Abstract
A mutation was discovered in the Pribnow box of the ampC promoter in a clinical Escherichia coli strain. This −11 C-to-T transition created a perfect homology with the −10 consensus sequence. The new promoter was cloned upstream of the cat gene of pKK232-8 and induced a sixfold increase in promoter strength.
The production of the AmpC β-lactamase in Escherichia coli is not inducible, as are most class C cephalosporinases, but is constitutive because of the absence of the ampR regulator gene. Consequently, in E. coli the level of transcription of the ampC gene depends mostly on the strength of the ampC promoter (8, 10).
E. coli promoters harbor two hexamers of conserved sequences, the −35 region and the −10 one, called the Pribnow box, which play an important role in the level of gene transcription (6). The −35 consensus sequence TTGACA and the −10 consensus sequence TATAAT have been defined. The closer to the consensus the sequences are, the stronger the promoter. The interbox distance also plays an important role in the transcription rate. Ideally, it is 17 bp long. In E. coli K-12, the ampC promoter differs from the consensus by one base in each box (−35 box, TTGTCA; −10 box, TACAAT) and the interbox distance is only 16 bp long. On the other hand, ampC promoter specificity is determined by the presence of an attenuator, which is a transcription terminator for the frd operon with a hairpin structure and which decreases the efficiency of the RNA polymerase and leads to low-level production of the enzyme (1, 4).
Clinical E. coli strains overproducing AmpC cephalosporinase have been described. Most of them presented a −42 mutation that created a new stronger and relocated upstream −35 box. −32 mutations creating a consensus −35 box have also been described (3).
In this paper, we report a clinical E. coli strain presenting a mutation in the Pribnow box, leading to the −10 consensus sequence.
(This study was presented at the 41st Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 16 to 19 December 2001.)
E. coli strain 99004202 was isolated from a blood culture. MICs for various β-lactams (Table 1) were determined by the E-test method (AES Laboratory, Combourg, France). The presence of a TEM-type β-lactamase was detected in this strain using TEM-specific primers TEM A (5′GACTGGATGGAGGCGGAT3′) and TEM B (5′CAATGCTTAATCAGTGAGGC3′), amplifying a 239-bp fragment. Cephalosporinase activity was measured using cephalothin as a substrate in a Beckman DU 7500 spectrophotometer and showed a fivefold increase for strain 99004202 compared with E. coli strain ATCC 25922.
TABLE 1.
MICs of several β-lactams for E. coli strain 99004202
| Antibiotic | MIC (mg/liter) |
|---|---|
| Amoxicillin | >256 |
| Amoxicillin-clavulanic acida | 64 |
| Ticarcillin | >256 |
| Ticarcillin-clavulanic acida | 64 |
| Piperacillin | 32 |
| Piperacillin-tazobactamb | 2 |
| Cephalothin | 64 |
| Cefuroxime | 32 |
| Cefoxitin | 4 |
| Cefotaxime | 0.12 |
| Ceftazidime | 0.5 |
| Imipenem | 0.06 |
Clavulanic acid was added at 2 mg/liter.
Tazobactam was added at 4 mg/liter.
Primers AB1 (5′GATCGTTCTGCCGCTGTG3′) and AmpC2 (5′GGGCAGCAAATGTGGAGCAA3′) were used to amplify a 271-bp fragment from the E. coli ampC promoter containing the −35 box, the −10 box, and the attenuator (3). PCR amplification was performed using a Perkin-Elmer 480 DNA thermal cycler (Perkin-Elmer Applied Biosystems, Cergy-Pontoise, France) in a final volume of 50 μl containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgC12, 200 μM (each) nucleotides, 0.5 μM (each) primer, 2.5 U of Taq DNA polymerase (Promega, Charbonnières, France), and 10 ng of target DNA. After 90 s of denaturation at 94°C, 30 PCR cycles were performed, each consisting of 30 s of denaturation at 94°C, 30 s of annealing at 57°C, and 60 s of extension at 72°C, followed by a final extension step of 7 min at 72°C. The PCR fragment was purified and sequenced with the ABI PRISM dye terminator cycle sequencing ready reaction kit (Perkin-Elmer). Sequence analysis was performed on an ABI377 DNA sequencer (Perkin-Elmer).
