Abstract
The abundance and activity of methane-oxidizing bacteria (MOB) in the water column were investigated in three lakes with different contents of nutrients and humic substances. The abundance of MOB was determined by analysis of group-specific phospholipid fatty acids from type I and type II MOB, and in situ activity was measured with a 14CH4 transformation method. The fatty acid analyses indicated that type I MOB most similar to species of Methylomonas, Methylomicrobium, and Methylosarcina made a substantial contribution (up to 41%) to the total bacterial biomass, whereas fatty acids from type II MOB generally had very low concentrations. The MOB biomass and oxidation activity were positively correlated and were highest in the hypo- and metalimnion during summer stratification, whereas under ice during winter, maxima occurred close to the sediments. The methanotroph biomass-specific oxidation rate (V) ranged from 0.001 to 2.77 mg CH4-C mg−1 C day−1 and was positively correlated with methane concentration, suggesting that methane supply largely determined the activity and biomass distribution of MOB. Our results demonstrate that type I MOB often are a large component of pelagic bacterial communities in temperate lakes. They represent a potentially important pathway for reentry of carbon and energy into pelagic food webs that would otherwise be lost as evasion of CH4.
Microbiological oxidation of methane takes place in many kinds of aquatic systems and soils (28, 32, 33, 58) and plays an important role in the global budget of this greenhouse gas (15, 43). It can proceed both under aerobic and anaerobic conditions, but the latter seems to be extensive primarily in waters with high ionic strength, e.g., saline lakes and marine systems (29, 31, 55). Many strains capable of aerobic methane oxidation have been isolated and characterized, and phylogenetically all belong to either the γ-proteobacteria (commonly referred to as “type I” methanotrophs) or the α-proteobacteria (“type II” methanotrophs) (10, 12, 24).
Aerobic methane-oxidizing bacteria (MOB) in the water columns of lakes consume a significant part of the methane produced in the sediment or in anaerobic layers of water (46, 54). Besides metabolizing methane that would otherwise be emitted to the atmosphere, they represent a route for reentry of carbon back into the food-webs (2, 26, 30, 46). In spite of this, direct studies of MOB populations in the pelagic systems of lakes are rare and, with the exception of a small oxbow lake (45) and a tropical hydropower reservoir (20, 21), very little is known about the abundance and population structure of pelagic methanotrophs in lakes.
The aerobic methanotrophic bacteria contain some phospholipid fatty acids (PLFAs) that seem to be unique for this group of organisms (16:1ω8c in type I MOB and 18:1ω8c in type II MOB (10, 40). Analysis of these PLFAs has been shown to be a very sensitive tool for the direct study of populations of type I and type II MOB in samples from several different kinds of ecosystems (7, 9, 16, 39, 51).
In the present study we use PLFA analysis to characterize the populations of MOB in the water columns of lakes and estimate the relative contribution of MOB to total water column bacteria. The efficiency of the PLFA method to determine bacterial abundance was evaluated by comparison with direct cell counts using flow cytometry. By combining the measurements of abundance of MOB with their activity estimated with a 14CH4 transformation method, we were able to estimate the in situ biomass-specific activity of this particular group of bacteria. Samples were collected during summer and winter conditions in three temperate lakes differing in nutrient status and concentrations of dissolved organic matter.
MATERIALS AND METHODS
Study lakes.
Three lakes in south central Sweden were sampled: Mårn (64°95′96N, 15°04′15W), eutrophic with intermediate water color; Illersjön (64°94′96N, 14°52′66W), eutrophic with low water color; and Lillsjön (65°04′01N, 15°19′83W), oligotrophic with high water color (for details about water chemistry, see reference 4). All lakes were sampled twice in 1999 and 2000: once in summer during stratified conditions and once from ice in winter.
Temperature, oxygen, and water sampling.
Profiles of temperature and oxygen concentrations were obtained with an oxygen probe (Orion model 35). Four to seven depths were sampled, ranging from 0.5 m below the surface to 0.5 m above the sediments. Water samples were pumped from each depth with a submersible pump (Amazon 10; Awimex International AB, Sweden).
Analysis of PLFAs.
Duplicate subsamples of 40 to 250 ml were filtered using 47-mm 0.2-μm-pore-size filters (polyvinylidene difluoride; Millipore) and frozen awaiting further analysis. The methods for extraction of total lipids, solid-phase extraction of the polar fraction, transformation of the fatty acids into fatty acid methyl esters (FAMEs) by alkaline methanolysis, dimethyl disulfide derivatization of monounsaturated PLFAs, and identification and quantification of FAMEs with GC-FID and GC-MS have been described previously (38, 52, 57). It was not possible to analyze all replicate filters for PLFAs. For three water samples where the duplicate filters were analyzed, the coefficient of variation for the total PLFA concentration varied from 11 to 23%.
Biomass and abundance.
