two solutions will be discussed that probe in more detail the qualitative predictions noted in Table 1. Namely, the effect of altering pyruvate kinase activity and a flux in the HMP pathway will be presented.
A representative Metabologica solution for a high folic-acid-yielding scenario is shown in Fig. 3. This solution indicates that highly attenuated PYK flux (r5 = 0) may be associated with a metabolically feasible and high folic-acid-yielding scenario. While network trafficking models do not directly account for mass action or kinetic effects, a possible beneficial consequence can be envisioned when PYK activity is reduced. The concentration of the folic acid precursors, PEP and 3GP, may increase due to a downstream blockage in consumption, and this increase could stimulate folic acid synthesis. Accordingly, the modeling results and envisioned mass action consequences suggest that a strategy based on decreasing PYK flux may work in B. subtilis as it did when implemented in E. coli (31).
FIG. 3.
One output from Metabologica that shows a metabolite trafficking pattern that is predicted by linear programming to confer the highest carbon yield for folic acid production. Flux values shown correspond to mmol/h g cell (basis growth rate is 0.4 h−1). The maximized yield solutions also correspond to minimized pyruvate kinase flux (r5). Metabolite abbreviations not found in the text are GAP (glyceraldehyde-3-phosphate), S7P (sedoheptulose-7-phosphate), PGP (1,3-biphosphoglycerate), OAA (oxaloacetate), Succ (succinate), aa (amino acid), na (nucleic acids), and c (carbon dioxide).
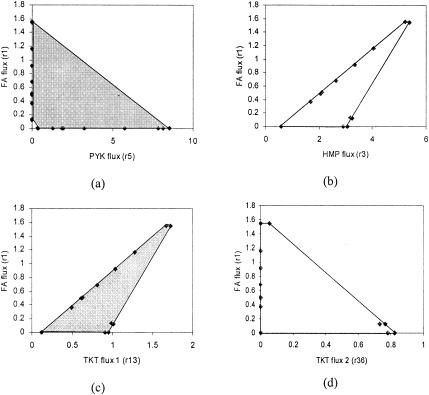
The use of phase plane-like graphical displays (26) provides a means for further exploring the effect of pyk and other mutations. One such phase plane is shown in Fig. 4a, where PYK-catalyzed flux is the x axis and folic acid flux is the y axis. Any point inside the bounded region is a feasible solution. The maximum folic acid synthesis rate increases linearly as the PYK-catalyzed flux decreases to zero. The geometry of the phase plane boundary also suggests that as PYK flux decreases to zero, solutions that yield high folic acid productivity still exist.
FIG. 4.
Phase planes for folic acid production. (a) Folic acid formation rate (Fig. 3, r1) versus PYK flux (r5); (b) folic acid formation rate (r1) versus HMP flux (r3); (c) folic acid formation rate (r1) versus transketolase flux converting R5P and X5P to GAP and S7P (r16); and (d) folic acid formation rate (r1) versus transketolase flux converting E4P and X5P to F6P and G3P (r36).
The effect of altering the net flux through the hexose monophosphate pathway on folic acid synthesis is shown in another phase plane (Fig. 4b). In general, increasing the HMP flux increases folic acid production. The net HMP flux may be directly increased through HMP enzyme amplification (e.g., glucose-6-phosphate isomerase and glucose-6-phosphate 1-dehydrogenase) and indirectly via PYK attenuation (20, 21). However, as noted earlier, the relationship between amplifying specific HMP pathway enzymes such as transketolase and folic acid production may be complex due to the tradeoffs between positive and negative consequences. Amplification of tktA gene from E. coli, which encodes one of the transketolase isozymes, could increase the beneficial reaction r13 and the deleterious reaction r36 at the same time (Fig. 3). The possible tradeoffs are apparent in the phase planes shown in Fig. 4c and Fig. 4d.