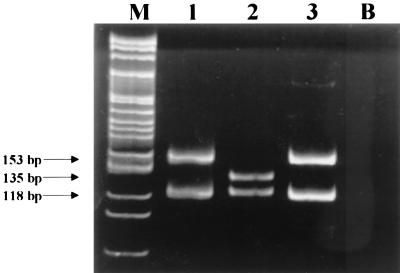
The sequence of the promoter from strain 99004202 is presented in Fig. 1 and compared with the E. coli K-12 ampC promoter (7) and the promoter of a previously synthesized mutant, 96010266M (3), which only differs by the −11 mutation from the promoter of strain 99004202. This mutation can easily be detected by MaeIII digestion. The restriction was performed at 55°C for 1 h in a final volume of 25 μl containing 1 U of MaeIII (Roche, Meylan, France) in 40 mM Tris-HCl-550 mM NaCl-12 mM MgCl2-14 mM 2-mercaptoethanol (pH 8.2). Indeed, the −11 transition abolished one of the two restriction sites present in the PCR fragment and the variation in fragment size was detected on an 8% acrylamide gel (Fig. 2).
FIG. 1.
ampC promoter sequence of E. coli K-12 (7), mutant strain 96010266M (3), and clinical isolate 99004202, a moderate-level AmpC producer.
FIG. 2.
Detection of the −11 mutation in the ampC gene promoter of E. coli by MaeIII digestion. Lanes: M, molecular size marker; 1 and 3, E. coli strain 99004202 promoter; 2, strain ATCC 25922 promoter; B, blank.
In order to study the role of the −11 mutation, both promoters were then compared in a gene reporter study. The 271-bp fragments of the ampC promoter from strains 99004202 and 96010266M were cloned in the post-PCR cloning vector pGEM-T easy (Promega). The plasmid DNA was digested with EcoRI and the fragment containing the ampC promoter was recovered from a low-melting-point agarose gel (Promega), blunt-ended with the Klenow fragment of E. coli DNA polymerase I (Promega), and ligated into the SmaI restriction site of pKK232-8 (Amersham Pharmacia Biotech, Uppsala, Sweden) previously treated with calf intestinal phosphatase (Promega). The pKK232-8 reporter plasmid carrying the chloramphenicol acetyltransferase (CAT) gene was used to clone the different ampC promoters of E. coli upstream of this CAT gene in order to study their strength. After overnight incubation at 16°C, the ligation products were used to transform JM109 highly competent cells (Promega). The sequence of the inserted fragment was verified using two primers from pKK232-8 (pKK1 [5′TGCGAAGCAACGGCCCGG3′] and pKK2 [5′AAGCTTGGCTGCAGGTCGA3′]), amplifying a 389-bp fragment containing the 271 bp from the ampC gene.
The chloramphenicol MIC from JM109 bacteria transformed with both constructions was determined by serial twofold dilutions in Mueller-Hinton agar (Difco Laboratories, Detroit, Mich.). Inocula of 104 CFU/spot from an 18-h culture in Mueller-Hinton broth were applied with a Steers multiple-inoculum replicator. After 18 h of incubation at 37°C, the chloramphenicol MIC was defined as the lowest concentration preventing bacterial growth on the plate.
CAT concentration from the different constructions was assayed in crude bacterial extracts prepared by sonication using a Branson sonifier 250 (Branson Ultrasonics, Danbury, Conn.) (intermittent exposure five times for 30 s each). Total protein concentration was determined by a pyrogallol red-molybdate method (Roche) (11). CAT enzyme was measured by a sandwich enzyme-linked immunosorbent assay test (Roche). Each measure was performed in triplicate. Untransformed JM109 bacteria and JM109 containing pKK232-8 without insert were used as controls.
The results of chloramphenicol MIC and CAT determinations are presented in Table 2. A 16-fold increase in chloramphenicol MIC and a 6-fold increase in CAT concentration were observed when the −11 mutation was present.
TABLE 2.
Comparison of the strength of the − 11 mutated ampC promoter from E. coli strain 99004202 with that of the ampC promoter from strain ATCC 25922 and that from the mutant 96010266M (3)
| Plasmid used to transform JM109 cells | Chloramphenicol MIC (mg/liter) | CAT level (pg/μg)a |
|---|---|---|
| None | 8 | 4 ± 0.5 |
| pKK232-8b | 8 | 4 ± 0.5 |
| pKK232-8 (ATCC 25922)c | 32 | 120 ± 32 |
| pKK232-8 (99004202)d | 512 | 761 ± 41 |
| pKK232-8 (96010266M)e | 32 | 95 ± 8 |
Mean ± standard deviation of three determinations of CAT concentration.
pKK232-8 without any insert.
pKK232-8 containing the ampC promoter from strain ATCC 25922 (low-level AmpC producer).
pKK232-8 containing the ampC promoter from strain 99004202 (moderate-level AmpC producer).
pKK232-8 containing the ampC promoter from the mutant 96010266M (3).