The total bacterial biomass and carbon content were determined from the concentrations of the PLFAs 14:0, i15:0, a15:0, 15:0, all 16:1, 16:0, i17:0, 17:0, 18:1ω7, 18:1ω8, and cy19:0, which are all of general bacterial origin (34, 60), assuming a total bacterial content of PLFA of 100 μmol g−1 dry weight (dw) of cells and that carbon constituted 50% of the dw (59). The bacterial abundance (i.e., the cell numbers) could then be calculated from bacterial cell volumes in the lakes and the relation between the dw and cell volume for bacterial cells according to the method of Loferer-Krössbacher et al. (36) (dw = 435 × V 0.86 fg). The average bacterial cell volume in the water samples, estimated by image analyzed fluorescence microscopy (5), was 0.12 μm3 for epi- and hypolimnion in Illersjön and Mårn, ranging from 0.08 to 0.13 μm3.
The biomass and cell numbers of MOB were calculated from the concentrations of 16:1ω8c and 18:1ω8c, considered to occur almost exclusively in type I and II MOB, respectively. The same conversion factors were used, and we assumed that 16:1ω8c and 18:1ω8c constituted 25 and 50% of the PLFAs in type I and type II MOB, respectively (10, 40, 61).
The total bacterial abundance based on the PLFA analysis was compared to an independent estimate of bacterial abundance, obtained by using flow cytometry (Becton Dickinson FACSCalibur; CellQuest 3.1 software) after staining with Syto 13 (Molecular Probes) according to the method of del Giorgio et al. (18).
Methane concentrations.
For each depth sampled, four replicate 330-ml infusion bottles were completely filled with lake water, followed by preservation with NaOH raising pH to above 11 (47). Methane concentrations were analyzed by a headspace equilibration method (3).
In situ methane oxidation.
The rates of methane oxidation in lake water were measured with a 14CH4 incorporation method as described in detail previously (3). In brief, water samples were amended with 14CH4 and incubated at actual depths for 4 to 8 h. The incubations were terminated by NaOH addition. In the lab, subsamples of 20 to 30 ml were filtered through 0.22-μm-pore-size mixed cellulose ester filters (Millipore GSWP). The filtrates were purged with air to remove labeled methane, and the radioactivity of filters and filtrates was measured by liquid scintillation counting. The total radioactivity in these fractions represents the total oxidation of methane as the sum of incorporation into cell biomass, substances released as dissolved organic carbon, and dissolved inorganic carbon from respiration.
Statistical analyses.
The relationships between bacterial cell numbers determined with the two independent methods (flow cytometry versus calculations based on PLFA concentrations) were investigated with single linear regression analysis. Both single and multiple linear regression analyses were used to evaluate relationships between methane oxidation and environmental parameters and were performed by using Kaleidagraph 3.6.2 (Synergy Software) and JMP 3.1.5 (SAS Institute, Inc.) software, respectively.
RESULTS
In all, 23 different PLFAs could be identified and quantified, including trans isomers of monounsaturated fatty acids. The total PLFA concentration ranged from 3.8 to 56 pmol ml−1, with the highest concentrations toward the lake surface and the sediment, in most profiles.
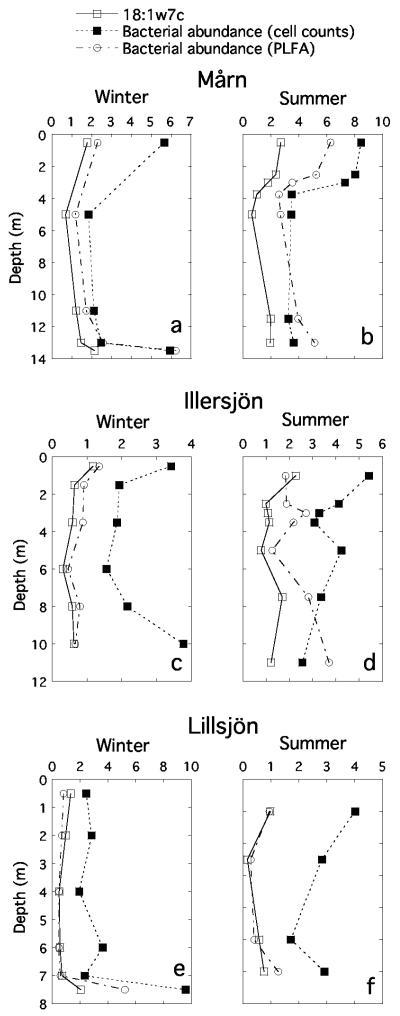
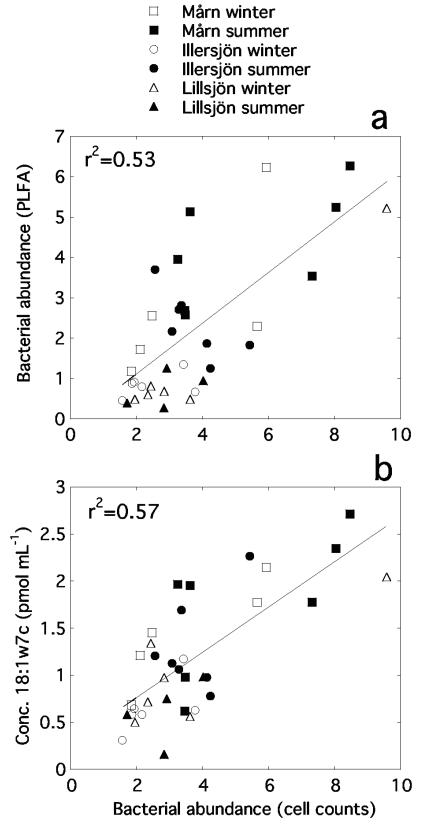
The two independent methods to determine total bacterial abundance showed the same trends within the lakes, but the absolute abundances differed among the lakes (Fig. 1). Treatment of all data points in one single regression resulted in a strong linear relationship (Fig. 2a; r2 = 0.53, P < 0.0001). Some individual PLFAs were also positively related to the bacterial abundance measured with flow cytometry, most strongly for 18:1ω7c, which gave an even better indication of bacterial abundance than the total concentration of bacterial PLFAs (Fig. 1 and 2b; r2 = 0.57, P < 0.0001).
