Abstract
Pyoluteorin is a chlorinated polyketide antibiotic secreted by the rhizosphere bacterium Pseudomonas fluorescens Pf-5. Genes encoding enzymes and transcriptional regulators involved in pyoluteorin production are clustered in the genome of Pf-5. Sequence analysis of genes adjacent to the known pyoluteorin biosynthetic gene cluster revealed the presence of an ABC transporter system. We disrupted two putative ABC transporter genes by inserting transcriptional fusions to an ice nucleation reporter gene. Mutations in pltI and pltJ, which are predicted to encode a membrane fusion protein and an ATP-binding cassette of the ABC transporter, respectively, greatly reduced pyoluteorin production by Pf-5. During the transition from exponential growth to stationary phase, populations of a pltI mutant were lower than those of a pltI+ strain in a culture medium containing pyoluteorin, suggesting a role for the transport system in efflux and the resistance of Pf-5 to the antibiotic. Although pltI or pltJ mutant strains displayed low pyoluteorin production, they did not accumulate proportionately more of the antibiotic intracellularly, indicating that pltI and pltJ do not encode an exclusive exporter for pyoluteorin. Transcription of the putative pyoluteorin efflux genes pltI and pltJ was enhanced by exogenous pyoluteorin. These new observations parallel an earlier finding that pyoluteorin enhances the transcription of pyoluteorin biosynthesis genes and pyoluteorin production in Pf-5. This report provides evidence of a coordination of pyoluteorin production and the transcription of genes encoding a linked transport apparatus, wherein each requires the other for optimal expression.
Pseudomonas fluorescens Pf-5 is a rhizosphere-inhabiting bacterium that suppresses soilborne plant diseases, in part by producing a spectrum of antibiotics that are toxic to plant pathogens. The antibiotics produced by Pf-5 include pyrrolnitrin, a chlorinated aromatic compound, and two polyketides, pyoluteorin and 2,4-diacetylphloroglucinol. Pyoluteorin, the focus of the present study, is toxic to seed- and root-rotting oomycetes (19, 28) and certain gram-negative and gram-positive bacteria (2).
The pyoluteorin biosynthetic gene cluster contains nine structural genes that are required for pyoluteorin biosynthesis (32, 33) and two genes that encode transcriptional regulators. pltR encodes a LysR-type transcriptional activator required for pyoluteorin production and the transcription of pyoluteorin biosynthetic genes (32). A second regulator linked to the pyoluteorin biosynthesis gene cluster, pltZ, was recently described for Pseudomonas sp. strain M18 (20). PltZ, a member of the TetR family of transcriptional repressors, inhibits pyoluteorin production and the expression of a pyoluteorin biosynthetic gene (20).
In addition to its established extracellular role as an antibiotic, pyoluteorin was recently shown to function as an autoinducer, enhancing pyoluteorin production and biosynthetic gene expression by Pf-5 (7). Furthermore, pyoluteorin serves as an intercellular signal between distinct populations of bacterial cells coinhabiting the rhizosphere (7). Established positive autoregulation systems, such as LuxI/LuxR, rely on diffusion or secretion and reuptake of the signal molecule across the membrane and include an intracellular signal receptor(s) that mediates signal transduction within the cell. Neither a pyoluteorin receptor nor a mechanism for pyoluteorin transport across the bacterial membrane is known, but genes encoding proteins required for the transport of a secondary metabolite across the cell membrane are often linked to genes encoding enzymatic functions for biosynthesis (27).
The present study focuses on a cluster of genes linked to the pyoluteorin biosynthetic gene cluster whose nucleotide sequences are similar to those of ATP-binding cassette (ABC) export proteins, a class of ATP-powered, multisubunit membrane transporters (4, 29, 37). Also within the cluster are open reading frames (ORFs) encoding a putative periplasmic membrane fusion protein and an outer membrane channel. We demonstrate that the transporter gene cluster is necessary for optimal pyoluteorin production by Pf-5. Furthermore, the transcription of pltI and pltJ, two genes with putative transporter functions, is positively regulated by exogenous pyoluteorin. The results herein provide evidence of the coordination of pyoluteorin production and efflux.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used for this study are listed in Table 1. Escherichia coli strains were cultured routinely in Luria-Bertani (LB) medium (36) and maintained on LB agar. Pf-5 and derivative strains were grown on King's medium B (KMB) (22). The following antibiotics were included in growth media as required: spectinomycin, 50 μg/ml; streptomycin, 100 μg/ml; kanamycin, 50 μg/ml; gentamicin, 40 μg/ml (P. fluorescens) or 15 μg/ml (E. coli); and tetracycline, 200 μg/ml (P. fluorescens) or 20 μg/ml (E. coli).
