Abstract
Viable microbes have been detected beneath several geographically distant glaciers underlain by different lithologies, but comparisons of their microbial communities have not previously been made. This study compared the microbial community compositions of samples from two glaciers overlying differing bedrock. Bulk meltwater chemistry indicates that sulfide oxidation and carbonate dissolution account for 90% of the solute flux from Bench Glacier, Alaska, whereas gypsum/anhydrite and carbonate dissolution accounts for the majority of the flux from John Evans Glacier, Ellesmere Island, Nunavut, Canada. The microbial communities were examined using two techniques: clone libraries and dot blot hybridization of 16S rRNA genes. Two hundred twenty-seven clones containing amplified 16S rRNA genes were prepared from subglacial samples, and the gene sequences were analyzed phylogenetically. Although some phylogenetic groups, including the Betaproteobacteria, were abundant in clone libraries from both glaciers, other well-represented groups were found at only one glacier. Group-specific oligonucleotide probes were developed for two phylogenetic clusters that were of particular interest because of their abundance or inferred biochemical capabilities. These probes were used in quantitative dot blot hybridization assays with a range of samples from the two glaciers. In addition to shared phyla at both glaciers, each glacier also harbored a subglacial microbial population that correlated with the observed aqueous geochemistry. These results are consistent with the hypothesis that microbial activity is an important contributor to the solute flux from glaciers.
Subglacial systems were, until recently, thought to be abiotic (see, e.g., references 16 and 25). However, increasing evidence points to a significant role for microbes in mediating dissolution and oxidation of minerals in sediments beneath ice masses (“subglacial weathering”), where liquid water is present (51, 54, 60, 63), and even within glacial ice (8, 9, 48, 50). Studies of subglacial water chemistry at Haut Glacier d'Arolla, Switzerland, have revealed active redox processes, e.g., nitrate removal and sulfide oxidation, including anoxic oxidation of iron sulfides by Fe(III) (7, 59, 60). Tranter and colleagues argue that the anoxic conditions in subglacial systems indicate that weathering beneath glaciers is likely microbially mediated, similar to processes in other environments (see. e.g., references 42 and 45). Similarly, Wadham et al. (63) used stable isotope measurements of subglacial waters at Finsterwalderbreen, Svalbard, to infer microbial sulfate reduction. These ideas are supported by studies that have shown that microbial populations are widespread in subglacial environments (see, e.g., references 51 and 54).
Microbes are abundant at the water-rock-ice interface beneath valley glaciers at Haut Glacier d'Arolla and Tsanflueron (>106 cells ml−1 in some subglacial waters) (51), at Fox and Franz-Josef Glaciers (14) and at John Evans Glacier (JEG), Ellesmere Island (54). Significant microbial populations are also present in subglacial lakes (15, 28, 49), with an estimated 105 to 106 cells ml−1 in the Antarctic subglacial Lake Vostok (49). Evidence for microbial activity in situ in these subglacial environments is also accumulating (14, 51, 54). For example, the debris-rich basal ice layers of JEG contain metabolically diverse microbes, including aerobic chemoheterotrophs, nitrate reducers, sulfate reducers, and methanogens, that could be enriched oligotrophically at low, near-in situ temperatures (0.3 to 4°C) (54).
The few studies of the composition of subglacial microbial communities to date (see e.g., references 14 and 54) have all been based on cultivation of microbes. However, it is generally accepted that most microbes are not yet culturable (2). PCR-based analysis of 16S rRNA genes from environmental samples provides significant insight into uncultivated microbial diversity in other environmental systems (58), but such studies have not yet been performed with subglacial samples.
Two primary factors likely influence the composition of microbial communities in subglacial environments: physical factors (i.e., cold, stable temperatures, and lack of light) that are common to all such environments and chemical factors (i.e., bedrock lithology/mineralogy and solute composition, including carbon sources and electron acceptors) that are unique to each environment. Therefore, those components of subglacial microbial communities that are influenced primarily by physical factors will have cosmopolitan distributions, whereas other components that are influenced primarily by chemical factors will be more endemic to individual sites. To test this hypothesis, water and basal ice samples from two glaciers with different bedrock lithologies were analyzed utilizing geochemical, isotopic, and molecular biological techniques. Phylogenetic analysis of the microbial component was used to infer relationships between community composition and in situ aqueous geochemistry.
MATERIALS AND METHODS
Field sites.
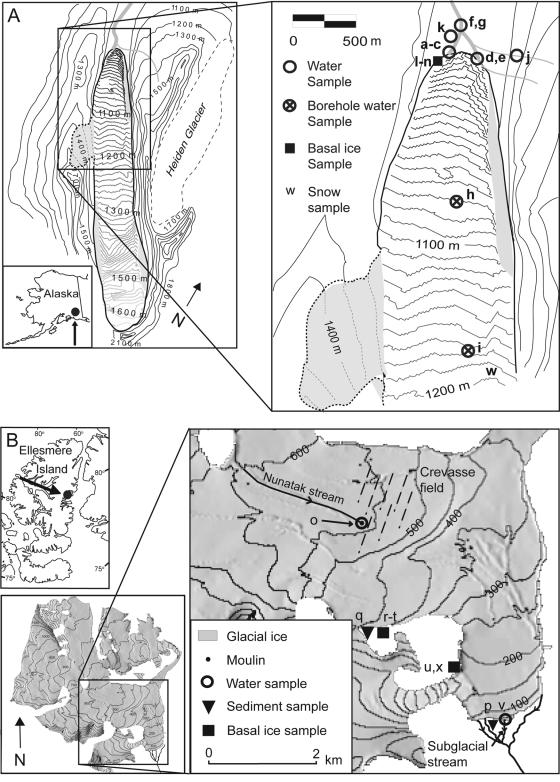
Bench Glacier (BG) is a small (7-km2) valley glacier located in the Chugach Range of south-central Alaska, at 61.03°N, 145.69′W, with an elevation range from 950 to 1,600 m (3, 4) (Fig. 1A). The basin is underlain by late-Cretaceous age meta-sediments of the Valdez Group (65). Sulfide mineralization is evident in some of the rocks underlying BG, as evidenced by rust stains and visible pyrite and chalcopyrite grains within rocks in the BG moraines. Sampling locations are noted in Fig. 1A.
FIG. 1.
Locations of sampling sites. (A) Bench Glacier. Ice topography from a Global Positioning System survey (39) is shown, with shading where the topography was not determined. Contour intervals are 10 m on ice and 100 m off ice. (B) John Evans Glacier. The contour interval is 100 m on ice. Letters indicate sampling sites. A detailed description of the sampling sites is in Materials and Methods.
JEG is a large valley glacier located at 79°40′N, 74°00′W, on the east coast of Ellesmere Island, Nunavut, Canada (Fig. 1B). It covers approximately 75% of a 220-km2 catchment, with a length of 15 km and an elevation range of 100 to 1500 m above sea level (55). The glacier is polythermal, with cold-based ice in the accumulation area and at the glacier margins where ice is thin and warm-based ice throughout the remainder of the ablation zone, where basal water is present (11). The glacier is underlain by an Ordovician/Silurian carbonate/evaporite sequence with a minor clastic component (30). Shale containing small amounts of pyrite outcrops near the glacier terminus. Sampling locations are noted in Fig. 1B.
Water, ice, and sediment sampling locations.
Subglacial stream samples (samples f and g) and basal ice samples (samples l to n) were collected at BG in July 1999 (for a detailed explanation of glacial water and ice types, see reference 54). West subglacial stream samples (samples a to c), east subglacial stream samples (samples d and e), and ice marginal stream samples (samples j and k) were collected at BG in May and June 2002. Boreholes were drilled to the bed of BG in May 2002 using a hot-water drill (see reference 22 for full details), and water samples were retrieved from the borehole with a 700-ml stainless steel point-source bailer equipped with Teflon ball stops (model 429; Solinst Canada Ltd.). The upper borehole water sample (sample i) was debris free and had a low solute concentration (electrical conductivity, ∼2 μS cm−1) and thus was interpreted to represent ice melt, i.e., supraglacial water. In contrast, the lower borehole sample (sample h) was heavily sediment laden and had a significantly higher solute concentration (electrical conductivity, ∼38 μS cm−1) and was interpreted to be a sample of subglacial water and sediments. One sample of snow (sample w) was collected at BG in a sterile 10-liter carboy and melted in the field in a water bath prior to processing. Sample j was from an ice marginal stream draining the valley wall (effectively a waterfall). Sample k was water draining though the proglacial sediments on the valley side and floor; it was sampled just prior to merging with the subglacial stream and is termed “groundwater.”