FIG. 1.
Total bacterial abundance in lake profiles (millions of cells ml−1), as estimated by direct cell counts and from concentrations of bacterial phospholipid fatty acids. For comparison, concentrations of the common bacterial fatty acid 18:1ω7c (pmol ml−1) are also shown. The profiles are from lakes Mårn (a and b), Illersjön (c and d), and Lillsjön (e and f).
FIG. 2.
Relationships between bacterial abundance estimated by direct cell counts and concentrations of phospholipid fatty acids in lakewater. Units for bacterial abundance are in millions of cells per milliliter.
The PLFA 16:1ω8c, considered to be unique for type I MOB, occurred in most samples. The concentrations correlated very closely to those of 16:1ω6c and 16:1ω5t but were more weakly related to other general bacterial PLFAs (Table 1). This is strong evidence that the two latter fatty acids also come from type I methanotrophs in these lakes. In contrast, the type II MOB signature fatty acid 18:1ω8c was only detected in roughly half of the samples, with no obvious distributional pattern and always in low concentrations. In the seven samples showing highest oxidation (2.6 to 37.5 mg CH4-C m−3 day−1), the biomass of type II MOB was only 0.0 to 4.8% (mean 2.2%) of that of type I. In 11 of the 20 samples where the type II signature 18:1ω8c was detected, the type II biomass was above 10% of the type I, but in all of these samples the oxidation rate was very low (below 0.15 mg CH4-C m−3 day−1).
TABLE 1.
Correlation coefficients (r) between concentrations of typical methanotrophic (16:1ω8c, 16:1ω6c, and 16:1ω5t) and general bacterial (16:0, 18:1ω7c, 16:1ω7c, and total bacterial) phospholipid fatty acids in lake water
| Fatty acid |
r for fatty acid:
|
|||||
|---|---|---|---|---|---|---|
| 16:1ω6c | 16:1ω5t | 16:0 | 18:1ω7c | 16:1ω7c | Total bacterial | |
| 16:1ω8c | 0.98 | 0.98 | 0.24 | 0.38 | 0.82 | 0.55 |
| 16:1ω6c | 0.99 | 0.19 | 0.35 | 0.80 | 0.52 | |
| 16:1ω5t | 0.20 | 0.34 | 0.81 | 0.52 | ||
| 16:0 | 0.79 | 0.66 | 0.92 | |||
| 18:1ω7c | 0.74 | 0.85 | ||||
| 16:1ω7c | 0.88 | |||||
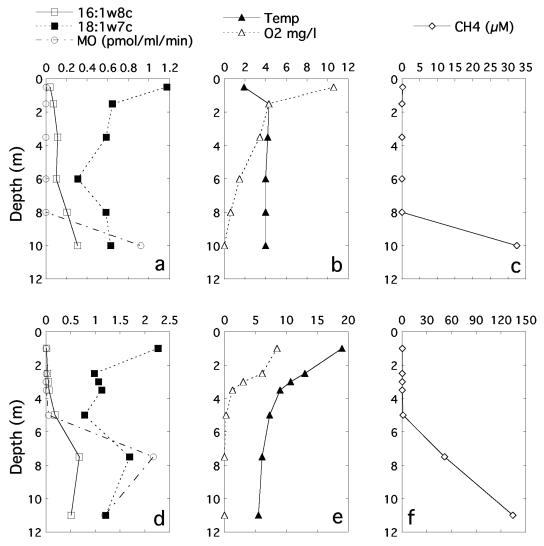
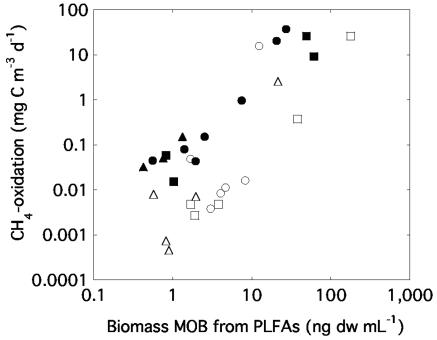
Because the fatty acid composition implied that type I MOB strongly dominated the MOB communities, we estimated the total biomass of methanotrophs from the concentration of 16:1ω8c. In all individual samples, the biomass ranged from 0.0 to 178 ng dw ml−1. By comparing the contribution of 16:1ω8c to the total concentration of bacterial fatty acids, we estimate that type I MOB account for up to 41% of the total bacterial biomass. During summer conditions the MOB biomass generally paralleled the CH4 consumption rate (exemplified with Illersjön in Fig. 3), being highest in the hypolimnion. During winter, the MOB abundance gradually increased toward the sediment, while the oxidation rate was very low, with the exception of the samples taken closest to the sediment (Fig. 3). Generally for all three lakes, the oxidation rate ranged from 0.0004 to 37.5 mg CH4-C m−3 day−1. Six samples showing an especially high oxidation rate, all had low oxygen concentrations (<25 μM O2), high methane concentrations (all except one above 33 μM CH4), and low temperature (3.0 to 6.1°C). The general positive relationship between MOB abundance and total oxidation, estimated with independent methods, is illustrated by a single regression with all data (Fig. 4, y = 2.0 + 0.18x; r2 = 0.37, P = 0.0003; n = 34).