The sequence of the −35 and −10 hexamers and the interbox distance are crucial for efficient binding of RNA polymerase, influencing the level of transcription of the gene. Furthermore, an interbox distance of 16 bp and the presence of a hairpin attenuator structure contribute to low-level transcription of the gene. Mutations have already been detected that create a −35 consensus sequence, TTGTCA. The new ampC promoter we described showed a mutation in the Pribnow box, leading to the −10 consensus sequence. Mutations in the Pribnow box have also been described in the promoters of other β-lactamase genes. In Shigella flexneri, the mechanism of hyperproduction of TEM-1 β-lactamase was found to be related to a point mutation in the Pribnow box (2). The same phenomenon was observed for the chromosomal β-lactamase of Klebsiella oxytoca (5). Recently, Rice et al. demonstrated by site-directed mutagenesis that a single base pair change in the Pribnow box resulted in a substantial increase in the ampicillin MIC of Klebsiella pneumoniae overproducing SHV-1 (12).
However, numerous ampC promoters have already been sequenced, and the −11 mutation in AmpC overproducers seems to be relatively rare compared to the −42 one. We demonstrated that a −11 substitution induced only a moderate increase of the promoter strength (6×), leading to a moderate increase in β-lactam MICs. These results confirmed those obtained in 1981 by Jaurin and Grundström with selected in vitro strains showing only a 7-fold increase in MICs with this mutation compared to a 21-fold increase with the −32 one (7). Low et al., studying the evolution of E. coli strains from multiple infected liver cysts, detected highly resistant strains (MIC for ceftazidime = 32 mg/liter) that also presented the −11 mutation in the ampC promoter, but it was always associated with one or two mutations in the attenuator loop and the exact effect of each one could not be determined (9).
Note that in the promoter of strain 99004202 the −11 mutation was associated with different sequence polymorphisms previously detected in the −42 and −32 mutated ampC promoters, at the −88, −82, −18, −1, and +58 positions. It seems that ampC promoter mutations increasing the transcription rate occur on a specific genetic framework.
However, a great variability can be observed in the ampC promoters from enzyme overproducers, and, particularly, the role of mutations described in the attenuator hairpin and insertions increasing the interbox distance need to be studied in detail.
REFERENCES
- 1.Bergström, S., F. P. Lindberg, O. Olsson, and S. Normark. 1983. Comparison of the overlapping frd and ampC operons of Escherichia coli with the corresponding DNA sequences in other gram-negative bacteria. J. Bacteriol. 155:1297-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caniça, M. M., C. Y. Lu, R. Krihnamoorthy, and G. C. Paul. 1997. Molecular diversity and evolution of blaTEM genes encoding β-lactamases resistant to clavulanic acid in clinical E. coli. J. Mol. Evol. 44:57-65. [DOI] [PubMed] [Google Scholar]
- 3.Caroff, N., E. Espaze, D. Gautreau, H. Richet, and A. Reynaud. 2000. Analysis of the effects of −42 and −32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing AmpC. J. Antimicrob. Chemother. 45:783-788. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T. 1984. Molecular and genetic aspects of the fumarate reductase of Escherichia coli. Biochem. Soc. Trans. 12:237-238. [DOI] [PubMed] [Google Scholar]
- 5.Fournier, B., A. Gravel, D. C. Hooper, and P. H. Roy. 1999. Strength and regulation of the different promoters for chromosomal Klebsiella oxytoca. Antimicrob. Agents Chemother. 43:850-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawley, D., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaurin, B., and T. Grundström. 1981. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of β-lactamases of the penicillinase type. Proc. Natl. Acad. Sci. USA 78:4897-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaurin, B., T. Grundström, and S. Normark. 1982. Sequence elements determining ampC promoter strength in E. coli. EMBO J. 1:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Low, A. S., F. M. McKenzie, I. M. Gould, and I. R. Booth. 2001. Protected environments allow parallel evolution of a bacterial pathogen in a patient subjected to long-term antibiotic therapy. Mol. Microbiol. 42:619-630. [DOI] [PubMed] [Google Scholar]
- 10.Nelson, E. C., and B. G. Elisha. 1999. Molecular basis of AmpC hyperproduction in clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 43:957-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orsonneau, J. L., P. Douet, C. Massoube, P. Lustenberger, and S. Bernard. 1989. An improved pyrogallol red-molybdate method for determining total urinary protein. Clin. Chem. 35:2233-2236. [PubMed] [Google Scholar]
- 12.Rice, L. B., L. L. Carias, A. M. Hujer, M. Bonafede, R. Hutton, C. Hoyen, and R. A. Bonomo. 2000. High-level expression of chromosomally encoded SHV-1 β-lactamase and an outer membrane protein change confer resistance to ceftazidime and piperacillin-tazobactam in a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]