Basal ice samples (samples x, r to t, and u) were collected from the ice margin at JEG in May 1999, July 2001, and July 2002, respectively (Fig. 1B). Supraglacial (sample o) and subglacial (sample v) water samples were collected in June 2002. The proglacial sediment sample (sample p) was collected as the finer fraction of gravels from the cut bank of a proglacial stream ∼100 m downstream from the glacier terminus. The subglacial sediment sample (sample q) was collected from beneath the glacier at an exposure close to where basal ice samples (samples r to t) were collected.
Water, ice, and sediment sampling for microbiological analyses.
All stream water samples at both BG and JEG were collected directly from the stream in autoclaved polypropylene bottles that had been prerinsed at least three times with the stream water. The bailer was cleaned and disinfected prior to borehole water sampling at BG in 2002 by immersion in 99% isopropanol. BG basal ice samples (samples l to n) were chipped from the glacier terminus with an ice pick into an aluminum pan; both tools were sterilized with 98% ethanol and flame. Samples were transferred to acid-cleaned, autoclave-sterilized 2-liter Nalgene bottles and allowed to melt in a snowbank. Basal ice sampling at JEG followed the procedures outlined by Skidmore et al. (54), and sediment sampling followed the procedures outlined in reference 6.
Direct microscopic cell counts for BG water samples collected in 2002.
Aliquots of 275 ml of water or melted snow were added to furnace-combusted (550°C) medium bottles containing 25 ml of filter-sterilized 37% formaldehyde in the field, mixed, and stored for transport at 4°C. Samples were incubated with the DNA-staining dye 4,6-diamino-2-phenylindole (DAPI), filtered, and subjected to cell counting as previously described (31, 47), with the exception that the volume of water examined varied from 1 to 25 ml, depending on the cell concentration. A procedural field blank consisting of 275 ml of sterile Nanopure water (0.2-μm filtered, followed by autoclaving) was examined in parallel.
Ion chemistry.
Stream water chemistry sampling programs for the subglacial outlet stream were conducted during the summer melt seasons in 1995, 1996, 1999, 2000, and 2002 at BG and in 1994, 1996, 1998, 1999, and 2000 at JEG. Water samples were collected twice daily at BG at the same location as samples f and g during field campaigns of 5 days (1995) to 6 weeks (2000) and daily at JEG at the same location as sample v during field campaigns of 3 weeks (1998) to 8 weeks (1994). Water samples were filtered in the field through 0.45-μm cellulose nitrate filters and split into two aliquots, one for cation analysis and one for anion analysis. Details on analytical methods for pH, alkalinity, and major cation (Mg, Ca, Na, and K) and anion (Cl, NO3, and SO4) concentrations are described elsewhere (4, 55).
Determination of DOC concentrations in BG samples collected in 2002.
Water samples for dissolved organic carbon (DOC) were collected at BG in 2002 as described previously (53). DOC concentrations were determined on a Shimadzu 5000 total organic carbon analyzer following the methods described previously (33). Sample blanks contained <0.1 ppm, and precision and accuracy were better than ±3% for all samples.
Genomic DNA extraction. (i) BG.
Melt water with sediment from either stream samples or melted basal ice was filtered in the field in July 1999 through a 10-μm-pore-size Nitex filter, and the filtrate was then filtered through 0.2-μm-pore-size Supor filters (VWR). One to 4 liters of stream water, borehole water, or snowmelt was filtered onto sterile 0.2-μm filters (Metricel; Gelman) using a peristaltic pump in the field in 2002. Isopropanol (95%) was pumped through the filtering system between samples to prevent cross contamination. A procedural blank was also prepared by filtering 3 liters of sterile Nanopure water through the same apparatus in the field. All filters were stored in sealed plastic bags with 5 ml lysis buffer (20 mM EDTA, 400 mM NaCl, 0.75 M sucrose, and 50 mM Tris HCl [pH 9.0], prepared in filter-sterilized water) (19), frozen (in a liquid N2 dry shipper [Fisher] for the 1999 samples or in a snowbank for the 2002 samples) for transport to the laboratory, and stored at −80°C until processing.
(ii) JEG.
Basal ice samples from JEG in 1999, 2001, and 2002 were collected in sterile bags as described by Skidmore et al. (54), shipped frozen, melted in sterile beakers in the laboratory, and filtered through 0.2-μm Supor filters (VWR). Filters were stored in sealable bags with 5 ml lysis buffer (19) and frozen at −80°C. Sediment and water samples collected in 2002 followed the protocols outlined previously (6).
Total cellular nucleic acids were extracted from the filters by use of a combination of procedures optimized for small sample sizes as previously described (19), with the exception of JEG samples collected in 2002, which were extracted via a modified bead beater technique (6).
Clone library construction and analysis.
PCR primers S-D-Bact-0008-a-A-19 and S-D-Bact-1492-a-A-21 (17) were used to amplify bacterial 16S rRNA genes from a BG glacier outlet stream (sample 6) and a JEG basal ice sample (sample B), both collected and archived in 1999. The PCR conditions used were 1 min of denaturation at 95°C, 2 min of annealing at 55°C, and 3 min of elongation at 72°C for 35 cycles in an MJ Research thermal cycler. After a final 10-min extension at 72°C, the product was purified using a gel extraction kit (QIAGEN, Chatsworth, CA). Amplification products were cloned into the plasmid vector pCR2.1 by TA cloning (Invitrogen, Carlsbad, CA). Grouping into operational taxonomic unit (OTUs) by restriction fragment length polymorphism (RFLP) analysis, sequencing, and phylogenetic analysis were carried out as described previously (35). All sequences from the same OTU were found to be >97% identical, indicating that they were the same species (56). Amplification with Archaea-biased primers 21F and 958R (12) did not consistently provide a PCR product for any sample (data not shown), so here we focus on Bacteria.
Oligonucleotide probe design and DNA:DNA dot blot hybridization.
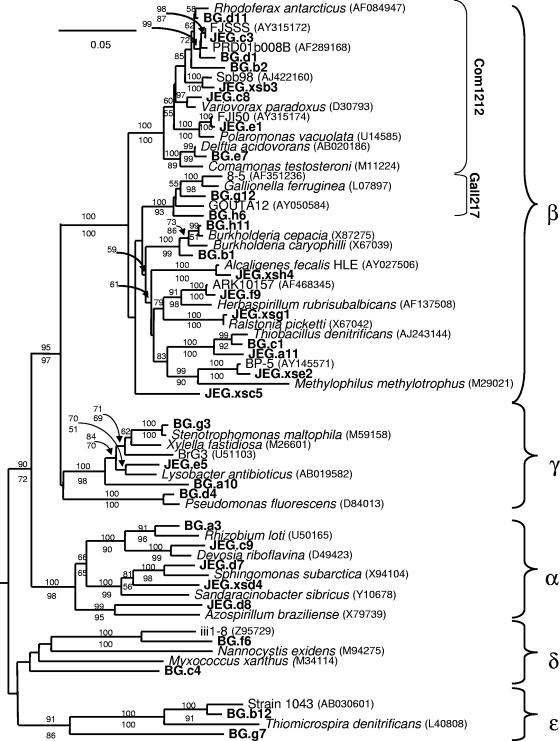
Oligonucleotide probes Gall217 (5′-GGCCACUCCGAAAGCGA-3′; melting temperature [Tm], 52°C) and Com1212 (5′-GUGUGUAGCCCCACCUAU-3′; Tm, 48°C) were designed using ARB (38) (http://www.arb-home.de/) to target the group of sequences indicated in Fig. 2. The “Com1212” group was predominant in both the BG and JEG clone libraries, and the “Gall217” group was prominent in the BG clone library but was not detected in the JEG clone library. PCR-amplified 16S rRNA gene inserts of two clones from the clone library for each of the oligonucleotide probes were used as positive controls on the blots and to determine the empirical Tms of the probes (19). No cross-hybridization was detected between the Gall217 and Com1212 probes.
FIG. 2.