FIG. 3.
Depth profiles of methanotrophic biomass (expressed as concentration of the signature PLFA 16:1ω8c, to facilitate comparison with 18:1ω7c), in situ methane oxidation rate, the concentration of the general bacterial fatty acid 18:1ω7c, and abiotic factors in Illersjön. (a to c) Winter conditions under ice; (d to f) stratified summer conditions. Note the different scales for winter and summer.
FIG. 4.
Relationship between the biomass of methanotrophic bacteria estimated with phospholipid fatty acid analysis and the in situ methane oxidation activity measured with a 14CH4 transformation method (logarithmic scales). Symbols are as described in Fig. 2.
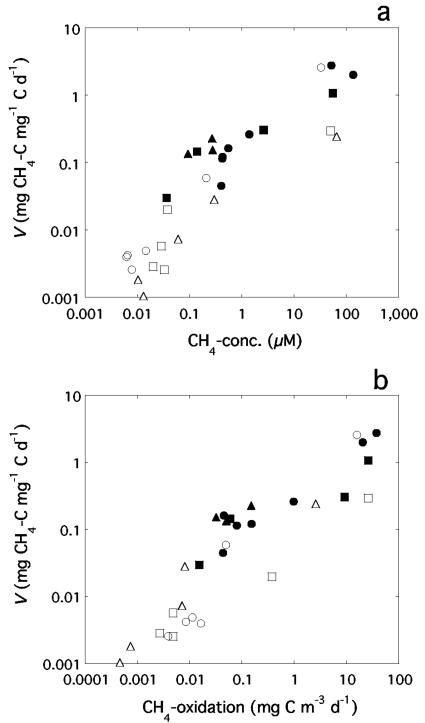
By dividing the in situ methane oxidation with the MOB biomass we obtained estimates of the biomass-specific activity (V) of this group of bacteria in lake water. The total range was 0.001 to 2.77 mg CH4-C mg−1 C day−1 (corresponding to 0.03 to 80 nmol mg−1 dw min−1), with the highest value representing a daily consumption of methane carbon corresponding to more than twice the standing stock of methanotroph carbon biomass. V was positively correlated with CH4-concentration (Fig. 5a, y = 0.14 + 0.017x, r2 = 0.47, P < 0.0001; n = 30) but also with the total methane oxidation (Fig. 5b). There was no clear relationship between V and temperature, except that the four samples with outstandingly high V all had low in situ temperatures of 4.0 to 6.1°C. Similarly, V was not related to oxygen concentration, except that it was quite low in the high-activity samples. These relationships were confirmed by a multiple linear regression with V as a dependent variable and concentrations of methane and oxygen and water temperature as independent variables. This regression once again demonstrated that methane concentration accounts for a substantial share of the variance (r2 = 0.49, P = 0.0007), whereas the addition of either oxygen concentration, temperature, or both as dependent variables increased the explained variance marginally but was not statistically significant (P ≥ 0.27).
FIG. 5.
Relationships of methane concentration (a) and absolute methane oxidation rate (b) in lake water with V, the methanotrophic biomass-specific methane oxidation activity (logarithmic scales). Symbols are as described in Fig. 2.
DISCUSSION
Total bacterial abundance.
Estimates from direct counts of cells and from concentrations of PLFAs correlated strongly (Fig. 1 and 2). Our data thus demonstrate that PLFAs can be used to estimate bacterial standing stock in lake waters. In all three lakes, the two methods yielded similar patterns of depth distribution, although the ratio of cell numbers from PLFAs to flow cytometry differed slightly among lakes. However, it should be kept in mind that the absolute levels of bacterial biomass derived from the two methods depend on choice of conversion factors. These factors are rough averages that surely differ among bacterial groups and are dependent on environmental conditions. It should also be noted that, since data on bacterial cell sizes were not available for all individual samples, we used a single average for all samples. Bacterial cell sizes may differ substantially depending on lake, depth, and sampling occasion (14). Such differences in cell size would affect the general ratio of abundances estimated with the fluorescence microscopy and PLFA methods and influence individual samples to a different degree as well. However, given the uncertainties inherent in the conversion factors, the general correlation between total bacterial abundance estimates from flow cytometry and PLFA concentrations still appears robust (Fig. 2).
Biomass and activity of methanotrophic bacteria.
The strong covariation of 16:1ω8c with both 16:1ω5t and 16:1ω6c supports that all three come from the same group of bacteria. Whereas the former is considered specific to MOB, the latter two are often present simultaneously with 16:1ω8c in methanotrophs. Therefore, they can be used as “qualifying” fatty acids (10, 40, 61) for type I MOB, but they do occur in some other bacteria as well. The simultaneous presence and the similarities in concentration ratios for these three PLFAs between the lake waters and known isolates of MOB suggest that the methanotrophs in these lakes are most similar to strains of Methylomonas, Methylomicrobium, and Methylosarcina (10, 12, 61).