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain genotype or plasmid | Strain no. | Descriptiona | Reference |
|---|---|---|---|
| Strains | |||
| P. fluorescens | |||
| Pf-5 | JL4474 | Rhizosphere isolate | 18 |
| Pf-5 pltB::inaZ | JL4389 | pltB::Tn3-nice derivative of Pf-5; Plt− Ina+ Kmr | 23 |
| Pf-5 pltR::aacC1 | JL4563 | pltR::aacC1 derivative of Pf-5; Plt− Gmr | 32 |
| Pf-5 pltF::aacC1 | JL4629 | pltF::aacC1 derivative of Pf-5; Plt− Gmr | This study |
| Pf-5 pltJ::inaZ | JL4671 | pltJ::inaZ derivative of Pf-5; Ina+ Kmr | This study |
| Pf-5 pltI::inaZ | JL4673 | pltI::inaZ derivative of Pf-5; Ina+ Kmr | This study |
| Pf-5 pltR::aacC1 pltJ::inaZ | JL4677 | pltR::aacC1 pltJ::inaZ derivative of Pf-5; Ina+ Gmr Kmr | This study |
| Pf-5 pltR::aacC1 pltI::inaZ | JL4679 | pltR::aacC1 pltI::inaZ derivative of Pf-5; Ina+ Gmr Kmr | This study |
| Pf-5 pltF::aacC1 pltJ::inaZ | JL4683 | pltF::aacC1 pltJ::inaZ derivative of Pf-5; Ina+ Gmr Kmr | This study |
| Pf-5 pltF::aacC1 pltI::inaZ | JL4685 | pltF::aacC1 pltI::inaZ derivative of Pf-5; Ina+ Gmr Kmr | This study |
| E. coli | |||
| S17-1 | Res− Mod+recA Tra+ | 40 | |
| DH5α | F−endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 φ80dlacZΔM15 λ− | 36 | |
| Plasmids | |||
| pRK415 | IncP1 replicon, polylinker of pUC19; Mob+ Tcr | 21 | |
| pRK2013 | Mobilizing plasmid; Tra+ Kmr | 11 | |
| pUC19 | ColE1 replicon; Apr | 36 | |
| pTn3-nice | pMB8 replicon, contains Tn3-nice; Apr Kmr | 23 | |
| pJEL1938 | 29 kb of the pyoluteorin biosynthetic gene cluster from Pf-5 cloned into pLAFR3; Tcr | 23 | |
| pJEL6198 | 1.7-kb PstI fragment containing pltF cloned from pJEL1938 into pUC19; Apr | This study | |
| pJEL6199 | pltF::aacC1 in pUC19; Apr Gmr | This study | |
| pJEL6200 | pltF::aacC1 in pRK415; Tcr Gmr Mob+ | This study | |
| pJEL6274 | 3.4-kb SmaI-HindIII fragment from pJEL1938 encompassing pltI and pltJ, cloned into pUC19; Apr | This study | |
| pJEL6279 | 6.2-kb fragment containing the inaZ reporter gene and nptI from pTn3-nice, cloned into pUC19; Apr Kmr Ina+ | 6 | |
| pJEL6285 | pltJ::inaZ fusion (at SphI site of pltJ) in pUC19; Kmr Apr | This study | |
| pJEL6287 | pltI::inaZ fusion in pUC19; Kmr Apr | This study | |
| pJEL6299 | pltJ::inaZ fusion in pRK415; Kmr Tcr Mob+ | This study | |
| pJEL6300 | pltI::inaZ fusion in pRK415; Kmr Tcr Mob+ | This study |
Apr, Gmr, Kmr, and Tcr indicate resistance to ampicillin, gentamicin, kanamycin, and tetracycline, respectively. Plt− indicates a lack of pyoluteorin production. Ina+ indicates ice nucleation activity at −5°C.
For experiments evaluating antibiotic production or gene transcription, cultures were grown at 20°C with shaking (200 rpm) in nutrient broth (pH 6.8; Difco Laboratories, Detroit, Mich.) containing 0.5% (wt/vol) glycerol (NBGly). Inoculums of Pf-5 and derivative strains were derived from overnight cultures grown with shaking (200 rpm) at 27°C in KMB broth, modified by omitting glycerol and substituting 1% (wt/vol) glucose (KBGlu) to repress pyoluteorin production. Each inoculum was washed twice in sterile KBGlu prior to inoculation and resuspended in 5 ml of NBGly to an optical density at 600 nm of 0.1. Where indicated in Results, purified pyoluteorin dissolved in methanol was added at a rate of 4 μg pyoluteorin/ml culture medium immediately prior to inoculation with Pf-5 or a derivative strain. An equivalent volume of methanol was added to negative controls. Bacterial population densities were determined by spreading serial dilutions of cultures onto KMB and counting CFU on the plates after 48 h of incubation at 27°C. For each experiment, triplicate 5-ml cultures were grown and evaluated; each experiment was performed at least twice, and the results from a representative experiment are presented.
Nucleotide sequencing.
The nucleotide sequence of the pyoluteorin biosynthetic gene cluster was extended beyond the 3′ end of pltG (32). Overlapping fragments of the region 3′ of pltG were subcloned from pJEL1938 into pUC19, and the resultant plasmids were used as templates for sequencing. Sequencing was initiated using primers complementary to the pUC19 multiple cloning site and continued by primer walking. Additional sequence data were obtained from the genomic sequence of Pf-5 (GenBank accession number CP000076). Codon preference analysis (45) was used to identify ORFs. The predicted amino acid sequence of each gene was examined for patterns, profiles, and motifs listed in the Pfam (3), ProSite (39), and InterPro (47) databases for predictions about functionality. Transmembrane regions were predicted by the dense alignment surface (DAS) method (9) and the “positive inside” rule (8).
Derivation of the pltF::aacC1 mutation.
The pltF gene, which is predicted to encode an acyl-coenzyme A synthetase necessary for precursor activation during the induction of pyoluteorin biosynthesis (32), was mutagenized by insertion of the gentamicin resistance-conferring gene aacC1. Briefly, a 1.7-kb PstI fragment containing pltF and flanking sequences was cloned into pUC19 to yield pJEL6198, and a 2.0-kb SmaI fragment containing aacC1 (30) was ligated into a blunted BsmI site located between nucleotides (nt) 937 and 942 of the pltF gene to yield pJEL6199. Nucleotide sequencing confirmed the integrity of pJEL6199. The 3.7-kb HindIII-KpnI fragment containing pltF::aacC1 was isolated from pJEL6199 and cloned into pRK415, an unstable plasmid in Pf-5, to construct pJEL6200.
Derivation of pltI::inaZ and pltJ::inaZ transcriptional fusions.
Transcriptional fusions of the promoterless ice nucleation reporter gene inaZ (25) within pltI and pltJ were constructed as follows. A 3.4-kb SmaI-HindIII fragment containing pltI and pltJ was subcloned from pJEL1938 into pUC19, resulting in pJEL6274. A 6.2-kb BamHI-AflIII fragment from pJEL6279 (containing the promoterless inaZ gene paired with the kanamycin resistance gene nptI) was inserted via blunt-ended ligation into the SphI site of pltJ (nt 1154) in pJEL6274 to construct pJEL6285 or into the BsaA1 site of pltI (nt 771) in pJEL6274 to construct pJEL6287. These transcriptional fusions were moved into pRK415, a plasmid useful for marker-exchange mutagenesis of Pf-5, by digesting pJEL6285 and pJEL6287 at the unique NdeI site in each plasmid and inserting the linearized plasmids into the unique EcoRI site of pRK415 via blunt-ended ligation, thereby creating pJEL6299 and pJEL6300, respectively.
Marker-exchange mutagenesis.