Phylogenetic analysis of proteobacterial 16S rRNA gene clones from JEG and BG. This phylogenetic tree was rooted with Bacteroides fragilis (GenBank accession number M61006). A mask of 917 nucleotides, including all nonambiguously aligned positions, spanning nearly the full length of the 16S rRNA gene, was included. The clusters which the Com1212 and Gall217 probes were designed to detect are indicated. Bootstrap values (100 replications) generated by the neighbor-joining method are shown above relevant nodes, and those generated by maximum-parsimony analysis are shown below the nodes. Only bootstrap values above 50 are shown. Sequences from formally named isolates are in italic, sequences from environmental gene clones and unnamed isolates are in plain text, and sequences from the subglacial samples are in boldface. GenBank accession numbers of the sequences from other studies are included.
Bacterial 16S rRNA genes were amplified from the genomic DNA extracted from water, ice, and sediment samples (samples a to v) and the procedural blank from BG by using the Bacteria-biased primers 27F and 1512R (17) and the PCR conditions described above. To minimize PCR bias, multiple PCRs with the minimum number of cycles yielding a product were combined (46, 57). Dot blot construction, probe labeling with [γ-32P]dATP, hybridization, and signal normalization to the Bacteria-biased probe Eub338 (5′-GCTGCCTCCCGTAGGAGT-3′) (1) for individual probes were performed as previously described (19). Blots were imaged using a Typhoon PhosphorImager, and spot intensity was quantified using ImageQuant software (Amersham Biosciences, NJ). One hundred nanograms of amplified DNA was applied per spot for natural populations, and 50 to 100 ng per spot was applied for positive and negative controls. A Methanococcus voltae 16S rRNA gene clone and the full procedural blank were used as negative controls. No hybridization signal to either of these negative controls was detected. Duplicate field samples for BG (sample a) and JEG (sample o) (see Table 5) underwent all of the procedural stages as for the other samples and showed good agreement in terms of probe hybridization signal, demonstrating internal consistency in the analytical procedure (data not shown). PCR duplicates of samples r and v (see Table 5) also showed good agreement, indicating low PCR variability (data not shown). All samples and the blanks were analyzed in triplicate.
TABLE 5.
Signal ratios from dot blot hybridization assays of DNA amplified from BG and JEG samples
| Glacier and sample(s)a | Yr | Sample type | Signal ratiob
|
|
|---|---|---|---|---|
| Com1212/Eub338 | Gall217/Eub338 | |||
| BG | ||||
| a-e | 2002 | Subglacial stream | 0.12 ± 0.03 | 0.18 ± 0.02 |
| f, g | 1999 | Subglacial stream | 0.18-0.23 | 0.09-0.11 |
| h | 2002 | Lower borehole, subglacial water | 0.08 | 0.11 |
| i | 2002 | Upper borehole, supraglacial water | 0.05 | BDLc |
| j, k | 2002 | Marginal stream | 0.06-0.12 | BDL-0.02 |
| l-n | 1999 | Basal ice | 0.12 ± 0.01 | BDL |
| JEG | ||||
| o | 2002 | Supraglacial stream | 0.06 | BDL |
| p | 2002 | Proglacial sediment | 0.03 | BDL |
| q | 2001 | Subglacial sediment | 0.17 | BDL |
| r-t | 2001 | Basal ice | 0.14 ± 0.02 | BDL |
| u | 2002 | Basal ice | 0.19 | BDL |
| v | 2002 | Subglacial stream | 0.60 | BDL |
For sample locations, see Fig. 1. Sample f was used to produce the BG clone library.
For all samples with n of ≥3, means and standard deviations are shown. For samples with n of 2, the range is shown.
BDL, below detection limit (ratio of <0.01).
Nucleotide sequence accession numbers.
Sequences were deposited in GenBank under accession numbers DQ228359 to DQ228420.
RESULTS
Water chemistry.
Sulfate and bicarbonate dominate the anionic weathering products in the subglacial stream at both glaciers, and calcium is the dominant cation (4, 23, 53, 55; S. P. Anderson, unpublished data). The Ca/SO4 ratio and the S ratio [SO4/(SO4 + HCO3)] are indicators of the dominant solute-producing weathering processes and can be used to distinguish between sulfide oxidation and gypsum dissolution as major weathering processes (Table 1). The stoichiometry for pyrite (a common sulfide) oxidation coupled to carbonate dissolution is
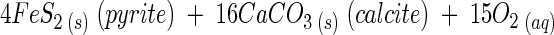 |
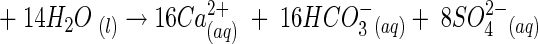 |
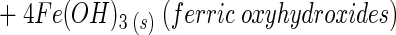 |
which produces a Ca/SO4 ratio of 2 and an S ratio [SO4/(SO4 + HCO3)] of 0.5 for concentrations in milliequivalents liter−1. Gypsum (CaSO4 · 2H2O) dissolution produces a Ca/SO4 ratio of 1. Calcite (CaCO3) dissolution by either hydrolysis or carbonic acid weathering adds Ca and HCO3 to solution and thus increases the Ca/SO4 ratio and lowers the S ratio, if all other things are held equal.
TABLE 1.
Subglacial stream chemistry data for BG (sample location f) and JEG (sample location v)
| Glacier | Yr (reference) | Ratioa
|
n | |||
|---|---|---|---|---|---|---|
| Ca/SO4
|
S
|
|||||
| Mean | SD | Mean | SD | |||
| JEG | 1994 (55) | 0.95 | 0.15 | 0.89 | 0.02 | 46 |
| 1996 (55) | 1.04 | 0.09 | 0.83 | 0.04 | 10 | |
| 1998 (53) | 1.41 | 0.34 | 0.66 | 0.13 | 11 | |
| 1999 (53) | 1.14 | 0.14 | 0.77 | 0.08 | 7 | |
| 2000 (23) | 1.34 | 0.12 | 0.64 | 0.06 | 13 | |
| Mean | 1.09 | 0.25 | 0.81 | 0.12 | 87 | |
| BG | 1995 (4) | 1.80 | 0.10 | 0.44 | 0.02 | 19 |
| 1996 (4) | 2.12 | 0.22 | 0.35 | 0.03 | 23 | |
| 1999 (S.P.A.)b | 1.74 | 0.28 | 0.49 | 0.07 | 114 | |
| 2000 (S.P.A.) | 2.81 | 0.24 | 0.63 | 0.03 | 74 | |
| 2002 (S.P.A.) | 2.25 | 0.29 | 0.51 | 0.06 | 30 | |
| Mean | 2.21 | 0.47 | 0.56 | 0.09 | 260 | |
All ratios were calculated using concentrations in milliequivalents liter−1. Sratio, SO4/(SO4 + HCO3).
S. P. Anderson, unpublished data.
Parametric (Welch's unpaired t test) and nonparametric (Mann-Whitney test) tests of the Ca/SO4 and S ratios (P < 0.001) indicate that they are the result of different weathering processes at BG and JEG. The average Ca/SO4 ratios and S ratios for 5 years of subglacial stream data at BG are approximately 2 (2.21 ± 0.47; n = 260) and 0.5 (0.56 ± 0.09; n = 260), respectively (Table 1). This strongly suggests that coupled sulfide oxidation and carbonate dissolution is the major weathering process at BG, confirming results previously reported by Anderson et al. (4). By contrast, Ca/SO4 ratios are much closer to 1 at JEG (1.09 ± 0.25; n = 87), strongly suggesting that gypsum dissolution is the dominant source of sulfate (Table 1). Sulfur isotope analysis on a sequence of 1998 subglacial waters, bedrock gypsum, and pyrite demonstrates that 86 to 100% of the SO42− in subglacial waters at JEG is from gypsum dissolution (53), supporting the interpretation from the geochemical data. The mean S ratio at JEG (0.81 ± 0.12; n = 87) indicates significant input of SO4 relative to HCO3, and the mean JEG Ca/SO4 ratio of >1 indicates an excess of calcium over SO4. The source for this Ca and HCO3 is likely from the dissolution of carbonate minerals, largely calcite. Thus, the dominant weathering processes at JEG are gypsum and carbonate dissolution.
Microbial abundance and DOC.