In all samples with significant methane oxidation activity, the ratio between the concentrations of the ω8c fatty acids from type I and type II methanotrophs suggest that the biomasses of type I MOB were substantially higher than those of type II MOB. Moreover, across all samples the biomass of type II seemed to be unrelated to the oxidation activity. Still, the relevance of type II methanotrophs for methane oxidation cannot be completely ruled out, since it was recently shown that acidophilic type II MOB (genera Methylocella and Methylocapsa) from peatlands lack 18:1ω8c (17).
There are few other studies with which the dominance of type I methanotrophs in the water columns of the lakes that we studied can be compared. However, in line with our results, two studies of MOB populations in pelagic freshwater systems (23, 45) and two in sediments (16, 41) all suggest predominance of type I over type II methanotrophs in lakes, although they represent very different lake ecosystems. However, it cannot be concluded that type I dominance is universal in freshwater bodies, since Dumestre et al. (20, 21), using PLFA analyses and a PCR/DGGE approach, found that type II MOB completely dominated methanotroph communities in a tropical water reservoir. Nevertheless, the available information from freshwater systems suggest that either main group of MOB firmly dominates. This seems to be in contrast to wetland soils, where it has been repeatedly found that the two groups often have similar population sizes (27, 37, 51, 56).
The total range of the biomass-specific methane oxidation rates in these lakes, V, was 0.03 to 80 nmol mg−1 dw min−1. These values fall below or in the lower range of published data for Vmax in laboratory cultures of type I MOB, with a reported range of roughly 50 up to 600 nmol mg−1 dw min−1 (13, 25, 49, 50). This literature range is based on MOB growing under nonlimiting nutrient conditions in the laboratory. However, taking, for example, the low temperatures of the samples exhibiting the highest V in our study lakes into account, our estimates of V generally appear as quite realistic for type I methanotrophic bacteria. We are not aware of any previous estimates of the biomass-specific V for methane oxidation under natural conditions.
In terms of carbon transformation, V ranged from 0.001 to 2.77 mg CH4-C mg−1 C day−1. It was lowest in the epilimnion in summer and at intermediate depths under ice in winter, at methane concentrations below 0.1 μM. The strong positive correlation of V with methane concentration, but much weaker with temperature and O2-concentration (Fig. 5 and the multiple regression studies), suggests that methane concentration has the overriding influence on the activity and biomass distribution of methanotrophic bacteria in these three lakes, similar to the situation reported by Liikanen et al. (35).
The peaks in activity and biomass of MOB occurred at very low O2 concentrations, for several deep water samples below 5.7 μM, representing the detection limit. Some other studies have also reported methane oxidation in lake water or sediments in the absence of O2, or at least at very low concentrations (1, 35, 42, 54, 62). Anaerobic oxidation of methane coupled to reduction of sulfate is well-known from various marine sediments (6, 19, 55) and saline lakes (29, 31). In order to get a rough idea whether the low O2 concentrations were sufficient to support the methane oxidation rates, we calculated turnover times for O2 based on the oxygen demand indicated by the measured methane oxidation rates. In the cases where O2 was at or below the detection limit, 5.7 μM was used. The standing stock of oxygen had to be replaced within 1.9 to 4.8 days (with one extreme of 34 days) in order to support aerobic methane oxidation. These turnover times are substantially longer than the incubation times in our 14CH4 uptake assay (4 to 8 h), supporting that methane oxidation was aerobic, provided that the affinity to O2 of the MOB was at least comparable to the affinities in the nonmethanotrophic heterotrophic bacteria, which has been demonstrated previously (44).
The largest MOB biomasses and activity in the lakes always occurred at a temperature below 7°C, suggesting that psychrotrophic or psychrophilic methanotrophs may have been involved. Several type I methanotrophs with a growth optimum at 15°C or lower have been isolated (11, 32, 53). In addition, in studies in a soil and a biofilter on landfills (8, 22) low temperatures favored the development of type I methanotroph populations over type II populations, whereas at 20°C or higher, either both groups or preferably type II populations grew. Together with our data, these studies suggest that low temperature selects for dominance of type I methanotrophic bacteria under several different kinds of environmental conditions.
In conclusion, our results demonstrate that MOB may account for a substantial fraction of water column bacterial biomass and carbon transformations. Thus, through grazing on bacterial communities by zooplankton that are able to tolerate O2 concentrations close to anoxia (48), they may constitute an important link for reentry of carbon into aquatic food webs.
Acknowledgments
We thank Liis Kärme, Sylvia Rönn, Jenny Grönesjö, David Kvick, and Dan Lindmark for invaluable technical and practical assistance. Håkan Olsson kindly provided information about the sampled lakes. We also thank Gunnar Börjesson and Bo Svensson for generously sharing their knowledge.
This study was funded by grants from the Swedish Research Council to L.J.T.