In summary, Metabologica indicated that for the mutations summarized in Table 1, each mutation would not be lethal in that stoichiometrically feasible and mass/energy-integrated solutions exist. In other cases (e.g., tktA), tradeoffs were identified that would be interesting to experimentally probe.
Growth and folic acid yield for an inducible pyruvate kinase mutant.
Based on our prior work with E. coli (31) and the aforementioned modeling results, the folic acid production of a pyk knockout of B. subtilis was first explored. When the PYK flux is zero, the modeling results suggest that such a pyk knockout is viable (Fig. 3). We found that such a mutant is indeed viable, but the growth is slow in glucose minimal medium (doubling time exceeds 6 h) (this work and reference 8). In contrast, a pyk knockout of E. coli grows nearly as fast as the wild type (29). The difference in the growth rates of B. subtilis and E. coli can be attributed to different anaplerotic routes to oxaloacetate formation are used by the two species as was explained in a prior publication (8).
To allow the growth rate of B. subtilis to be varied between that of a knockout and wild-type phenotype, a strain of B. subtilis was endowed with inducible pyk [pyk(Ind)]. Here, the aim was to explore the effect of pyk expression on folic acid yield as well as to investigate whether a high-yielding (amount per cell) and productive strain (amount per time) could be developed. The strain is designated BSZT0402 (Table 2).
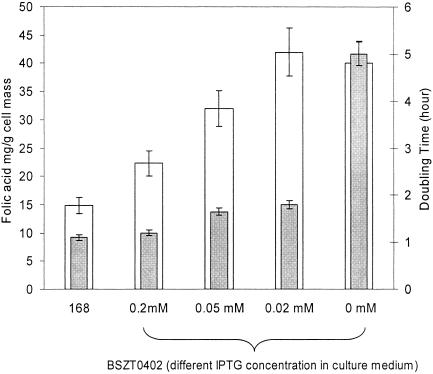
Figure 5 shows the folic acid yield and doubling time exhibited by BSZT0402 when different levels of IPTG are provided in glucose minimal medium. The yield and doubling time characteristics of BSZT0402 are also contrasted to those exhibited by B. subtilis 168 (trpC2). When 0.2 mM IPTG is present, the growth rate of BSZT0402 is essentially equal to that of B. subtilis 168 (trpC2), whereas the growth of BSZT0402 is very slow when no IPTG is added. Both this result and enzyme activity measurements (not shown) indicate that significant control via IPTG induction exists over pyk expression in BSZT0402 (2).
FIG. 5.
How folic acid yield (white bar) and doubling time (gray bar) depend on IPTG concentration in strain BSZT0402 [trpC2, pyk(Ind)], which possesses IPTG-inducible pyruvate kinase. The parent strain's (B. subtilis 168) folic acid yield and doubling time are included for comparison.
The advantage of inducible control of pyk is evident in the case when the IPTG concentration was set to 0.02 mM. The severalfold enhancement in folic acid yield of BSZT0402 compared to B. subtilis 168 (trpC2) is similar to that exhibited by the pyk knockout. However, the doubling time of BSZT0402 is severalfold smaller than the knockout. Consequently, when pyk induction is controlled, a threefold increase in folic acid yield can be obtained while sustaining 60% of the parent strain's growth rate. Reasonable growth rate, in turn, translates into increased bioprocess productivity.
Folic acid yield for other B. subtilis mutants.
The folic acid yields and growth rates exhibited by 1A96 (pheA1 trpC2), BSZT0410 {trpC2 overexpressed aroH [aroH(Over)]}, BSZT0412 [trpC2 aroH(Over) tktA(Over)], and BSZT0408 (trpC2 mtrB::erm) were measured in glucose minimal medium. The yields and growth rates are compared to each other and those of B. subtilis 168 (trpC2) in Table 3. The cell mass concentration at the time of sampling and the yield on the basis of the total volume of cells and spent medium are also listed in Table 3. Amplifying aroH (BSZT0410) and disrupting mtrB (BSZT0408) both increase the folic acid yield by about 80%. Blocking phenylalanine production (1A96) increases the folic acid yield by about 100%. In contrast, amplifying the tktA gene (BSZT0412) reduced the folic acid yield by 46% compared to the parent strain (BSZT0410).