Direct cell counts of the planktonic portion of the microbial community ranged from 6.6 × 104 to 3.7 × 105 cells ml−1 in the snowmelt, ice marginal and subglacial streams, and borehole waters at BG in 2002, indicating that there is low but significant microbial biomass in these environments. The lowest and highest counts recorded were in the snowmelt and the east subglacial stream, respectively. DOC concentrations range from 0.4 to 7.4 mg liter−1, indicating that an available organic carbon source exists in these same environments. There is a moderate correlation between cell counts and DOC for all the environments (r2 = 0.64). The lowest DOC value was in the snowmelt, and the highest DOC value was from the ice marginal stream (sample k), which is likely the closest measured representative of “groundwater” in the catchment. While samples for direct cell counts were not taken from JEG, viable microbes have been cultured from supraglacial and subglacial waters and melted basal ice samples (54). DOC concentrations at JEG were 1.2 mg liter−1 in basal ice and 0.29 mg liter−1 in the glacier ice (54) and ranged from 0.11 to 0.47 mg liter−1 (n = 54) and 0.12 to 0.43 mg liter−1 (n = 47) in the supraglacial and subglacial streams, respectively, in 2001 (5). The DOC values from BG and JEG are similar to those reported for other glaciated environments; for example, 0.14 to 0.77 mg liter−1 (n = 61) was the range over three summer melt seasons, from 1998 to 2000, at Bow Glacier, Alberta, Canada (33), and there were 0.51 mg liter−1 in accretion ice above Lake Vostok, Antarctica (49), and 0.11 mg liter−1 in the snowpack on the summit of the Greenland Ice Sheet (62).
Clone library composition.
A total of 227 16S rRNA gene clones (94 from BG and 133 from JEG) were preliminarily grouped into OTUs by RFLP analysis. OTU groups were confirmed by partial sequencing of 16S rRNA genes from representative clones. Seventeen and 29 OTUs were found in the BG and JEG clone libraries, respectively. Accumulation curves for each site reached an asymptote, indicating that most of the species richness present in the extracted DNA was sampled in these clone libraries (26). This finding was confirmed by the Chao1 nonparametric diversity estimator (10), which predicted 25 ± 8.3 and 30 ± 1.5 OTUs at BG and JEG, respectively (analyses were performed at http://www2.biology.ualberta.ca/jbrzusto/rarefact.php; data not shown). Therefore, an estimated 68% of the OTU diversity was detected at BG and 97% of such diversity was detected at JEG. It is unusual for a clone library to sample so much of the total estimated diversity (26, 29); therefore, these two subglacial systems appear to have low species richness. Although prevalence in clone libraries and PCR products is not a direct indication of abundance in the original environment due to potential biases in sample collection, cell lysis, DNA extraction, and PCR (46, 57), some correlation may be expected between abundance in the original sample and prevalence in clone libraries.
The BG clone library was almost completely dominated by Proteobacteria (97% of clones); only three other “phylum”-level groups were present, and these were represented by only single clones (Table 2). Betaproteobacteria were the dominant group at BG (68% of clones), but Alphaproteobacteria (4.2% of clones), Gammaproteobacteria (10.6% of clones), Deltaproteobacteria (3.2% of clones), and Epsilonproteobacteria (10.6% of clones) were also present (Table 2). BLAST searches and phylogenetic analyses showed that many sequences from BG (27.6% of clones) (Table 3) were associated with a betaproteobacterial cluster of sequences that includes Polaromonas vacuolata, Rhodoferax spp., and Comamonas spp. (designated “Com1212” in Fig. 2). Another cluster of betaproteobacterial sequences (designated “Gall217” in Fig. 2) related to Gallionella ferruginea (a stalked, neutrophilic, iron-oxidizing bacterium [21]), were highly abundant at BG (27.6% of clones) (Table 3). Relatives of betaproteobacterial Thiobacillus spp., some isolates of which are involved in acidic iron oxidation and/or neutrophilic sulfur oxidation (36), were also present at significant levels (7.4% of clones). The remaining betaproteobacterial clones (5.3% of clones) were related to the metabolically diverse Burkholderia genus (Table 3) (37).
TABLE 2.
Distribution of clones in the clone libraries for JEG (133 clones) and BG (94 clones) by major phylogenetic group
| Phylogenetic group | No. of clones
|
% of clone library
|
||
|---|---|---|---|---|
| JEG | BG | JEG | BG | |
| Alphaproteobacteria | 10 | 4 | 7.5 | 4.3 |
| Betaproteobacteria | 74 | 64 | 55.6 | 68.1 |
| Gammaproteobacteria | 2 | 10 | 1.5 | 10.6 |
| Deltaproteobacteria | 0 | 3 | 0.0 | 3.2 |
| Epsilonproteobacteria | 0 | 10 | 0.0 | 10.6 |
| Cytophaga/Flavobacterium/Bacteroides | 33 | 1 | 24.8 | 1.1 |
| Holophaga/Acidobacteria | 2 | 1 | 1.5 | 1.1 |
| Spirochaeta | 0 | 1 | 0.0 | 1.1 |
| Planctomycetales | 2 | 0 | 1.5 | 0.0 |
| Actinobacteria | 7 | 0 | 5.3 | 0.0 |
| Verrucomicrobium | 3 | 0 | 2.3 | 0.0 |
TABLE 3.
Taxonomic affiliations of representative clones from the RFLP groups for the clone libraries from BG, determined by phylogenetic analysis of 16S rRNA gene sequences
| Representative clone(s)a | No. of clones | Phylogenetic clusterb | Nearest neighborc (accession no.) | % Similarityd | Comment |
|---|---|---|---|---|---|
| BG.A3, BG.F8 | 4 | Alphaproteobacteria | Rhizobium sp. strain RM1-2001 (AF331662) | 97 | Isolated from arsenic-contaminated mine tailings |
| BG.B9, BG.H6 | 23 | Betaproteobacteria (“Gall217”) | JG36-GS-54 (AJ582038) | 95 | Heavy-metal-contaminated environment clone |
| BG.G12, BG.F2 | 3 | Betaproteobacteria (“Gall217”) | GOUTA12 (AY050584) | 95 | Monochlorobenzene-contaminated groundwater clone |
| BG.A7, BG.A8, BG.E7, BG.F11 | 19 | Betaproteobacteria (“Com1212”) | Delftia acidovorans (AF181575) | 99 | Related to S0-oxidizing isolates |
| BG.D1 | 3 | Betaproteobacteria (“Com1212”) | PRD01b008B (AF289168) | 97 | Freshwater bacterial clone |
| BG.D11 | 1 | Betaproteobacteria (“Com1212”) | Rhodoferax ferrireducens (AF435948) | 98 | Psychrotolerant, facultatively acetate-oxidizing, Fe(III)-reducing isolate |
| BG.B2 | 3 | Betaproteobacteria (“Com1212”) | Rhodoferax ferrireducens (AF435948) | 96 | Psychrotolerant, facultatively acetate-oxidizing, Fe(III)-reducing isolate |
| BG.C1 | 7 | Betaproteobacteria | P3OB-54 (AF414582) | 98 | U(VI)-reducing community clone |
| BG.H11, BG.B1 | 5 | Betaproteobacteria | Burkholderia cepacia LMG12614 (AF311970) | 99 | Isolate from cystic fibrosis sputum |
| BG.G3 | 4 | Gammaproteobacteria | Stenotrophomonas maltophilia strain e-a21 (AJ293470) | 98 | Isolate from sewage treatment plant |
| BG.D4 | 4 | Gammaproteobacteria | Pseudomonas synxantha (AF267911) | 99 | Isolated from pine tree buds |
| BG.A10 | 2 | Gammaproteobacteria | HTB10 (AF418945) | 94 | Freshwater aquifer metal-rich particle clone |
| BG.C4 | 3 | Deltaproteobacteria | Hyd24-27 (AJ535224) | 90 | Marine sediment clone |
| BG.B12, BG.B6 | 10 | Epsilonproteobacteria | RA9C8 (AF407391) | 99 | Monochlorobenzene-contaminated groundwater clone |
| BG.C5 | 1 | CFB | GKS2-106 (AJ290025) | 98 | Lake Gossenkoellesee clone |
| BG.F6 | 1 | Holophaga/Acidobacteria | DS-18 (AY289393) | 91 | Soil clone |
| BG.H3 | 1 | Spirochaeta | Leptospira fainei (Y19243) | 91 | Isolated from human patient with Weil's disease |
Fully, bidirectionally sequenced clones representing HhaI RFLP groupings.
Phylum-level grouping. Names in parentheses are groupings shown in Fig. 2.