REFERENCES
- 1.Abramochkina, F. N., L. V. Bezrukova, A. V. Koshelev, V. F. Gal'chenko, and M. V. Ivanov. 1987. Microbial oxidation of methane in a body of fresh water. Microbiology 56:375-382. [Google Scholar]
- 2.Bastviken, D., J. Ejlertsson, I. Sundh, and L. Tranvik. 2003. Methane as a source of carbon and energy for lake pelagic food webs. Ecology 84:969-981. [Google Scholar]
- 3.Bastviken, D., J. Ejlertsson, and L. Tranvik. 2002. Measurement of methane oxidation in lakes: a comparison of methods. Environ. Sci. Technol. 36: 3354-3361. [DOI] [PubMed] [Google Scholar]
- 4.Bastviken, D., J. Ejlertsson, and L. Tranvik. 2001. Similar bacterial growth on dissolved organic matter in anoxic and oxic lake water. Aquat. Microb. Ecol. 24:41-49. [Google Scholar]
- 5.Bertilsson, S., R. Stepanauskas, R. Cuadros-Hansson, W. Granéli, J. Wikner, and L. Tranvik. 1999. Photochemically induced changes in bioavailable carbon and nitrogen pools in a boreal watershed. Aquat. Microb. Ecol. 19:47-56. [Google Scholar]
- 6.Blumenberg, M., R. Seifert, J. Reitner, T. Pape, and W. Michaelis. 2004. Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc. Natl. Acad. Sci. 101:11111-11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boon, P. I., P. Virtue, and P. D. Nichols. 1996. Microbial consortia in wetland sediments: a biomarker analysis of the effects of hydrological regime, vegetation and season on benthic microbes. Mar. Freshwater Res. 47:27-41. [Google Scholar]
- 8.Börjesson, G., I. Sundh, and B. Svensson. 2004. Microbial oxidation of CH4 at different temperatures in landfill cover soils. FEMS Microbiol. Ecol. 48:305-312. [DOI] [PubMed] [Google Scholar]
- 9.Börjesson, G., I. Sundh, A. Tunlid, and B. H. Svensson. 1998. Methane oxidation in landfill cover soils, as revealed by potential oxidation measurements and phospholipid fatty acid analyses. Soil. Biol. Biochem. 30:1423-1433. [Google Scholar]
- 10.Bowman, J. P. 2000. The methanotrophs: the families Methylococcaceae and Methylocystaceae. .In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. [Online] Springer-Verlag, New York, N.Y.http://141.150.157.117:8080/prokPUB/chaprender/jsp/showchap.jsp?chapnum=249.
- 11.Bowman, J. P., S. A. McCammon, and J. H. Skerratt. 1997. Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology 143:1451-1459. [DOI] [PubMed] [Google Scholar]
- 12.Bowman, J. P., L. I. Sly, P. D. Nichols, and A. C. Hayward. 1993. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int. J. Syst. Bacteriol. 43:735-753. [Google Scholar]
- 13.Carlsen, H. N., L. Joergensen, and H. Degn. 1991. Inhibition by ammonia of methane utilization in Methylococcus capsulatus (Bath). Appl. Microbiol. Biotechnol. 35:124-127. [Google Scholar]
- 14.Cole, J. J., M. L. Pace, N. F. Caraco, and G. S. Steinhart. 1993. Bacterial biomass and cell size distributions in lakes: more and larger cells in anoxic waters. Limnol. Oceanogr. 38:1627-1632. [Google Scholar]
- 15.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costello, A. M., A. J. Auman, J. L. Macalady, K. M. Scow, and M. E. Lidstrom. 2002. Estimation of methanotroph abundance in a freshwater lake sediment. Environ. Microbiol. 4:443-450. [DOI] [PubMed] [Google Scholar]
- 17.Dedysh, S. N., V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, W. Liesack, and J. M. Tiedje. 2002. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 52:251-261. [DOI] [PubMed] [Google Scholar]
- 18.del Giorgio, P. A., D. B. Bird, Y. T. Prairie, and D. Planas. 1996. Flow cytometric determination of bacterial abundance in lake plankton with the green nucleic acid stain Syto 13. Limnol. Oceanogr. 41:783-789. [Google Scholar]
- 19.Devol, A. H., J. J. Anderson, K. Kuivila, and J. W. Murray. 1984. A model for coupled sulfate reduction and methane oxidation in the Saanich Inlet. Geochim. Cosmochim. Acta 48:993-1004. [Google Scholar]
- 20.Dumestre, J.-F., E. O. Casamayor, R. Massana, and C. Pedrós-Alió. 2001. Changes in bacterial and archaeal assemblages in an equatorial river induced by the water eutrophication of Petit Saut dam reservoir (French Guiana). Aquat. Microb. Ecol. 26:209-221. [Google Scholar]
- 21.Dumestre, J.-F., A. Vaquer, P. Gosse, S. Richard, and L. Labroue. 1999. Bacterial ecology of a young equatorial hydroelectric reservoir (Petit Saut, French Guiana). Hydrobiologia 400:75-83. [Google Scholar]
- 22.Gebert, J., A. Groengroeft, and G. Miehlich. 2003. Kinetics of microbial landfill methane oxidation in biofilters. Waste Management 23:609-619. [DOI] [PubMed] [Google Scholar]
- 23.Hanson, R. S., B. J. Bratina, and G. A. Brusseau. 1993. Phylogeny and ecology of methylotrophic bacteria, p. 285-302. In J. C. Murrell and D. P. Kelly (ed.), Microbial growth on C1 compounds. Proceedings of the 7th International Symposium. Intercept, Ltd., Andover, United Kingdom.