The pltF::aacC1 mutation and pltI::inaZ and pltJ::inaZ transcriptional fusions were introduced into the genome of Pf-5 (or derivative strains) via marker-exchange mutagenesis. Plasmids containing disrupted genes were introduced into Pf-5 or derivative strains by conjugation, and transconjugants were selected on KMB containing streptomycin (to counterselect against E. coli) together with tetracycline and gentamicin (to select for pJEL6200) or tetracycline and kanamycin (to select for pJEL6299 or pJEL6300). Single colonies were inoculated into KMB broth lacking tetracycline to release selection for pRK415 but containing gentamicin or kanamycin to maintain selection for the mutagenic antibiotic resistance cassettes. After 3 days, serial dilutions from these cultures were spread onto KMB containing streptomycin and kanamycin (for pltI::inaZ and pltJ::inaZ) or streptomycin and gentamicin (for pltF::aacC1). Single colonies from these plates were patched onto KMB containing either kanamycin or gentamicin, as appropriate, and onto replica plates also containing tetracycline. The presence of inaZ in the expected location in the genomes of tetracycline-sensitive strains was verified by PCR using primers specific to inaZ and pltI or pltJ or primers pltF-f and pltF-r (Table 2). Further verification was done by Southern analysis using biotinylated probes from the wild-type pltI and pltJ sequences or by sequencing of the amplification product for the pltF::aacC1 insertion.
TABLE 2.
Oligonucleotide primers used for this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| Forward primers | |
| pltA-f | CGAGAAAGCCAACTTCCC |
| pltF-f | GTGTTTCCTGGTTTCTCC |
| pltZ-f | ATCTATCACCGCCAGCATC |
| pltI-f1 | GTGTTTCCTGGTTTCTCC |
| pltI-f2 | GGTGGAAACGGAAGACCT |
| pltJ-f | GGGAACTGTGGAGCATCATCG |
| pltK-f | ATGCGTGACAAGAGCAACC |
| pltN-f | ATTGCTGTTCGGCTACGG |
| PltO-f | CTGGTGGCAGGTGTTTGG |
| pltP-f | CGAAATGCTCGGCTACACC |
| orf1-f | GCCCACACCTATTCCAACC |
| Reverse primers | |
| inaZ-r | CCAACGCCTTGTCGAGATTCAT |
| pltA-r | TTCAGGCGGTAATAGAGCG |
| pltF-r | CCAACGCCTTGTCGAGATTCAT |
| pltZ-r | ACGCAGAACCAGCAGAAG |
| pltI-r | AGGTCTTCCGTTTCCACC |
| pltJ-r1 | ATCGTCGTGCCGCTTTGC |
| pltJ-r2 | CGATGATGCTCCACAGTTCCC |
| pltK-r | AAGAGGAACCCCGAGAGC |
| pltN-r | GCAGCGGATCGAGATAGG |
| pltO-r | CGCTGGTATTCGTTGTGG |
| pltP-r | AAAACAGCGGTCCAGAAACC |
| orf1-r | TCCCTGCTCGACGTACTGC |
Transcript analysis using RT-PCR.
Reverse transcription-PCRs (RT-PCRs) of RNAs isolated from Pf-5 and Pf-5 pltI::inaZ were done to investigate the transcriptional organization of the pltIJKNOP cluster. In two separate experiments, RNAs were extracted using an RNAqueous kit (Ambion Inc., Austin, TX) from three replicate cultures of each strain harvested 8 h after inoculation and subjected to LiCl precipitation followed by DNase treatment (DNA-free or Turbo DNA-free; Ambion Inc., Austin, TX) until PCR amplification with pltA-f and pltA-r (Table 2) yielded no detectable product. RNA samples were further purified by using a MegaClear kit (Ambion Inc., Austin, TX) and ammonium acetate-ethanol precipitation prior to reverse transcription with SuperScript II (Invitrogen, Carlsbad, CA). Following reverse transcription, RNAs were hydrolyzed with 2.5 M NaOH, and samples were neutralized with 2 M HEPES free acid. cDNAs were purified by ammonium acetate-ethanol precipitation. Samples processed in parallel without reverse transcriptase were used as negative controls. Genomic DNA, cDNAs, and negative controls from each replicate culture were subjected to PCR using primers for pltI, pltJ, pltK, pltN, pltO, pltP, and orf1 (Table 2). PCR determinations of cotranscribed genes were done by pairing the forward primer of the upstream gene with the reverse primer of the downstream gene. PCR analysis of pltI was done using pltI-f1 and pltI-r1, that of pltJ was done with pltJ-f and pltJ-r1, and that of the pltIJ intergenic region was done with pltI-f2 and pltJ-r2. ThermalAce DNA polymerase (Invitrogen, Carlsbad, CA) or Takara LA Taq (Takara Bio, Inc., Shiga, Japan) was used according to the manufacturer's directions.
Antibiotic quantification.
Culture supernatants were extracted for the recovery of pyoluteorin and analyzed by high-performance liquid chromatography as described previously (7). Quantification was done by comparison to a standard curve generated from authentic compounds. The detection limit was 0.02 μg/ml of bacterial cultures. The recovery of pyoluteorin using the extraction procedure described above averaged 70%. In experiments evaluating the influence of exogenous pyoluteorin on gene expression by Pf-5, concentrations of pyoluteorin resulting from amendment of the culture medium were estimated over time from cultures of a pyoluteorin-deficient strain (Pf-5 pltB::inaZ). Concentrations of pyoluteorin presented in the Results are mean values for three replicate cultures. The experiment was done twice with the same results, and results from one experiment are presented.
Growth of Pf-5 and derivatives in the presence of pyoluteorin.
Cultures of Pf-5 and Pf-5 pltI::inaZ were grown at 20°C with shaking (200 rpm) in NBGly and NBGly amended with 12 μg/ml pyoluteorin, which represents the high end within the range of concentrations produced by Pf-5 in culture (7). Inoculums for experimental cultures of Pf-5 and derivative strains were derived from overnight cultures grown with shaking (200 rpm) at 27°C in KBGlu to repress pyoluteorin production. Each inoculum was washed twice in sterile 20 mM phosphate buffer (pH 7.0) prior to inoculation and resuspended in 5 ml of NBGly to an optical density at 600 nm of 0.02. Bacterial population densities were determined at 0, 9, 12, 15, 20, and 24 h by spreading serial dilutions of cultures onto KMB and counting CFU on the plates after incubation at 27°C. For each experiment, triplicate 5-ml cultures were grown and evaluated. Each experiment was performed at least twice, and the results from a representative experiment are presented.
Transcriptional activity from pyoluteorin biosynthetic genes.
Transcriptional fusions of a promoterless inaZ gene to several pyoluteorin biosynthetic genes of Pf-5 were constructed previously (23). The ice nucleation activity (INA) from strains carrying the inaZ reporter gene was assessed using a droplet-freezing assay, which provides a quantitative assessment of transcriptional activity (26). In studies measuring INA following pyoluteorin addition, INA peaked at 12 h, and measurements taken between 12 and 48 h were similar. After confirming this observation for the pltI::inaZ and pltJ::inaZ strains, INA was measured only at 0, 2, and 48 h to assess the onset and maximum levels of induction. For each experiment, triplicate 5-ml cultures were grown and evaluated. Each experiment was performed at least twice, and the results from a representative experiment are presented.