TABLE 3.
Folic acid yields of wild-type and engineered B. subtilis in glucose minimum medium (5 g/liter)
| Strain name | Description | Doubling time (h) | Folic acid mass yield (μg/g cell) | Dry cell mass concn (g/liter) | Folic acid volumetric yield (μg/liter) |
|---|---|---|---|---|---|
| I68 | trpC2 | 1.08 ± 0.08 | 14.7 ± 0.7 | 1.95 ± 0.05 | 28.7 |
| IA96 | trpC2 pheA | 1.08 ± 0.08 | 30.2 ± 2.0 | 1.98 ± 0.04 | 59.8 |
| BSZT0410 | trpC2 aroH(Over) | 1.15 ± 0.08 | 26.8 ± 1.3 | 1.85 ± 0.05 | 49.6 |
| BSZT0412 | trpC2 aroH(Over) tktA(Over) | 1.40 ± 0.1 | 14.3 ± 0.7 | 1.60 ± 0.05 | 22.9 |
| BSZT0408 | trpC2 mtrB::erm | 1.20 ± 0.1 | 26.3 ± 1.5 | 1.78 ± 0.05 | 46.8 |
| BSZT0419 | trpC2 mtrB::cm, pyk(Ind) | 1.50 ± 0.1a | 56.0 ± 4.0 | 1.75 ± 0.05 | 98 |
| BSZT0425 | trpC2 aroH(Over) pyk(Ind) | 1.25 ± 0.1a | 60.0 ± 4.0 | 1.90 ± 0.06 | 114 |
| BSZT0437 | trpC2 aroH(Over) mtrB::cm pyk(Ind) | 1.60 ± 0.2a | 113 ± 11.0 | 1.45 ± 0.05 | 163 |
A total of 0.05 mM IPTG was added to the culture at the time of inoculation.
Combinations of mutation strategies.
The different mutation strategies that introduced inducible expression of pyk, aroH amplification, and mtrB disruption were combined to explore the synergistic effects. The folic acid yields of BSTZ0419 [trpC2 mtrB::cm pyk(Ind)], BSTZ0425 [trpC2 aroH(Over) pyk(Ind)], and BSTZ0437 [trpC2 aroH(Over) mtrB::cm pyk(Ind)] are listed in Table 3 along with doubling times, cell yields, and volumetric yields. The combination of the three mutations in BSTZ0437 resulted in an eightfold increased yield of folic acid compared to B. subtilis 168. Moreover, the growth rate and carbon yield of BSTZ0437 are both about 70% of the starting strain 168 when 0.05 mM IPTG is added to induce pyruvate kinase expression.
DISCUSSION
Previously, eliminating pyruvate kinase flux was investigated as a means for increasing folic acid synthesis in E. coli (31). Computational work presented here (Fig. 3 and 4) indicates that attenuating PYK flux to a near-zero level may also be a fruitful starting point for enhancing folic acid production by B. subtilis. Moreover, controlling pyk activity at a low value via IPTG induction may overcome the low growth rate limitation of a pyk knockout (8). Overall, we found that when provided with a low level of IPTG (0.02 mM), an inducible pyk mutant, BSTZ0402, exhibits a threefold higher folic acid yield compared to the parent strain B. subtilis 168. The doubling time of BSTZ0402 was found to be 1.8 h as opposed to 6 h or more for a pyk knockout. When IPTG concentration is increased, the BSTZ0402 mutant increasingly exhibits the parental strain phenotype in terms of growth rate and lessened folic acid yield.