- 24.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harwood, J. H., and S. J. Pirt. 1972. Quantitative aspects of growth of the methane oxidizing bacterium Methylococcus capsulatus on methane in shake flask and continuous chemostat culture. J. Appl. Bacteriol. 35:597-607. [DOI] [PubMed] [Google Scholar]
- 26.Hessen, D., and K. Nygaard. 1992. Bacterial transfer of methane and detritus; implications for the pelagic carbon budget and gaseous release. Ergebn. Limnol. 37:139-148. [Google Scholar]
- 27.Horz, H.-P., A. S. Raghubashi, J. Heyer, C. Kammann, R. Conrad, and P. F. Dunfield. 2002. Activity and community structure of methane-oxidizing bacteria in a wet meadow soil. FEMS Microbiol. Ecol. 41:247-257. [DOI] [PubMed] [Google Scholar]
- 28.Hutchens, E., S. Radajewski, M. G. Dumont, I. R. McDonald, and J. C. Murrell. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6:111-120. [DOI] [PubMed] [Google Scholar]
- 29.Iversen, N., R. S. Oremland, and M. J. Klug. 1987. Big Soda Lake (Nevada). 3. Pelagic methanogenesis and anaerobic methane oxidation. Limnol. Oceanogr. 32:804-814. [Google Scholar]
- 30.Jones, R. I., J. Grey, D. Sleep, and L. Arvola. 1999. Stable isotope analysis of zooplankton carbon nutrition in humic lakes. Oikos 86:97-104. [Google Scholar]
- 31.Joye, S. B., T. L. Connell, L. G. Miller, R. S. Oremland, and R. S. Jellison. 1999. Oxidation of ammonia and methane in an alkaline, saline lake. Limnol. Oceanogr. 44:178-188. [Google Scholar]
- 32.Kalyuzhnaya, M. G., V. N. Khmelenina, S. Kotelnikova, L. Holmquist, K. Pedersen, and Y. A. Trotsenko. 1999. Methylomonas scandinavica sp. nov., a new methanotrophic psychrotrophic bacterium isolated from deep igneous rock ground water of Sweden. Syst. Appl. Microbiol. 22:565-572. [DOI] [PubMed] [Google Scholar]
- 33.King, G. M. 1992. Ecological aspects of methane oxidation, a key determinant of global methane dynamics. Adv. Microb. Ecol. 12:431-468. [Google Scholar]
- 34.Klamer, M., and E. Bååth. 1998. Microbial community dynamics during composting of straw material studied using phospholipid fatty acid analysis. FEMS Microbiol. Ecol. 27:9-20. [Google Scholar]
- 35.Liikanen, A., J. T. Huttunen, K. Valli, and P. J. Martikainen. 2002. Methane cycling in the sediment and water column of mid-boreal hyper-eutrophic Lake Kevätön, Finland. Arch. Hydrobiol. 154:585-603. [Google Scholar]
- 36.Loferer-Krössbacher, M., J. Klima, and R. Psenner. 1998. Determination of bacterial cell dry mass by transmission electron microscopy and densitometric image analysis. Appl. Environ. Microbiol. 64:688-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macalady, J. L., A. M. S. McMillan, A. F. Dickens, S. C. Tyler, and K. M. Scow. 2002. Population dynamics of type I and II methanotrophic bacteria in rice soils. Environ. Microbiol. 4:148-157. [DOI] [PubMed] [Google Scholar]
- 38.Nichols, P. D., J. B. Guckert, and D. C. White. 1986. Determination of monounsaturated fatty acid double-bond position and geometry for microbial monocultures and complex consortia by capillary GC-MS of their dimethyl disulphide adducts. J. Microbiol. Methods 5:49-55. [Google Scholar]
- 39.Nichols, P. D., M. Henson, C. P. Antworth, J. Parsons, J. T. Wilson, and D. C. White. 1987. Detection of a microbial consortium, including type II methanotrophs, by use of phospholipid fatty acids in an aerobic halogenated hydrocarbon-degrading soil column enriched with natural gas. Environ. Toxicol. Chem. 6:89-97. [Google Scholar]
- 40.Nichols, P. D., G. A. Smith, C. P. Antworth, R. S. Hanson, and D. C. White. 1985. Phospholipid and lipopolysaccharide normal and hydroxy fatty acids as potential signatures for methane-oxidizing bacteria. FEMS Microbiol. Ecol. 31:327-335. [Google Scholar]
- 41.Nold, S. C., H. T. S. Boschker, R. Pel, and H. J. Laanbroek. 1999. Ammonium addition inhibits 13C-methane incorporation into methanotroph membrane lipids in a freshwater sediment. FEMS Microbiol. Ecol. 29:81-89. [Google Scholar]
- 42.Panganiban, A. T., Jr., T. E. Patt, W. Hart, and R. S. Hanson. 1979. Oxidation of methane in the absence of oxygen in lake water samples. Appl. Environ. Microbiol. 37:303-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reeburgh, W. S., S. C. Whalen, and M. J. Alperin. 1993. The role of methylotrophy in the global methane budget, p. 1-14. In J. C. Murrell, and D. P. Kelly (ed.), Microbial growth on C1 compounds. Proceedings of the 7th International Symposium. Intercept, Ltd., Andover, United Kingdom.