Statistical analysis.
Treatment means were compared using Student's t test following an analysis of variance. Two-tailed P values are reported. Statistical analyses were performed using JMP, version 3 (SAS Institute, Cary, NC).
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequences of pltZ and pltIJKNOP is AF081920.
RESULTS
Identification of regulatory and transport genes linked to the pyoluteorin biosynthetic gene cluster.
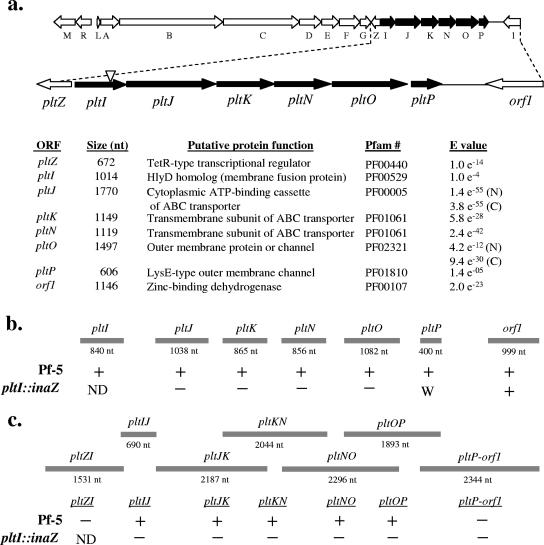
Sequence analysis of the region 3′ of pltG in the pyoluteorin gene cluster of Pf-5 revealed a set of ORFs whose predicted protein products function in regulation and transport (Fig. 1a).
FIG. 1.
(a) Physical map of pyoluteorin biosynthesis and transport gene cluster of P. fluorescens Pf-5. Dashed lines denote the enlarged region containing pltZ and the pltIJKNOP locus. Black arrows denote the direction and relative size of each gene in the locus. The location of the reporter gene inaZ insertion in the pltI::inaZ mutant is denoted by a triangle in the pltI gene. Listed below are genes, predicted protein products, Pfam designations, and expect (E) values for the consensus with Pfam models for each protein family. For pltJ and pltO, predicted amino- and carboxy-terminal domains are denoted by N and C, respectively. (b) RT-PCR analysis of the pltIJKNOP locus with oligonucleotide primer pairs internal to each gene. (c) RT-PCR analysis of the pltIJKNOP locus with oligonucleotide primer pairs spanning two contiguous genes. In panels b and c, the gray bars indicate the designations and predicted sizes of RT-PCR amplification products of RNAs isolated from Pf-5 and Pf-5 pltI::inaZ. +, detection of an RT-PCR product of the expected size; −, lack of detection; W, weak detection of the expected product; ND, the reaction was not done.
(i) pltZ.
Immediately downstream and convergently transcribed from pltG is a homolog of pltZ (70% identical at the amino acid level), a transcriptional regulator found within the syntenic pyoluteorin biosynthetic gene cluster of Pseudomonas sp. strain M18. In Pf-5, as in M18, PltZ is predicted to bear an N-terminal DNA binding motif that typifies the TetR family of transcriptional regulators.
(ii) pltI.
The predicted amino acid sequence encoded by pltI, which is adjacent to and divergently transcribed from pltZ, is similar across its entire length to the HlyD family of membrane fusion proteins involved in secretion. Members of the HlyD family are components of multidrug efflux pumps. As is typical for proteins in the HlyD family, a single transmembrane domain is predicted for PltI, between amino acid residues 6 and 21.
(iii) pltJ.
Adjacent to pltI and transcribed from the same strand is pltJ. pltJ overlaps the last four nucleotides of pltI and lacks a consensus Shine-Dalgarno sequence preceding the ATG start codon. PltJ contains two separate consensus ABC transporter ATP-binding motifs, both of which have the characteristic γ-phosphate-binding Walker A box (accession number PDOC00017), the magnesium-binding Walker B box, and a signature motif (previously termed the linker peptide and proposed to facilitate interactions with periplasmic substrate binding proteins for ABC importers) (4, 15, 16, 31, 37, 43). A single switch region, thought to be important for ATP hydrolysis (4), follows the Walker B box in the C-terminal ABC transporter motif. There are no apparent transmembrane domains, indicating that PltJ is likely to be cytoplasmic.
(iv) pltK and pltN.
Downstream from pltJ are two adjacent genes whose predicted amino acid sequences resemble consensus sequences for the transmembrane domain subunits of an ABC transporter apparatus. DAS analysis of PltK and PltN revealed six predicted transmembrane domains in each protein.
(v) pltO.
Downstream from pltN lies pltO, whose predicted amino acid sequence matches consensus models for a TolC-type outer membrane efflux protein. In keeping with other genes encoding outer membrane efflux proteins, the gene for PltO comprises two repeats, both of which match the consensus sequence for this protein family. In support of the membrane localization of PltO, an N-terminal transmembrane domain is predicted for PltO by DAS analysis.
(vi) pltP.
Adjacent to pltO is pltP, whose predicted protein product resembles consensus patterns for a LysE-type export protein. PltP is predicted to bear six transmembrane domains, the same number as the prototypical LysE translocator. PltP shares homology with CmaU (GenPept accession number AAC46034) from the coronatine biosynthesis pathway in Pseudomonas syringae pv. glycinea.
Pyoluteorin production by pltI and pltJ derivatives of Pf-5.
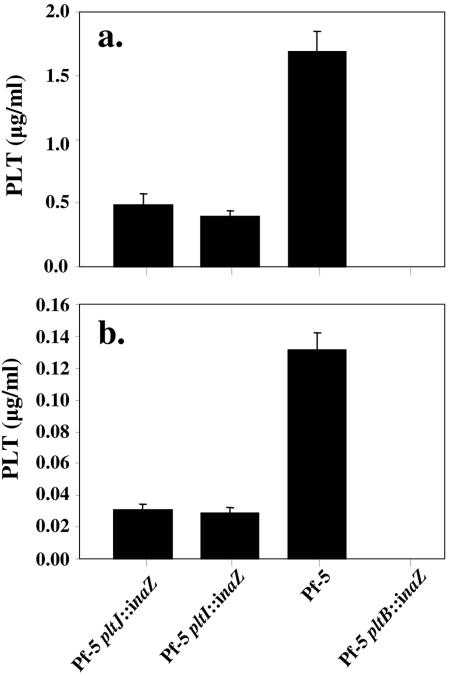
Derivatives of Pf-5 containing transcriptional fusions of the inaZ reporter gene to the chromosomal pltI and pltJ genes were derived via marker-exchange mutagenesis in Pf-5 (pyoluteorin-producing) and in pltR and pltF derivatives of Pf-5 (both pyoluteorin deficient). After 48 h of growth, 4.7 times more pyoluteorin was detected in extracts of culture supernatants from Pf-5 than in those from the pltI::inaZ or pltJ::inaZ mutant (P = 0.011) (Fig. 2). Similarly, pyoluteorin concentrations in cell extracts of Pf-5 were 5.2 times greater than those in cell extracts of the pltI or pltJ mutant (P = 0.009). Population sizes (not shown) did not differ significantly among strains at any time point (P = 0.43).