To more fully explore folic acid production, other mutations were also investigated. Adding extracellular para-aminobenzoic acid to B. subtilis cultures has been reported to increase folic acid production (U.S. patent 5,968,788). Amplifying E. coli aroH in B. subtilis could have a similar effect by increasing the rate of DAHP synthesis, which is a step in the PABA synthesis pathway (Fig. 1). There are three DAHP synthases in E. coli. One enzyme, aroH, is feedback inhibited by tryptophan. The other two DAHP synthases are Tyr and Phe sensitive. The aroH-encoded DAHP synthase remains 30% active even when the inhibitor tryptophan is present (3). The dosage of aroH is higher due to the copy number of plasmid pRB374 in B. subtilis. Although the enzyme activity was not measured, the results indicate that overexpressing aroH in BSZT0410 results in higher folic acid production (Table 3). Additionally, the strain BSZT0425 indicates that overexpressing aroH in tandem with low induction of pyk further increases the folic acid yield. The yield enhancement conceivably results from the elevated PEP pool that has been found in B. subtilis mutants with impaired pyruvate kinase activity (8).
BSZT0437 is the highest folic-acid-yielding mutant that was developed. This mutant exhibited eightfold increased folic acid yield compared to B. subtilis 168 (Table 3). This mutant combined control over pyk expression, overexpressed aroH, and increased expression of the genes encoded by the folate operon. Thus, with respect to the folic acid synthesis pathway, mutations that targeted upstream and downstream biosynthetic processes proved to be most effective. It is noteworthy that the doubling time of BSZT0437 was not excessively long. Using the data in Table 3, BSZT0437 exhibits about fourfold greater bioreactor volume productivity (μg folic acid/liter h) compared to B. subtilis 168. B. subtilis strains with attenuated pyruvate kinase activity also produce less acetate (8). Thus, apart from increased folic acid yield, another attribute is that BSZT0437 may prove easier to culture to a high cell concentration using fed-batch techniques.
Based on prior metabolic engineering work and the results that we obtained when the folic acid operon was relieved from negative control, the yield-enhancing effect of a mutation (e.g., pyk) localized far upstream in a biosynthetic pathway is not surprising. The response between biosynthetic gene amplification and product flux often proves to be nonlinear due to insufficient precursor supply, distributed flux control (13), or other reasons. In this work, the translational and transcriptional negative control of the folate operon was relieved by disrupting mtrB. Prior work provides some reference points that can be used to interpret the effect of overexpressing the genes in the folate operon. It has been reported that mostly 2.7-kb production occurs from the folate operon, which suggests that low transcription and translation activity normally occurs (27). Following mtrB disruption, it has been reported that the 5.7- and 8-kb transcription products increase eightfold. Additionally, encoded protein levels also increase by 25-fold (5). The increased folic acid yield displayed by the ΔmtrB mutant BSZT0408 (Table 3) is consistent with yield being related, in part, to operon expression. However, the elevation in yield was not directly proportional to the reported level of enhanced transcription or protein, which indicates that operon-encoded enzymes contribute to, but do not dominate, flux control.
Finally, the yields associated with the different mutants provide some leads on the how folic acid yield may be further increased in B. subtilis. Transketolase (tktA) amplification has been shown to increase aromatic amino acid production in E. coli (9). However, our network analysis shows that tktA amplification may or may not increase folic acid production (Fig. 4c and d). Overall, the results suggest that a small but finite level of tktA expression is required for optimal folic acid production. The results from BSZT0412 suggest that amplifying tktA in an overexpressed aroH context reduces folic acid production (Table 3). Higher tktA activity may increase DAHP synthesis, which is advantageous. However, a conceivable negative impact is that the concentration of a GTP precursor (ribose 5-phosphate) decreases. Thus, controlling tktA expression via an inducible promoter might be an avenue that merits further investigation.