- 44.Ren, T., J. A. Amaral, and R. Knowles. 1997. The response of methane consumption by pure cultures of methanotrophic bacteria to oxygen. Can. J. Microbiol. 43:925-928. [Google Scholar]
- 45.Ross, J. L., P. I. Boon, P. Ford, and B. T. Hart. 1997. Detection and quantification with 16S rRNA probes of planktonic methylotrophic bacteria in a floodplain lake. Microb. Ecol. 34:97-108. [DOI] [PubMed] [Google Scholar]
- 46.Rudd, J. W. M., and R. D. Hamilton. 1978. Methane cycling in a eutrophic shield lake and its effects on whole lake metabolism. Limnol. Oceanogr. 23:337-348. [Google Scholar]
- 47.Rudd, J. W. M., R. D. Hamilton, and N. E. R. Campbell. 1974. Measurement of microbial oxidation of methane in lake water. Limnol. Oceanogr. 19:519-524. [Google Scholar]
- 48.Salonen, K., and A. Lehtovaara. 1992. Migrations of haemoglobin-rich Daphnia longispina in a small, steeply stratified, humic lake with an anoxic hypolimnion. Hydrobiologia 229:271-288. [Google Scholar]
- 49.Sheehan, B. T., and M. J. Johnson. 1971. Production of bacterial cells from methane. Appl. Microbiol. 21:511-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, K. S., A. M. Costello, and M. E. Lidstrom. 1997. Methane and trichloroethylene oxidation by an estuarine methanotroph, Methylobacter sp. strain BB5.1. Appl. Environ. Microbiol. 63:4617-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sundh, I., P. Borgå, M. Nilsson, and B. H. Svensson. 1995. Estimation of cell numbers of methanotrophic bacteria in boreal peatlands based on analysis of specific phospholipid fatty acids. FEMS Microbiol. Ecol. 18:103-112. [Google Scholar]
- 52.Sundh, I., M. Nilsson, and P. Borgå. 1997. Variation in microbial community structure in two boreal peatlands as determined by analysis of phospholipid fatty acid profiles. Appl. Environ. Microbiol. 63:1476-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tourova, T. P., M. V. Omel'chenko, K. V. Fegeding, and L. V. Vasil'eva. 1999. The phylogenetic position of Methylobacter psychrophilus sp. nov. Microbiology 68:493-495. [Google Scholar]
- 54.Utsumi, M., Y. Nojiri, T. Nakamura, T. Nozava, A. Otsuki, N. Takamura, M. Watanabe, and H. Seki. 1998. Dynamics of dissolved methane and methane oxidation in dimictic Lake Nojiri during winter. Limnol. Oceanogr. 43:10-17. [Google Scholar]
- 55.Valentine, D. L. 2002. Biogeochemistry and microbial ecology of methane oxidation in anaerobic environments: a review. Antonie Leeuwenhoek 81:271-282. [DOI] [PubMed] [Google Scholar]
- 56.Vecherskaya, M. S., V. F. Galchenko, E. N. Sokolova, and V. A. Samarkin. 1993. Activity and species composition of aerobic methanotrophic communities in tundra soils. Curr. Microbiol. 27:181-184. [DOI] [PubMed] [Google Scholar]
- 57.Virtue, P., P. D. Nichols, and P. I. Boon. 1996. Simultaneous estimation of microbial phospholipid fatty acids and diether lipids by capillary gas chromatography. J. Microbiol. Methods 25:177-185. [Google Scholar]
- 58.Whalen, S. C., W. S. Reeburgh, and V. A. Barber. 1992. Oxidation of methane in boreal forest soils: a comparison of seven measures. Biogeochemistry 16:181-211. [Google Scholar]
- 59.White, D. C., R. J. Bobbie, J. S. Herron, J. D. King, and S. J. Morrison. 1979. Biochemical measurements of microbial mass and activity from environmental samples, p. 69-81. In J. W. Costerton and R. R. Colwell (ed.), Native aquatic bacteria: enumeration, activity, and ecology. American Society for Testing of Materials, West Conshohocken, Pa.
- 60.White, D. C., J. O. Stair, and D. B. Ringelberg. 1996. Quantitative comparisons of in situ microbial biodiversity by signature biomarker analysis. J. Ind. Microbiol. 17:185-196. [Google Scholar]
- 61.Wise, M. G., J. V. McArthur, and L. J. Shimkets. 2001. Methylosarcina fibrata gen. nov., sp. nov. and Methylosarcina quisquiliarum sp. nov., novel type I methanotrophs. Int. J. Syst. Evol. Microbiol. 51:611-621. [DOI] [PubMed] [Google Scholar]
- 62.Zehnder, A. J. B., and T. D. Brock. 1980. Anaerobic methane oxidation: occurrence and ecology. Appl. Environ. Microbiol. 39:194-204. [DOI] [PMC free article] [PubMed] [Google Scholar]