FIG. 2.
Pyoluteorin production by Pf-5 and by pltI::inaZ, pltJ::inaZ, and pltB::inaZ derivatives of Pf-5. Cultures were grown for 48 h in NBGly before being harvested and extracted, and supernatants (a) and cells (b) were used to determine pyoluteorin (PLT) concentrations. Panels a and b show data from the same experiment. Error bars denote 1 standard error.
Transcript analysis.
Sequence analysis predicts that pltI and pltJ reside in an operon of six genes that encode subunits of an ABC transport pore (Fig. 1a). To investigate the transcriptional organization of the pltIJKNOP locus, we utilized RT-PCR to evaluate RNAs isolated from Pf-5 and Pf-5 pltI::inaZ. Polycistronic transcripts were detected in Pf-5 using primers spanning contiguous genes in the locus. Portions of transcripts corresponding to pltIJ, pltJK, pltKN, pltNO, and pltOP were successfully amplified (Fig. 1c), providing evidence for the operonic organization of the pltIJKNOP locus. As expected, RT-PCR did not detect the presence of RNAs spanning the pltZ-pltI and pltP-orf1 regions, which are predicted to contain divergently and convergently transcribed genes, respectively.
RT-PCR analysis using primers internal to each gene (pltI, pltJ, pltK, pltN, pltO, and pltP) provided further evidence for the presence of a pltIJKNOP operon. Portions of transcripts corresponding to each gene were successfully amplified from Pf-5, but not from the pltI::inaZ mutant (Fig. 1b), indicating that the inaZ insertion had a polar effect on the transcription of downstream genes within the operon. However, following RT-PCR with pltP from the pltI::inaZ mutant, a very weak band was observed on agarose gels, suggesting a low level of pltP transcription in the pltI mutant.
Transcription of pltI and pltJ is reduced in Plt− derivatives of Pf-5.
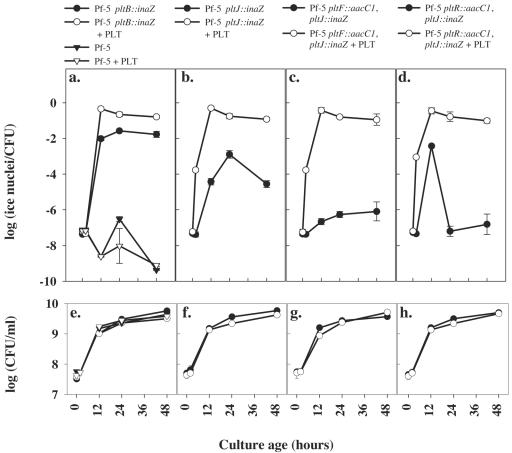
The transcription of pltI and pltJ was assessed over time, using the INA expressed in culture by the pltI::inaZ and pltJ::inaZ derivatives of Pf-5 (Fig. 3b and 4b). Although pltI and pltJ are predicted to lie within a single operon, INA expressed from pltI::inaZ (Fig. 3b) was lower than that expressed from pltJ::inaZ (Fig. 4b). The apparent difference in transcription between the two genes is thought to reflect positional effects of the inaZ insertion. In support of this supposition, a second set of pltJ::inaZ mutants bearing an inaZ insertion at an upstream NruI site showed an INA essentially identical to that from the corresponding pltI::inaZ fusion in wild-type, pltR, and pltF mutant backgrounds (data not shown) (6). However, an unexplained increase in inaZ expression between 12 and 24 h observed during all experiments for pltJ::inaZ in a pltR mutant background (Fig. 4d) was not observed for the strain bearing a pltJ::inaZ fusion at the NruI site or for pltI::inaZ in a pltR mutant background (data not shown).
FIG. 3.
Transcriptional activity of the inaZ reporter gene measured from the parental strain Pf-5, which lacks inaZ (a); pltI::inaZ in the wild-type Pf-5 background (b); pltI::inaZ in a pltF background (c); and pltI::inaZ in a pltR background (d). Cultures were grown in NBGly with (open symbols) or without (filled symbols) an amendment of 4 μg/ml pyoluteorin added at the time of inoculation and were harvested at 0, 2, and 48 h for assessments of INA (a to d) and population densities (e to h). Population sizes did not differ significantly between treatments at any time point. Error bars denote 1 standard error.
FIG. 4.
Transcriptional activity from pltB::inaZ in the wild-type Pf-5 background (a), pltJ::inaZ in the wild-type Pf-5 background (b), pltJ::inaZ in a pltF background (c), and pltJ::inaZ in a pltR background (d). The parental strain Pf-5 (Ina−; triangles) was included as a negative control (a). Cultures were grown in NBGly with (open symbols) or without (filled symbols) an amendment of 4 μg/ml pyoluteorin added at the time of inoculation and were harvested at 0, 2, 12, 24, and 48 h for assessments of INA (a to d) and population densities (e to h). Error bars denote 1 standard error.
The expression of biosynthetic genes in the pyoluteorin gene cluster of Pf-5 is dependent on the transcriptional regulator PltR (32). To determine if the putative transport genes are also regulated by PltR, we assessed pltI and pltJ expression in a pltR mutant background. After 48 h of growth, the INA expressed from both the pltI::inaZ and pltJ::inaZ fusions was approximately 100-fold lower in the pltR background than in the Pf-5 wild-type background (Fig. 3 and 4) (P ≤ 0.025). In a pltF background, the expression of pltI and pltJ was similarly reduced, in contrast to their expression in the wild-type background (Fig. 3 and 4). At no time were population sizes significantly different among strains; thus, growth differences among strains did not account for differences in INA.
Transcription of pltI and pltJ is enhanced by exogenous pyoluteorin.