Another possible avenue for further metabolic engineering is suggested by the yield exhibited by strain 1A96, which is a phenylalanine auxotroph. Blocking phenylalanine synthesis increased folic acid production by about 100% (Table 3). Developing other strains with blocked chorismate-consuming pathways, such as a tyrosine auxotroph, might lead to increased folic acid production. Finally, it may be worthwhile to develop a leaky mutant (e.g., a biotin autotroph) of BSZT0437. Once high cell density has been attained and the initial biotin supplied is exhausted, a productive phase of folic acid production may ensue that is liberated from feedback inhibition effects. Such a mutant is now under investigation.
Acknowledgments
This work was supported in part by grants BES-0224603 and BES-0118961 from the National Science Foundation. Publication of this work does not imply endorsement by the National Science Foundation.
REFERENCES
- 1.Aymerich, S., G. Gonzy-Treboul, and M. Steinmetz. 1986. 5′-Noncoding region sacR is the target of all identified regulation affecting the levansucrase gene in Bacillus subtilis. J. Bacteriol. 166:993-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhavsar, A. P., X. Zhao, and E. D. Brown. 2001. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl. Environ. Microbiol. 67:403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camakaris, J., and J. Pittard. 1974. Purification and properties of 3-deoxy-d-arabinoheptulosonic acid-7-phosphate synthetase (trp) from Escherichia coli. J. Bacteriol. 120:590-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, R., V. Hatzimanikatis, W. Yap, P. W. Postma, and J. E. Bailey. 1997. Metabolic consequences of phosphotransferase (PTS) mutation in a phenylalanine-producing recombinant Escherichia coli. Biotechnol. Prog. 13:768-775. [DOI] [PubMed] [Google Scholar]
- 5.de Saizieu, A., P. Vankan, C. Vockler, and A. P. van Loon. 1997. The trp RNA-binding attenuation protein (TRAP) regulates the steady-state levels of transcripts of the Bacillus subtilis folate operon. Microbiology 143:979-989. [DOI] [PubMed] [Google Scholar]
- 6.De Wulf, P., and E. J. Vandamme. 1997. Production of D-ribose by fermentation. Appl. Microbiol. Biotechnol. 48:141-148. [DOI] [PubMed] [Google Scholar]
- 7.Eichler, K., C. Vockler, R. Carballido-Lopez, and A. P. G. M. van Loon. 1997. Investigation of the biotechnological potential of folic acid biosynthesis in Bacillus subtilis, p. 46. Abstr. 9th International Conference on Bacilli, Lausanne, Switzerland.
- 8.Fry, B., T. Zhu, M. M. Domach, C. Phalakornkule, R. Koepsel, and M. M. Ataai. 2000. Characterization of growth and acid formation in a Bacillus subtilis pyruvate kinase mutant. Appl. Environ. Microbiol. 66:4045-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gosset, G., J. Yong-Xiao, and A. Berry. 1996. A direct comparison of approaches for increasing carbon flow to aromatic biosynthesis in Escherichia coli. J. Ind. Microbiol. 17:47-52. [DOI] [PubMed] [Google Scholar]
- 10.Hintze, D. N., and J. L. Farmer. 1975. Identification of poly-γ-glutamyl chain lengths in folates of Bacillus subtilis. J. Bacteriol. 124:1236-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 12.Ishii, K., and I. Shiio. 1973. Regulation of purine nucleotide synthesis in Bacillus subtilis. Agric. Biol. Chem. 37:287-296. [Google Scholar]
- 13.Kascer, H., and J. A. Burns. 1973. The control of flux. Symp. Soc. Exp. Biol. 27:65-104. [PubMed] [Google Scholar]