Most closely related sequence in the GenBank public database, based upon BLASTN search.
Percent similarity to nearest neighbor, based upon BLASTN search.
Another prevalent group in the BG clone library (11% of clones) (Table 2) included Epsilonproteobacteria related to sulfur-oxidizing symbionts of vent fauna (Fig. 2). Other proteobacterial groups (Fig. 2) included relatives of alphaproteobacterial Rhizobium spp. (4.2% of clones), Pseudomonas spp., and other members of the Gammaproteobacteria (10.6% of clones) and Deltaproteobacteria (3.2% of clones) (Table 2). None of the deltaproteobacterial clones were affiliated with the deltaproteobacterial sulfate-reducing bacteria (SRB). One clone each from the bacterial phyla Cytophaga/Flavobacterium/Bacteroides (CFB) and Holophaga/Acidobacteria, which are most similar to environmental gene clones, were present (Tables 2 and 3). Intriguingly, several clones had strong bootstrap support for affiliation with proteobacterial subphyla but were only distantly related to gene sequences available in public databases (e.g., clones BG.a10, BG.c4, and BG.g7) (Fig. 2). One clone (BG.h3) had no strong support for affiliation with any bacterial phylum and was not included in Fig. 2; its nearest neighbor is Leptospira fainei, a member of the bacterial phylum Spirochaeta (Table 3).
Proteobacteria were also highly represented in the JEG clone library (64.6% of clones), and Betaproteobacteria were predominant (55.6% of clones) (Fig. 2; Table 2). No Delta- or Epsilonproteobacteria were observed in this clone library. Some betaproteobacterial clones in the JEG library were affiliated with some of the same groups as those in the BG library. Similar to the BG library, members of the “Com1212” group were extremely abundant (44.4% of clones); however, relatives of Thiobacillus spp., while present, were far less well represented in the JEG library (2.2% of clones) (Tables 3 and 4). Notably, no clones related to Gallionella ferruginea, a dominant group at BG (see above), were present in the JEG clone library. Other betaproteobacterial clones from JEG were related to diverse isolates and environmental gene sequences (Fig. 2). An association of two particularly intriguing clones (represented by JEG.xsc5) with the Betaproteobacteria is well supported (100 of 100 bootstraps with both neighbor-joining and maximum-parsimony approaches); however, they do not strongly affiliate with any gene sequences available in public databases (Fig. 2). Alphaproteobacteria related to various genera (Fig. 2) were well represented in the JEG clone library (7.5% of clones) (Table 2). Gammaproteobacteria were also present, but at low abundance (1.5% of clones).
TABLE 4.
Taxonomic affiliations of representative clones from the RFLP groups for the clone libraries from JEG, determined by phylogenetic analysis of 16S rRNA gene sequences
| Representative clone(s)a | No. of clones | Phylogenetic clusterb | Nearest neighborc (accession no.) | % Similarityd | Comment |
|---|---|---|---|---|---|
| JEG.C9 | 5 | Alphaproteobacteria | SM1E02 (AF445680) | 98 | Related to Antarctic and Arctic isolates |
| JEG.D8 | 2 | Alphaproteobacteria | JG30-KF-C3 (AJ536857) | 95 | Uranium waste pile clone |
| JEG.XSD4 | 2 | Alphaproteobacteria | Sphingomonas sp. clone KL-2-4-7 (AF408323) | 93 | Clone from clean-room facility |
| JEG.D7 | 1 | Alphaproteobacteria | HTB9 (AF418947) | 96 | Freshwater aquifer metal-rich particle clone |
| JEG.XSB3, JEG.A12, JEG.A1 | 21 | Betaproteobacteria (“Com1212”) | Spb98 (AJ422160) | 98 | Related to Antarctic lake mat clone |
| JEG.E1 | 16 | Betaproteobacteria (“Com1212”) | GKS16 (AJ224987) | 99 | High mountain lake clone |
| JEG.G5 | 3 | Betaproteobacteria (“Com1212”) | GKS16 (AJ224987) | 98 | High mountain lake clone |
| JEG.C3, JEG.B8 | 14 | Betaproteobacteria (“Com1212”) | PRD01b008B (AF289168) | 98 | Typical freshwater bacterial clone |
| JEG.C8 | 5 | Betaproteobacteria (“Com1212”) | Variovorax sp. strain WDL1 (AF538929) | 97 | Linuron-degrading soil isolate |
| JEG.XSE2 | 3 | Betaproteobacteria | PRD01a011B (AF289159) | 95 | Typical freshwater bacterial clone |
| JEG.A11 | 3 | Betaproteobacteria | Thiobacillus sp. clone 44a-B2-21 (AY082471) | 96 | ZnS-precipitating biofilm clone |
| JEG.XSG1 | 3 | Betaproteobacteria | Ralstonia detusculanense (AF280433) | 99 | Isolated from heavy water |
| JEG.F9 | 2 | Betaproteobacteria | ARKMP80 (AY198106) | 99 | Arctic sea ice clone |
| JEG.XSH4 | 2 | Betaproteobacteria | BW6 (AF394171) | 99 | Related to thiosulfate-oxidizing soil clone |
| JEG.XSC5, JEG.XSC6 | 2 | Betaproteobacteria | JG36-GS-10 (AJ582037) | 95 | Uranium waste pile clone |
| JEG.E5 | 2 | Gammaproteobacteria | Gitt-GS-126 (AJ582198) | 97 | Heavy-metal-contaminated environment clone |
| JEG.XSH1 | 3 | Actinobacteria | Sporichthya polymorpha (AB025317) | 93 | Related to Antarctic cryptoendolith community clone |
| JEG.A2 | 2 | Actinobacteria | 911-1 (AY326619) | 93 | Soil clone |
| JEG.A3 | 1 | Actinobacteria | ML316M-15 (AF454303) | 93 | Related to freshwater aquifer metal-rich particle clone |
| JEG.XSB2 | 1 | Actinobacteria | Rhodococcus erythropolis strain NVI (AY147846) | 99 | Isolated from Atlantic salmon |
| JEG.E12, JEG.G1 | 11 | CFB | Flavobacterium limicola (AB075232) | 97 | Psychrophilic isolate from freshwater sediments |
| JEG.F1, JEG.D11 | 8 | CFB | Flavobacterium frigoris (AJ557887) | 97 | Psychrophilic isolate from Antarctic lake microbial mat |
| JEG.B6 | 6 | CFB | FJS5 (AY315161) | 97 | Franz-Josef Glacier isolate |
| JEG.C5 | 6 | CFB | GKS2-106 (AJ290025) | 98 | High mountain lake clone |
| JEG.B12 | 2 | CFB | PRD01a005B (AF289153) | 95 | Typical freshwater bacterial clone |
| JEG.XSF2 | 2 | Holophaga/Acidobacteria | 1091-1 (AY326536) | 96 | Soil clone |
| JEG.D6 | 1 | Loosely affiliated with Planctomycetales | LB3-13 (AF173818) | 91 | Antarctic lake mat clone |
| JEG.XSF4 | 1 | Planctomycetales | SC-I-11 (AJ252614) | 98 | Soil clone |
| JEG.B7 | 3 | Verrucomicrobium | C019 (AF013522) | 91 | Soil clone |
Fully, bidirectionally sequenced clones representing HhaI RFLP groupings.
Phylum-level grouping. Names in parentheses are groupings shown in Fig. 2.
Most closely related sequence in the GenBank public database, based upon BLASTN search.
Percent similarity to nearest neighbor, based upon BLASTN search.
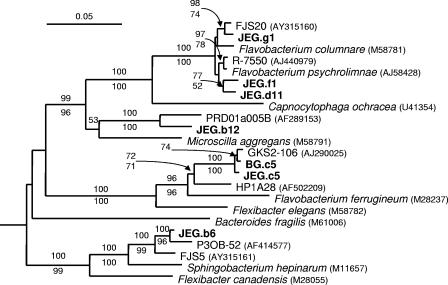
The JEG clone library composition was more diverse in phyla other than the Proteobacteria than was the BG clone library. Members of the CFB (24.8% of clones), which were rare in the BG clone library (see above), and Actinobacteria (5.3% of clones), which were absent in the BG clone library, were well represented (Table 2; Fig. 3). Other phylum-level groups, including the Planctomycetales (1.5% of clones), Holophaga/Acidobacteria (1.5% of clones), and Verrucomicrobium (2.2% of clones), were present and represented by only one or a few clones (Tables 2 and 4). One clone, distantly related to sequences obtained from an Antarctic lake microbial mat (20), was not strongly associated with any known bacterial phyla but was loosely affiliated with the Planctomycetales (Table 4).