Because pltR and pltF mutants both exhibit a Plt− phenotype, we hypothesized that the effects of pltR and pltF mutations on pltI and pltJ transcription might be due to a requirement for pyoluteorin. To test this hypothesis, the transcription of pltI and pltJ was compared in a nonamended culture medium and in a culture medium amended with 4 μg/ml pyoluteorin at the time of inoculation. Amendment with pyoluteorin restored INA expression to both pltI::inaZ (Fig. 3) and pltJ::inaZ (Fig. 4) derivatives. By 48 h, the INA expressed by the pltI::inaZ and pltJ::inaZ derivative strains in pyoluteorin-amended cultures was 1,000-fold higher (P ≤ 0.004) than that expressed in nonamended medium, and INA enhancement was independent (P = 0.78) of the genetic background (Fig. 3 and 4).
Slight differences in population sizes were observed between pyoluteorin-amended and nonamended cultures at some time points (Fig. 3 and 4). However, the values reported and analyzed for INA were calculated on a per-CFU basis; therefore, the small differences in population size did not contribute significantly to differences in INA.
The influence of pyoluteorin on the transcription of the biosynthetic gene pltB differed temporally from its influence on the transcription of the transport genes pltI and pltJ. At 2 h, INAs expressed by pltB::inaZ derivatives in pyoluteorin-amended and nonamended medium were not significantly different (Fig. 4a; P = 0.95), whereas the INA expressed from pltI::inaZ and pltJ::inaZ fusions at 2 h (Fig. 3b to d and 4b to d) was already enhanced at least 100-fold or 1,000-fold, respectively, by pyoluteorin amendment, in all genetic backgrounds (P ≤ 0.0004).
Differential effect of pyoluteorin on populations of Pf-5 and a pltI mutant.
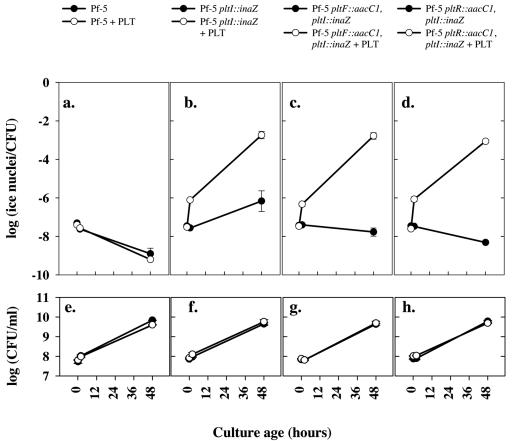
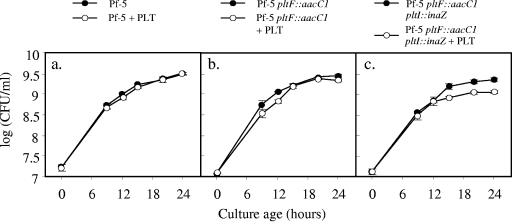
Toxic secondary metabolites such as pyoluteorin require some form of self-resistance. One common mechanism for providing self-resistance is efflux of the toxin via membrane transport. To evaluate the role of the pltIJKNOP transport locus in pyoluteorin resistance, we compared the growth of Pf-5 pltF::aacC1 (lacking pyoluteorin production) and Pf-5 pltF::aacC1 pltI::inaZ (defective both in pyoluteorin production and in the transport locus) in medium amended with pyoluteorin at 12 μg/ml. This concentration represents the high end within the range of pyoluteorin concentrations produced by Pf-5 in culture (7) and exceeds by twofold the MIC of pyoluteorin against E. coli (2), which is sensitive to the antibiotic. This concentration of pyoluteorin had little effect on the population of any of the three strains during the exponential growth phase (Fig. 5 and data not shown), but a significant effect on populations of Pf-5 pltF::aacC1 pltI::inaZ was observed as cells entered stationary phase (18- to 24-h time points; Fig. 5). In the medium amended with 12 μg/ml pyoluteorin, populations of Pf-5 pltF::aacC1 pltI::inaZ were ca. 0.3 log unit smaller than those in nonamended medium. In contrast, this concentration of pyoluteorin had no significant effect on populations of Pf-5. These data suggest a role for the pltIJKNO locus in the response of Pf-5 to pyoluteorin but also demonstrate that other factors are responsible for the pyoluteorin resistance of the bacterium.
FIG. 5.
Influence of pyoluteorin on growth of Pf-5 and derivatives deficient in pyoluteorin biosynthesis and transport. Pf-5 (a), Pf-5 pltF::aacC1 (deficient in pyoluteorin biosynthesis) (b), and Pf-5 pltF::aacC pltI::inaZ (deficient in pyoluteorin biosynthesis and transport) (c) were grown in NBGly medium (filled circles) or NBGly amended with 12 μg/ml pyoluteorin (open circles). Population sizes were estimated periodically by dilution plating of samples from three replicate cultures. Error bars denote 1 standard error.
DISCUSSION
This report provides evidence that a transport system encoded by the pltIJKNO cluster linked to the pyoluteorin biosynthetic locus is necessary for optimal pyoluteorin production by P. fluorescens Pf-5. The predicted protein products of pltIJKNO are likely to form an ABC efflux system characterized by paired cytoplasmic ATP-binding domains and paired transmembrane domains. The predicted PltJ polypeptide contains two fused ATP-binding domains within a single open reading frame. This arrangement is not uncommon for ABC transporters, whose ATP-binding domains occur in nearly every possible combination of fused and individual subunits (4). The adjacent genes pltK and pltN each have ABC transporter transmembrane domains; PltK and PltN could heterodimerize to form the paired transmembrane subunits characteristic of ABC transporters. We propose that the PltJKN ABC transporter in the inner membrane is linked to the outer membrane channel protein PltO by the membrane fusion protein PltI. ABC transporters whose cargoes are small molecules commonly utilize membrane fusion proteins as accessory factors (14), a function analogous to the proposed role of PltI as an accessory factor in the PltIJKNO transport system.
ABC transporters can serve as substrate importers or substrate exporters. Several lines of evidence suggest that the PltIJKNO transporter functions in the efflux of pyoluteorin. First, the locus shares the greatest sequence similarity with other ABC export systems: the gene cluster does not encode a periplasmic binding protein, which forms an integral part of ABC importers, and the gene cluster contains homologues of a membrane fusion protein and an outer membrane channel, which are associated with export systems. Second, its proximity to pyoluteorin biosynthesis genes and the induction of pltI and pltJ transcription by pyoluteorin indicate that pltIJKNO is very likely to function in pyoluteorin export. In many antibiotic efflux systems, the inducer of gene expression is also the substrate of transport (13). Finally, the transport system contributed partially to the resistance of Pf-5 to pyoluteorin in culture: upon entry into stationary phase, populations of a pltI mutant were reduced by the addition of pyoluteorin to the culture medium, whereas populations of Pf-5 were not reduced by the antibiotic. Therefore, this report provides compelling indirect evidence supporting a role for PltIJKNO in pyoluteorin efflux. Further studies are needed to provide direct evidence of the identity of the entity transported by PltIJKNO.