- 14.Lee, J., A. Goel, M. M. Ataai, and M. M. Domach. 1997. Supply-side analysis of growth of Bacillus subtilis on glucose citrate medium: feasible network alternatives and yield optimality. Appl. Environ. Microbiol. 63:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, S., C. Phalakornkule, M. M. Ataai, M. M. Domach, and I. E. Grossmann. 2000. Recursive MILP model for finding all the alternate optima in LP models for metabolic networks. Comput. Chem. Eng. 24:711-716. [Google Scholar]
- 16.Liao, J. C., and M. Oh. 1999. Toward predicting metabolic fluxes in metabolically engineered strains. Metab. Eng. 1:214-223. [DOI] [PubMed] [Google Scholar]
- 17.Lin, M., and C. M. Young. 2000. Biosynthesis of folates by Streptococcus thermophilus and Lactobacillus delbrueckii ssp. bulgaricus. J. Food Drug Anal. 8:195-199. [Google Scholar]
- 18.Patnaik, R., and J. C. Liao. 1994. Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield. Appl. Environ. Microbiol. 60:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkins, J. B. A. Sloma, T. Hermann, K. Theriault, E. Zachgo, T. Erdenberger, N. Hannett, N. P. Chatterjee1, V. Williams, I. I., G. A. Rufo, Jr., R. Hatch, and J. Pero1. 1999. Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J. Ind. Microbiol. Biotech. 22:8-18. [Google Scholar]
- 20.Phalakornkule, C., B. Fry, T. Zhu, R. Kopesel, M. M. Ataai, and M. M. Domach. 2000. 13C NMR evidence for pyruvate kinase flux attenuation underlying suppressed acid formation in Bacillus subtilis. Biotechnol. Prog. 16:169-175. [DOI] [PubMed] [Google Scholar]
- 21.Ponce, E., N. Flores, A. Martinez, F. Bolivar, and F. Valle. 1998. Simulation of glucose catabolism through the pentose pathway by the absence of the two pyruvate kinase isoenzymes in Escherichia coli. Biotechnol. Bioeng. 58:292-295. [DOI] [PubMed] [Google Scholar]
- 22.Prágai, Z., and C. R. Harwood. 2000. YsxC, a putative GTP-binding protein essential for the growth of Bacillus subtilis 168. J. Bacteriol. 182:6819-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauer, U., D. C. Cameron, and J. E. Bailey. 1998. Metabolic capacity of Bacillus subtilis for the production of purine nucleosides, riboflavin, and folic acid. Biotechnol. Bioeng. 59:227-238. [PubMed] [Google Scholar]
- 24.Sybesma, W., M. Starrenburg, M. Kleerebezem, I. Mierau, W. M. de Vos, and J. Hugenholtz. 2003. Increased production of folate by metabolic engineering of Lactococcus lactis. Appl. Environ. Microbiol. 69:3069-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 26.Varma, A., and B. O. Palsson. 1994. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 60:3724-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, M., A. de Saizieu, A. P. van Loon, and P. Gollnick. 1995. Translation of trpG in Bacillus subtilis is regulated by the trp RNA-binding attenuation protein (TRAP). J. Bacteriol. 177:4272-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasbin, R. E., G. A. Wilson, and F. E. Young. 1975. Transformation and transfection in lysogenic strains of Bacillus subtilis: evidence for selective induction of prophage in competent cells. J. Bacteriol. 121:296-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu, T., C. Phalakornkule, R. R. Koepsel, M. M. Domach, and M. M. Ataai. 2001. Cell growth and by-product formation in a pyruvate kinase mutant of E. coli. Biotechnol. Prog. 17:624-628. [DOI] [PubMed] [Google Scholar]
- 30.Zhu, T., C. Phalakornkule, R. R. Koepsel, M. M. Ataai, and M. M. Domach. 2003. A metabolic network analysis and NMR experiment design tool with user interface-driven model construction for depth-first search analysis. Metab. Eng. 5:74-85. [DOI] [PubMed] [Google Scholar]
- 31.Zhu, T., I. E. Grossmann, M. M. Domach, and M. M. Ataai. 2003. Metabolic engineering of folic acid production. Ferment. Biotechnol. ACS Symp. 862:207-219. [Google Scholar]