FIG. 3.
Phylogenetic analysis of Cytophaga/Flavobacterium/Bacteroides 16S rRNA gene clones from JEG and BG. This phylogenetic tree was rooted with Escherichia coli (GenBank accession number J01695). A mask of 1,074 nucleotides, including all nonambiguously aligned positions, spanning nearly the full length of the 16S rRNA gene, was included. Bootstrap values (100 replications) generated by the neighbor-joining method are shown above relevant nodes, and those generated by maximum-parsimony analysis are shown below the nodes. Only bootstrap values above 50 are shown. Sequences from formally named isolates are in italics, sequences from environmental gene clones and unnamed isolates are in plain text, and sequences from the subglacial samples are in boldface. GenBank accession numbers of the sequences from other studies are included.
Dot blot hybridization.
Probe Com1212 hybridized to 16S rRNA gene PCR products amplified from all samples at both glaciers, and probe Gall217 hybridized to 10 of 14 samples from BG but to none of the 8 samples at JEG (Table 5). We note that normalization to the Eub338 probe provides relative quantitation with the same probe but does not allow determination of the absolute abundance of genes in these environmental samples. Further, the dot blot hybridization procedure also utilizes PCR products generated by the same methods as for the clone library and thus is likely subject to the same potential biases as noted above. However, detailed examination of the results by environmental sample type (e.g., subglacial, ice marginal, sediment, basal ice, etc.) revealed interesting patterns at each glacier.
Hybridization to the Gall217 probe occurred in all the subglacial stream samples from BG (samples a to g), with a signal that was approximately an order of magnitude greater than that in the ice marginal stream samples (samples j and k) (Table 5). This suggests that conditions in the subglacial environment favor the growth of organisms detected by this probe. This conclusion was further supported by the BG borehole hybridization data (samples h and i). The Gall217 hybridization signal in the sample from the subglacial water from the lower borehole (sample h) was at least 11 times greater than that for the supraglacial water from the upper borehole (sample i). The lower borehole signal was similar in magnitude to that of the subglacial streams (samples a to g). Interestingly there was no hybridization signal with probe Gall217 in the basal ice samples (samples l to n) from BG, compared to the strong signal for the subglacial waters (samples a to h).
The JEG samples can be placed into three groups based on their hybridization to probe Com1212 (Table 5). In all cases, a strong signal was obtained with the Com1212 probe relative to Eub338. The supraglacial water sample (sample o) and the proglacial subsurface sediment sample (sample p) had a significantly lower signal for the Com1212 probe than the subglacial sediment sample (sample q) and basal ice samples (samples r to u), and the subglacial water sample (sample v) had a ∼3× higher signal for the Com1212 probe than the average for the basal ice samples. The hybridization signal for the Gall217 probe to all of the samples at JEG was below the detection limit, further demonstrating that the distribution of these sequences was limited to BG.
DISCUSSION
Subglacial microbial community composition.
Many 16S rRNA gene clones from BG were greater than 95% similar to sequences from JEG (data not shown), placing them in the same genus (56). This is somewhat surprising, since these samples were collected from geographically distant and geologically different catchments. The clone libraries of both of the BG and JEG subglacial communities were dominated by Proteobacteria, especially members of the Betaproteobacteria subgroup (Table 2). In particular, sequences found in the “Com1212” cluster (Fig. 2) were highly represented in both clone libraries and appear to be present in all of the glacial environments (i.e., subglacial streams, basal ice, supraglacial water, and marginal streams), although there are indications that this group is enriched in subglacial samples (Table 5). A similar dominance by Com1212 relatives was observed in clone libraries from soils collected at the terminus of a Peruvian high-altitude glacier (44) and a collection of isolates from subglacial samples of two New Zealand glaciers (14). 16S rRNA gene sequences or cultures from this cluster have been obtained from other low-temperature environments, including glacial ice cores (52), Antarctic marine (27) and lake (40) waters, and other uncultured organisms from cold or frozen environments (18, 20). Thus, the “Com1212” cluster appears to be ubiquitous in cold and frozen environments and may be predominant in many subglacial environments.
Despite these similarities between the two systems, there are also important differences in the clone library compositions of the two sites. It is intriguing that sequences closely related to Gallionella ferruginea or the genus Thiobacillus were highly abundant in the BG clone library but were rare (Thiobacillus relatives) or entirely absent (Gallionella ferruginea relatives) in the JEG clone library (Tables 3 and 4). This limited distribution is supported by the dot blot results: the Gall217 probe consistently gives hybridization signals above the detection limit only with subglacial samples from BG. Cultured members of both the Gallionella ferruginea and Thiobacillus clusters are involved in iron and/or sulfur cycling. Although physiology cannot be deduced from phylogeny, especially in the case where 16S rRNA gene sequence similarity is as low as approximately 95%, the presence of iron- and/or sulfur-oxidizing organisms at BG but not at JEG would be consistent with the chemical data from the two sites (see below). Assuming such a correlation, the data suggest that there were forcing factors on microbial community composition beyond the broad physical and chemical conditions.
Members of the CFB phylum were abundant in the JEG clone library but were rare in the BG clone library. It is intriguing that CFB are rare in BG, as they are numerically significant in nearly every aquatic and soil system examined (32, 64, 66). All cultivated members of this phylum are heterotrophic (32), and the high representation of CFB in the JEG clone library is consistent with an important role for heterotrophy in the subglacial system at JEG.
No clones were found to be related to known SRB at either glacier. Their absence is somewhat surprising, since sulfate is the dominant anion in the waters at both glaciers and sulfate-reducing bacteria have been cultured from basal ice and water at JEG in a previous study (54). The lack of SRB, however, may be an artifact of a heterogeneous spatial or temporal distribution of SRB, sampling errors, or the poor ability of clone libraries to detect rare (<1 to 2% of the population), but potentially geochemically significant, organisms.
The ubiquity of the “Com1212” group and the restricted distribution of the “Gall217” group in the clone libraries were confirmed by blot hybridization to PCR products of 16S rRNA genes amplified from DNA extracted from samples collected over several years (Table 5). At the broad scale there was a degree of interannual stability of the subglacial microbial communities at both BG and JEG. Such stability supports the hypothesis that there is an endogenous community specifically associated with the subglacial environment.
Hybridization to the Gall217 probe at BG in subglacial water samples (samples a to h) was approximately an order of magnitude greater than that in ice melt (supraglacial water) and ice marginal stream samples (samples i-k) (Table 5). Similarly, subglacial waters at JEG were significantly enriched in “Com1212” group 16S rRNA genes relative to the input supraglacial waters, basal ice, or subglacial sediments (Table 5). Interestingly, the proglacial sediment ∼100 m from the glacier terminus exhibited a low Com1212 signal similar to that of the supraglacial input waters and noticeably lower than that of the subglacial sediment, basal ice, or subglacial waters. Similarly, Nemergut et al. (44) observed a predominance of Betaproteobacteria related to “Com1212” in clone libraries of proglacial sediments at a Peruvian high-altitude glacier terminus and that this predominance decreases with increasing distances from the glacier terminus. Together, these data strongly suggest that the subglacial environment is selective for specific groups of organisms relative to supraglacial and proglacial (ice marginal) environments and furthermore that there is an endogenous subglacial microbial community that is distinct from other environments on and near the glacier.
Controls on microbial community composition.
Subglacial systems can be considered extreme environments in several respects. They are permanently dark, oligotrophic, and constantly cold (∼0°C). Microbes in these environments must be capable of regular freezing and thawing and possibly seasonal cycling between highly oxygenated and anoxic conditions (54, 61, 63). Such conditions may, in part, explain the low species richness we observed in the clone libraries from these sites (discussed above), since only a limited set of microbes are capable of surviving and thriving in such an environment. The extreme nature of these environments is consistent with the finding that several of the clones from each site are unrelated to sequences available in the public databases.