Mutations in pltI and pltJ greatly reduced the levels of pyoluteorin in culture supernatants of Pf-5, a finding consistent with a proposed role for pltIJKNO in pyoluteorin export. However, low extracellular pyoluteorin concentrations were not accompanied by pyoluteorin accumulation in the cells, as might be expected if PltIJKNO is required for pyoluteorin export. Rather, pyoluteorin concentrations in both cell and supernatant extracts of pltI and pltJ strains were similarly reduced relative to the wild type. The detection of pyoluteorin in culture supernatants of pltI and pltJ mutants probably indicates that PltIJKNO is not the sole mechanism for pyoluteorin export from the bacterial cell. Another observation supports the possibility that an alternative export mechanism for pyoluteorin exists: experiments comparing the self-resistance of Pf-5 and that of a pltI mutant to pyoluteorin indicated that PltIJKNO does not play a significant role in the self-resistance of Pf-5 to the antibiotic until the entry into stationary phase. Because the concentration of pyoluteorin used to amend the culture medium represents the high end of concentrations attained by wild-type Pf-5 under identical culture conditions, these results represent an authentic picture of the bacterium's self-resistance to pyoluteorin. We speculate that pltP, which is predicted to encode a LysE transporter, could function in pyoluteorin export. pltP is a member of the pltIJKNOP operon, but there could also be an alternative promoter for pltP, possibly located in the 65-bp intergenic region immediately 5′ of the gene. If so, PltP could export pyoluteorin from the cells of Pf-5 as well as from pltI or pltJ mutant derivatives. Alternatively, the pltIJKNOP operon could be weakly transcribed in the pltI mutant at levels below reliable detection by RT-PCR. Further studies are needed to identify alternative mechanisms for pyoluteorin export in Pf-5.
We describe herein the linked transport and biosynthetic genes for the autoinducer pyoluteorin, whereby optimal levels of autoinducer are achieved only if the transport genes are intact. Our observations resemble those reported for PhlE, a putative transporter of the antibiotic and autoinducer 2,4-diacetylphloroglucinol in P. fluorescens F113 (1). In a previous study, phlE mutants produced less 2,4-diacetylphloroglucinol than did the parental strain F113, and the ratios of antibiotic extracted from the cell to that in the culture supernatant were similar between the two strains. Therefore, neither pyoluteorin nor 2,4-diacetylphloroglucinol appears to accumulate in cells with defective export systems. Feedback inhibition of the antibiotics on the activity of enzymes catalyzing steps in biosynthesis could be one mechanism by which intracellular accumulation is prevented, although this possibility was not explored in this study. In the case of pyoluteorin, a reciprocal regulation of transport gene expression and pyoluteorin production was observed; mutations in pltI or pltJ repressed pyoluteorin accumulation, whereas enhanced levels of the antibiotic induced pltI and pltJ transcription. Similar reciprocal regulation has been shown for ABC transporters of inorganic phosphate (Pi) and maltose in Escherichia coli (5, 44) and for the ABC transporter that imports the autoinducer AI-2 in Salmonella enterica serovar Typhimurium (41, 42). In Pseudomonas aeruginosa, transport of the secondary metabolite pyoverdine (an iron-binding siderophore) was found to affect the expression of genes for pyoverdine biosynthesis (24) and transport (12, 34, 35) via the ferripyoverdine receptor and associated proteins (24, 35). Our study demonstrates that such reciprocal regulation also exists for another secondary metabolite in Pseudomonas spp., i.e., the antibiotic pyoluteorin.
In Pseudomonas sp. strain M18, pltZ serves as a transcriptional repressor of pyoluteorin biosynthetic genes, and a pltZ deletion results in enhanced pyoluteorin production (20). It is likely that pltZ plays a similar role in Pf-5, although this relationship has not been tested. TetR homologs are often involved in the cofactor-dependent regulation of genes, frequently linked, encoding biosynthesis or export functions for hydrophobic compounds (10, 17, 38, 46). Thus, it is also plausible that pltZ regulates the transcription of pltI and pltJ. In Azotobacter vinelandii, there is a cluster (encoding GenPept accession numbers ZP_00415237.1 through ZP_00415241) of colinear homologs of pltZ, pltI, pltJ, pltK, and pltN (but not pltG or pltO) sharing 45% and 65% identity at the amino acid level with the corresponding genes in Pf-5. The conserved synteny of pltZ and pltIJKNO in Pf-5 and A. vinelandii, a bacterium lacking pyoluteorin production, is consistent with the notion that PltZ serves as a regulatory component of the pltIJKNO gene cluster or its homolog in A. vinelandii. Elucidation of the mechanism by which pyoluteorin enhances the transcription of the transport genes pltI and pltJ is a subject for future study.
In conclusion, this report introduces a new aspect of pyoluteorin regulation, namely, its dependence on membrane transporter genes for optimal production. Expression of the transport genes pltI and pltJ is, in turn, highly sensitive to concentrations of extracellular pyoluteorin. This study provides the first evidence for reciprocal regulation of pyoluteorin production and a linked transport apparatus.
Acknowledgments
We thank Marcella Henkels and Brenda Shaffer for their extensive technical assistance and expertise and Brian Nowak-Thompson for providing purified pyoluteorin. We also thank the Center for Biotechnology and Gene Research at Oregon State University for sequencing support. We are grateful to Caroline Press, Sara Sawyer, and Virginia Stockwell for critical reviews of the manuscript.
This research was supported in part by a fellowship to M.B. from the U.S. Environmental Protection Agency’s Science to Achieve Results program (U-91582801-0) and by the Initiative for Future Agriculture and Food Systems of the USDA Cooperative State Research, Education and Extension Service (grant 2001-52100-11329).
REFERENCES
- 1.Abbas, A., J. E. McGuire, D. Crowley, C. Beysse, M. Dow, and F. O'Gara. 2004. The putative permease PhlE of Pseudomonas fluorescens F113 has a role in 2,4-diacetylphloroglucinol resistance and in general stress tolerance. Microbiology 150:2443-2450. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, D. M., R. E. Johnson, and U. J. Salvador. 1973. Pyrrole antibacterial agents. 1. Compounds related to pyoluteorin. J. Med. Chem. 16:1298-1300. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boos, W., and T. Eppler. 2001. Prokaryotic binding protein-dependent ABC transporters, p. 77-114. In G. Winkelmann (ed.), Microbial transport systems. Wiley VCH, Weinheim, Germany.
- 5.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodhagen, M. 2003. Ph.D. thesis. Oregon State University, Corvallis.
- 7.Brodhagen, M., M. D. Henkels, and J. E. Loper. 2004. Positive autoregulation and signaling properties of pyoluteorin, an antibiotic produced by the biological control organism Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 70:1758-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10: 685-686. [DOI] [PubMed] [Google Scholar]
- 9.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in procaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 10.Delaney, I., M. M. Sheehan, A. Fenton, S. Bardin, S. Aarons, and F. O'Gara. 2000. Regulation of production of the antifungal metabolite 2,4-diacetylphloroglucinol in Pseudomonas fluorescens F113: genetic analysis of phlF as a transcriptional repressor. Microbiology 146:537-546. [DOI] [PubMed] [Google Scholar]
- 11.Figurski, K. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gensberg, K., K. Hughes, and A. W. Smith. 1992. Siderophore-specific induction of iron uptake in Pseudomonas aeruginosa. J. Gen. Microbiol. 138:2381-2387. [DOI] [PubMed] [Google Scholar]
- 13.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harley, K. T., G. M. Djordjevic, T. T. Tseng, and M. H. Saier, Jr. 2000. Membrane fusion protein homologues in gram-positive bacteria. Mol. Microbiol. 36:516-517. [DOI] [PubMed] [Google Scholar]
- 15.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 16.Higgins, C. F. 2001. ABC transporters: physiology, structure, and mechanism—an overview. Res. Microbiol. 152:205-210. [DOI] [PubMed] [Google Scholar]
- 17.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 43:345-369. [DOI] [PubMed] [Google Scholar]
- 18.Howell, C. R., and R. D. Stipanovic. 1979. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69:480-482. [Google Scholar]
- 19.Howell, C. R., and R. D. Stipanovic. 1980. Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 70:712-715. [Google Scholar]
- 20.Huang, X., D. Zhu, Y. Ge, H. Hu, X. Zhang, and Y. Xu. 2004. Identification and characterization of pltZ, a gene involved in the repression of pyoluteorin biosynthesis in Pseudomonas sp. M18. FEMS Microbiol. Lett. 232:197-202. [DOI] [PubMed] [Google Scholar]
- 21.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 22.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 23.Kraus, J., and J. E. Loper. 1995. Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 61: 849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamont, I. L., P. A. Beare, U. Ochsner, A. I. Vasil, and M. L. Vasil. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 99:7072-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindgren, P. B., R. Frederick, A. G. Govindarajan, N. J. Panopoulos, B. J. Staskawicz, and S. E. Lindow. 1989. An ice nucleation reporter gene system: identification of inducible pathogenicity genes in Pseudomonas syringae pv. phaseolicola. EMBO J. 8:1291-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loper, J. E., and S. E. Lindow. 2001. Reporter gene systems useful in evaluating in situ gene expression by soil and plant-associated bacteria, p. 627-637. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 27.Martín, J. F., and P. Liras. 1989. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 43:173-206. [DOI] [PubMed] [Google Scholar]
- 28.Maurhofer, M., C. Keel, U. Schnider, C. Voisard, D. Haas, and G. Defago. 1992. Influence of enhanced antibiotic production in Pseudomonas fluorescens strain CHA0 on its disease suppressive capacity. Phytopathology 82:190-195. [Google Scholar]
- 29.Mendez, C., and J. A. Salas. 2001. The role of ABC transporters in antibiotic-producing organisms: drug secretion and resistance mechanisms. Res. Microbiol. 152:341-350. [DOI] [PubMed] [Google Scholar]
- 30.Murillo, J., H. J. Shen, D. Gerhold, A. Sharma, D. A. Cooksey, and N. T. Keen. 1994. Characterization of pPT23B, the plasmid involved in syringolide production by Pseudomonas syringae pv. tomato PT23. Plasmid 31:275-287. [DOI] [PubMed] [Google Scholar]
- 31.Nikaido, H., and J. A. Hall. 1998. Overview of bacterial ABC transporters. Methods Enzymol. 292:3-20. [DOI] [PubMed] [Google Scholar]
- 32.Nowak-Thompson, B., N. Chaney, J. S. Wing, S. J. Gould, and J. E. Loper. 1999. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J. Bacteriol. 181:2166-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowak-Thompson, B., S. J. Gould, and J. E. Loper. 1997. Identification and sequence analysis of the genes encoding a polyketide synthase required for pyoluteorin biosynthesis in Pseudomonas fluorescens Pf-5. Gene 204:17-24. [DOI] [PubMed] [Google Scholar]
- 34.Poole, K., S. Neshat, and D. Heinrichs. 1991. Pyoverdine-mediated iron transport in Pseudomonas aeruginosa: involvement of a high-molecular-mass outer membrane protein. FEMS Microbiol. Lett. 78:1-6. [PubMed] [Google Scholar]
- 35.Rédly, G. A., and K. Poole. 2003. Pyoverdine-mediated regulation of FpvA synthesis in Pseudomonas aeruginosa: involvement of a probable extracytoplasmic-function sigma factor, FpvI. J. Bacteriol. 185:1261-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22:1-20. [DOI] [PubMed] [Google Scholar]
- 38.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, C. Gigot-Gonnefoy, C. Reimmann, R. Notz, G. Défago, D. Haas, and C. Keel. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sigrist, C. J., L. Cerutti, N. Hulo, A. Gattider, L. Falquet, M. Pagni, A. Bairoch, and P. Bucher. 2002. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 3:265-274. [DOI] [PubMed] [Google Scholar]
- 40.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 41.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 42.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 43.Walker, J. E., M. Saraste, M. J. Runswick, and J. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wanner, B. L. 1995. Signal transduction and cross regulation in the Escherichia coli phosphate regulon by PhoR, CreC, and acetyl phosphate, p. 203-221. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 45.West, S. E. H., and B. H. Iglewski. 1988. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 16:9323-9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, K., L. Han, and L. C. Vining. 1995. Regulation of jadomycin B production in Streptomyces venezuelae ISP5230: involvement of a repressor gene, jadR2. J. Bacteriol. 177:6111-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]