The predominance of Betaproteobacteria in the “Com1212” group in cold and especially glaciated environments (discussed above) implies that the factor(s) which selects for this group is present in all of these environments. While the nature of this factor is unclear, it seems unlikely that any chemical factor is shared among all of these disparate environments. Most likely, organisms in the “Com1212” group thrive in subglacial systems due to adaptation to physical factors common to these environments (such as low, constant temperature; lack of light; or increased availability of mineral substrates due to increased surface area and exposure of fresh mineral surfaces during subglacial grinding and crushing).
Microbial activity has been noted as a major driving force behind mineral weathering and redox chemistry in many natural systems (see, e.g., references 13 and 43). However, links between redox chemistry and microbial populations have yet to be demonstrated for subglacial environments. Direct microscopic cell counts and DOC concentrations indicated the presence of significant microbial populations and available organic carbon, albeit at low concentrations, at both JEG and BG, supporting the possibility of a microbial influence on subglacial chemical weathering. As shown by our analysis of the Ca/SO4 and S ratios (Table 1) and confirmed at JEG by δ34S analysis (53), the sulfate is derived from different mineral weathering processes at the two glaciers. Sulfate at BG is derived from pyrite via sulfide oxidation (see the equation in “Water chemistry” in Results), whereas the sulfate at JEG is derived from gypsum dissolution. Thus, it is possible that these mineralogical differences are directly related to the compositions of the microbial communities in the two subglacial systems. The BG microbial system may be biased toward organisms that may affect sulfide and sulfur oxidation, such as Gallionella, Thiobacillus, and sulfur-oxidizing Epsilonproteobacteria, but the JEG community may be biased away from these groups. Thus, while we have not directly demonstrated a role for these microorganisms in subglacial chemical weathering processes, it is reasonable to assume that the correlation of microbial community composition and chemical weathering products is significant.
Wider relevance to other subglacial systems.
There are significant similarities between the subglacial water chemistry at BG (4; this study) and that at Haut Glacier d'Arolla, Switzerland (53), where subglacial microbial processes have also been inferred (see, e.g., references 51 and 60). This is interesting since the two glaciers are on different continents but both overlie meta-sedimentary rocks of a broadly similar composition, indicating that the water chemistry (and possibly the microbial community) is controlled by bedrock mineralogy. This suggests that the observations at BG are not unique and that, on a broader scale, microbes may be important in catalyzing mineral weathering reactions in subglacial environments. Other studies related to acid mine drainage document sulfide oxidation occurring at low (3°C) (34) and even subzero (−2°C) (41) temperatures, indicating that low temperatures do not preclude this reaction. Furthermore, the widespread predominance of the “Com1212” group in clone libraries (44; this study) and culture collections (14) from subglacial or recently deglaciated environments indicates that studies of individual subglacial microbial communities may have widespread implications for other subglacial environments.
Eleven percent of the continents is currently ice covered, and this proportion was 30% during Pleistocene glaciations and ∼100% during the long periods of pervasive low-latitude glaciation (“snowball Earth”) in the Neoproterozoic era (24). Hence, an understanding of the rates of sulfide oxidation in cryospheric systems has important ramifications for biogeochemical Fe and S cycling. However, the relative magnitude of biotic versus abiotic sulfide oxidation in subglacial environments remains to be determined.
Acknowledgments
We thank F. Carsey and the NASA Jet Propulsion Laboratory for funding transport of B. D. Lanoil to BG. BG fieldwork was funded by NSF grants EAR-9404465, EAR-9706180, and OPP-9818251 to S. P. Anderson. The JEG field program was supported by NSERC Discovery Grants to M. Sharp, a GSA grant to M. Skidmore, and logistical support from PCSP.
R. Anderson and M. Loso provided field assistance and logistical support at BG in 2002. We thank N. Humphrey and J. Harper for providing access to their boreholes at BG in 2002. J. Barker, A. Arendt, and R. Young provided field assistance at JEG. M. Bhatia kindly provided genomic DNA samples for JEG in 2002. We thank the Nunavut Research Institute and the peoples of Grise Fjord and Resolute Bay for permission to conduct fieldwork at JEG.
This is PCSP contribution no. 00604.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent oligonucleotide probing of whole cells for determinative, phylogenetic and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R. S., S. P. Anderson, K. C. MacGregor, E. D. Waddington, S. O'Neel, C. Riihimaki, and M. G. Loso. 2004. Strong feedbacks between hydrology and sliding of a small alpine glacier. J. Geophys. Res. 109:10.1029/2004JF000120. [Google Scholar]
- 4.Anderson, S. P., J. I. Drever, C. D. Frost, and P. Holden. 2000. Chemical weathering in the foreland of a retreating glacier. Geochim. Cosmochim. Acta 64:1173-1189. [Google Scholar]
- 5.Barker, J. D., M. J. Sharp, and R. J. Turner. Organic carbon abundance and dynamics in glacier systems. Submitted for publication.
- 6.Bhatia, M. 2004. Molecular characterization of bacterial communities associated with a high Arctic polythermal glacier. M.Sc. thesis. University of Alberta, Edmonton, Alberta, Canada.
- 7.Bottrell, S. H., and M. Tranter. 2002. Sulphide oxidation under partially anoxic conditions at the bed of the Haut Glacier d'Arolla, Switzerland. Hydrol. Process 16:2363-2368. [Google Scholar]
- 8.Campen, R. K., T. Sowers, and R. B. Alley. 2003. Evidence of microbial consortia metabolizing within a low-latitude mountain glacier. Geology 31:231-234. [Google Scholar]
- 9.Christner, B. C. 2002. Incorporation of DNA and protein precursors into macromolecules by Bacteria at −15°C. Appl. Environ. Microbiol. 68:6435-6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colwell, R. K., and J. A. Coddington. 1994. Estimating terrestrial biodiversity through extrapolation. Phil. Trans. R. Soc. London B 345:101-118. [DOI] [PubMed] [Google Scholar]
- 11.Copland, L., and M. Sharp. 2001. Mapping of thermal and hydrological conditions beneath a polythermal glacier with radioecho sounding. J. Glaciol. 47:232-242. [Google Scholar]
- 12.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrlich, H. L. 1998. Geomicrobiology: its significance for geology. Earth-Sci. Rev. 45:45-60. [Google Scholar]
- 14.Foght, J. M., J. Aislabie, S. Turner, C. E. Brown, J. Ryburn, D. J. Saul, and W. Lawson. 2004. Culturable bacteria in subglacial sediments and ice from two southern hemisphere glaciers. Microb. Ecol. 47:329-340. [DOI] [PubMed] [Google Scholar]
- 15.Gaidos, E. J., B. D. Lanoil, T. Thorsteinsson, A. Graham, M. L. Skidmore, S. Han, T. Rust, and B. Popp. 2004. A viable microbial community in a subglacial volcanic crater lake, Iceland. Astrobiology 4:327-344. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs, M. T., and L. R. Kump. 1994. Global chemical erosion during the last glacial maximum and the present: sensitivity to changes in lithology and hydrology. Paleoceanography 9:529-543. [Google Scholar]
- 17.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-201. In E. Stackebrandt and M. Goodfellow (ed.), Sequencing and hybridization techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 18.Glockner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon, D. A., and S. J. Giovannoni. 1996. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl. Environ. Microbiol. 62:1171-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon, D. A., J. C. Priscu, and S. J. Giovannoni. 2000. Origin and phylogeny of microbes living in permanent Antarctic lake ice. Microb. Ecol. 39:197-202. [DOI] [PubMed] [Google Scholar]
- 21.Hallbeck, L., F. Stahl, and K. Pedersen. 1993. Phylogeny and phenotypic characterization of the stalk-forming and iron-oxidizing bacterium Gallionella ferruginea. J. Gen. Microbiol. 139:1531-1535. [DOI] [PubMed] [Google Scholar]
- 22.Harper, J. T., N. F. Humphrey, T. W. Pfeffer, S. V. Huzurbazar, D. B. Bahr, and B. C. Welch. 2001. Spatial variability in the flow of a valley glacier: deformation of a large array of boreholes. J. Geophys. Res. Solid Earth 106:8547-8562. [Google Scholar]
- 23.Heppenstall, K. 2002. Chemical weathering in a glaciated carbonate catchment, Canadian High Arctic: implications for subglacial hydrology. M.Sc. thesis. University of Alberta, Edmonton, Alberta, Canada.
- 24.Hoffman, P. F., A. J. Kaufman, G. P. Halverson, and D. P. Schrag. 1998. A neoproterozoic snowball Earth. Science 281:1342-1346. [DOI] [PubMed] [Google Scholar]
- 25.Hubbard, B., and P. Nienow. 1997. Alpine subglacial hydrology: research status and implications for glacial geology. Quat. Sci. Rev. 16:939-956. [Google Scholar]
- 26.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irgens, R. L., J. J. Gosink, and J. T. Staley. 1996. Polaromonas vacuolata gen. nov., sp. nov., a psychrophilic, marine, gas vacuolate bacterium from Antarctica. Int. J. Syst. Bacteriol. 46:822-826. [DOI] [PubMed] [Google Scholar]
- 28.Karl, D. M., D. F. Bird, K. Bjorkman, T. Houlihan, R. Shackelford, and L. Tupas. 1999. Microorganisms in the accreted ice of Lake Vostok, Antarctica. Science 286:2144-2147. [DOI] [PubMed] [Google Scholar]
- 29.Kemp, P. F., and J. Y. Aller. 2004. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 47:161-177. [DOI] [PubMed] [Google Scholar]
- 30.Kerr, J. W. 1972. Map 1358A geology: Dobbin Bay, District of Franklin. Geological Survey of Canada, Ottawa, Canada.
- 31.Kirchman, D., J. Sigda, R. Kapuscinski, and R. Mitchell. 1982. Statistical analysis of the direct count method for evaluating bacteria. Appl. Environ. Microbiol. 44:376-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 33.Lafreniere, M. J., and M. J. Sharp. 2004. The concentration and fluorescence of dissolved organic carbon (DOC) in glacial and nonglacial catchments: interpreting hydrological flow routing and DOC sources. Arctic Antarctic Alpine Res. 36:156-165. [Google Scholar]
- 34.Langdahl, B. R., and K. Ingvorsen. 1997. Temperature characteristics of iron solubilisation and 14C carbon assimilation in ore samples from Citronen Fjord, North Greenland (83° northern latitude). FEMS Microbiol. Ecol. 23:275-283. [Google Scholar]
- 35.Lanoil, B. D., M. T. La Duc, S. T. Sweet, K. H. Nealson, and R. Sassen. 2001. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leduc, L. G., and G. D. Feroni. 1994. The chemotrophic bacterium Thiobacillus ferroxidans. FEMS Microbiol. Rev. 14:103-120. [Google Scholar]
- 37.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 38.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Bucher, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacGregor, K. R. 2002. Modeling and field constraints on glacier dynamics, erosion, and alpine landscape evolution. Ph.D. thesis. University of California, Santa Cruz.
- 40.Madigan, M. T., D. O. Jung, C. R. Woese, and L. A. Achenbach. 2000. Rhodoferax antarcticus sp. nov., a moderately psychrophilic purple nonsulfur bacterium isolated from an Antarctic microbial mat. Arch. Microbiol. 173:269-277. [DOI] [PubMed] [Google Scholar]
- 41.Meldrum, J. L., H. E. Jamieson, and L. D. Dyke. 2001. Oxidation of mine tailings from Rankin Inlet, Nunavut, at subzero temperatures. Can. Geotechnol. J. 38:957-966. [Google Scholar]
- 42.Mountfort, D. O., H. F. Kaspar, M. Downes, and R. A. Asher. 1999. Partitioning effects during terminal carbon and electron flow in sediments of a low-salinity meltwater pond near Bratina Island, McMurdo Ice Shelf, Antarctica. Appl. Environ. Microbiol. 65:5493-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nealson, K. H., and D. A. Stahl. 1997. Microorganisms and biogeochemical cycles: what can we learn from layered microbial communities?, p. 5-34. In K. H. Nealson and J. F. Banfield (ed.), Geomicrobiology: interactions between microbes and minerals, vol. 35. Mineralogical Society of America, Washington, D.C. [Google Scholar]
- 44.Nemergut, D. R., S. P. Anderson, C. C. Cleveland, A. P. Martin, A. E. Miller, A. Seimon, and S. K. Schmidt. Microbial community succession in unvegetated, recently-deglaciated soils. Submitted for publication. [DOI] [PubMed]
- 45.Nordstrom, D. K., and G. Southam. 1997. Geomicrobiology of sulfide mineral oxidation, p. 361-390. In K. H. Nealson and J. Banfield (ed.), Geomicrobiology: interactions between microbes and minerals, vol. 35. Mineralogical Society of America, Washington, D.C. [Google Scholar]
- 46.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identification and counting aquatic flora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 48.Price, P. B., and T. Sowers. 2004. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc. Natl. Acad. Sci. USA 101:4631-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Priscu, J. C., E. E. Adams, W. B. Lyons, M. A. Voytek, D. W. Mogk, R. L. Brown, C. P. McKay, C. D. Takacs, K. A. Welch, C. F. Wolf, J. D. Kirshtein, and R. Avci. 1999. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286:2141-2144. [DOI] [PubMed] [Google Scholar]
- 50.Priscu, J. C., and B. C. Christner. 2003. Earth's icy biosphere, p. 130-145. In A. Bull. (ed.), Microbial diversity and bioprospecting. ASM Press, Washington, D.C.
- 51.Sharp, M., J. Parkes, B. Cragg, I. J. Fairchild, H. Lamb, and M. Tranter. 1999. Widespread bacterial populations at glacier beds and their relationship to rock weathering and carbon cycling. Geology 27:107-110. [Google Scholar]
- 52.Sheridan, P. P., V. I. Miteva, and J. E. Brenchley. 2003. Phylogenetic analysis of anaerobic psychrophilic enrichment cultures obtained from a Greenland glacier ice core. Appl. Environ. Microbiol. 69:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skidmore, M. L. 2001. Hydrology, microbiology, and carbon cycling in a high Arctic glacial environment. Ph.D. thesis. University of Alberta, Edmonton, Alberta, Canada.
- 54.Skidmore, M. L., J. M. Foght, and M. J. Sharp. 2000. Microbial life beneath a high arctic glacier. Appl. Environ. Microbiol. 66:3214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skidmore, M. L., and M. J. Sharp. 1999. Drainage system behaviour of a High-Arctic polythermal glacier. Ann. Glaciol. 28:209-215. [Google Scholar]
- 56.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 57.Suzuki, M., and S. Giovannoni. 1995. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theron, J., and T. E. Cloete. 2000. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit. Rev. Microbiol. 26:37-57. [DOI] [PubMed] [Google Scholar]
- 59.Tranter, M., G. H. Brown, A. Hodson, A. Gurnell, and M. Sharp. 1994. Variations in the nitrate concentration of glacial runoff in alpine and arctic polar environments. Int. Assoc. Hydrol. Sci. Publ. 223:299-310. [Google Scholar]
- 60.Tranter, M., M. J. Sharp, H. R. Lamb, G. H. Brown, B. P. Hubbard, and I. C. Willis. 2002. Geochemical weathering at the bed of Haut Glacier d'Arolla, Switzerland—a new model. Hydrol. Process 16:959-993. [Google Scholar]
- 61.Tranter, M., M. Skidmore, and J. L. Wadham. 2005. Hydrological controls on microbial communities in subglacial environments. Hydrol. Process 19:995-998. [Google Scholar]
- 62.Twickler, M. S., M. J. Spencer, W. B. Lyons, and P. A. Mayewski. 1986. Measurement of organic carbon in polar snow samples. Nature 320:156-157. [Google Scholar]
- 63.Wadham, J. L., S. H. Bottrell, M. Tranter, and R. Raiswell. 2004. Stable isotope evidence for microbial sulphate reduction at the bed of a polythermal high Arctic glacier. Earth Planet. Sci. Lett. 219:341-355. [Google Scholar]
- 64.Weller, R., F. O. Glockner, and R. Amann. 2000. 16S rRNA-targeted oligonucleotide probes for the in situ detection of members of the phylum Cytophaga-Flavobacterium-Bacteroides. Syst. Appl. Microbiol. 23:107-114. [DOI] [PubMed] [Google Scholar]
- 65.Winkler, G. R., M. L. Silberman, A. Grantz, R. J. Miller, and E. M. MacKevett, Jr. 1981. Geologic map and summary geochronology of the Valdez quadrangle, southern Alaska. Open file report 80-892-A. U.S. Geological Survey, Reston, Va.
- 66.Zwart, G., B. C. Crump, M. P. K. V. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28: 141-155. [Google Scholar]