Abstract
We report here the development, validation, and use of three real-time PCR assays to quantify the abundance of the following three groups of tetracycline resistance genes: tet(A) and tet(C); tet(G); and tet genes encoding ribosomal protection proteins, including tet(M), tet(O), tetB(P), tet(Q), tet(S), tet(T), and tet(W). The assays were validated using known numbers of sample-derived tet gene templates added to microbiome DNA. These assays are both precise and accurate over at least 6 log tet gene copies. New tet gene variants were also identified from cloned tet amplicons as part of this study. The utility of these real-time PCR assays was demonstrated by quantifying the three tet gene groups present in bovine and swine manures, composts of swine manure, lagoons of hog house effluent, and samples from an Ekokan upflow biofilter system treating hog house effluent. The bovine manures were found to contain fewer copies of all three groups of tet genes than the swine manures. The composts of swine manures had substantially reduced tet gene abundance (up to 6 log), while lagoon storage or the upflow biofilter had little effect on tet gene abundance. These results suggest that the method of manure storage and treatment may have a substantial impact on the persistence and dissemination of tet genes in agricultural environments. These real-time PCR assays provide rapid, quantitative, cultivation-independent measurements of 10 major classes of tet genes, which should be useful for ecological studies of antibiotic resistance.
Studies of antimicrobial resistance (AR) have largely been limited to cultured bacterial pathogens, but it is now well recognized that AR is much more widespread in the bacterial world, including in commensal bacteria from the gut (1, 31, 35). Commensal bacteria resistant to antibiotics may also pose a potential threat to human health for several reasons. First, commensal bacteria and the AR genes they harbor can enter the human food chain, either through foods grown in fields fertilized with animal manure or wastewater or by other routes, such as meat or milk products (8, 13, 19, 35). Second, some commensal bacteria are also opportunistic pathogens (22, 34). Furthermore, due to their enormous abundance, commensal bacteria can serve as a reservoir of AR genes and probably contribute to AR gene transfer among bacteria, including pathogenic bacteria (42). The dynamics of AR in microbiomes is therefore a pertinent issue for microbial ecologists (3). The Scientific Advisory Panel for Facts about Antimicrobials in Animals and the Impact on Resistance emphasized that the ecology of AR in agriculture should be a research priority (12). In that context, AR present in all members (pathogenic or commensal, culturable or nonculturable) of microbiomes should be examined so that the true gene reservoirs of a particular AR, and their dynamics, can be assessed.
The application of PCR to detecting AR genes in both bacterial isolates and environmental samples has provided additional insights into the occurrence of AR in various environments (4, 5, 9, 15, 24). The demonstration that AR genes are detectable in groundwater samples collected downstream of livestock production environments and animal waste lagoons (4, 9) has further heightened concerns about the role of agriculture in the dissemination of AR (13, 32). The use of antibiotics in food-producing animals, especially at subtherapeutic levels, is also widely believed to contribute substantially to the increased prevalence of AR (10, 39, 43), but there are also conflicting opinions (14, 27). The lack of quantitative data pertaining to the relationship between the use of antibiotics in food-producing animals and the emergence, spread, and persistence of AR underpins this discrepancy of opinions. Quantitative methods of analysis which can quantify AR in entire microbiomes of animal manure and manure treatment facilities (such as farm lagoons and manure compost) would be very useful for understanding the microbial ecology of AR and for the development of strategies to mitigate AR.
Real-time PCR is now widely used in life science research and diagnosis because of its sensitivity, accuracy, precision, and high-throughput capacity (16, 18, 21). For instance, real-time PCR is now a method of choice in molecular diagnoses to detect and quantify pathogens (25, 28, 30) and in the enumeration of particular bacteria in environments (7, 33). Recently, real-time PCR was also reported for the quantification of a single tet gene, tet(Q), in several clinical plaque samples (23). We report here the development and validation of real-time PCR assays to quantify three of the major groups (including 10 classes) of tet genes: the tet(A) and tet(C) group, the tet(G) group, and the ribosomal protection protein (RPP) tet genes, including tet(M), tet(O), tetB(P), tet(Q), tet(S), tet(T), and tet(W). We chose tet genes because of their ubiquity in the environment, the availability of numerous tet gene sequences that facilitate the design of PCR primers, and the widespread use of tetracyclines in both the treatment of human infections and food-animal production. The utility of the real-time PCR assays was tested by quantifying the tet gene abundance in samples of swine and bovine manures, swine wastewater lagoons, an Ekokan upflow biofilter system, and composted swine manures. The diversity of tet genes recovered from one of the bovine manures was also examined.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica serovar Typhimurium strain 96-5227 carrying the tet(G) gene was kindly provided by Michael Mulvey (Health Canada, Winnipeg, Canada) and maintained on Luria-Bertani (LB) agar plates containing 10 μg/ml of tetracycline chloride. Escherichia coli strain TOPO10 carrying the tet(H) gene on plasmid pMHT1 was kindly provided by Stefan Schwarz (Institute of Animal Science and Animal Behavior, Celle, Germany). This strain, as well as E. coli strain XL1-Blue (Stratagene, La Jolla, CA) carrying tet(A) and E. coli strain HB101 (Promega, Madison, WI) carrying the tet(C) gene on plasmid pBR328, was grown on LB agar plates supplemented with 25 μg/ml of tetracycline chloride.
Microbiome samples and DNA extraction.
Fresh fecal samples were collected from 24 beef cattle kept at The Ohio State University farm. These fecal samples were randomly pooled into six composite samples of equal wet weight prior to DNA extraction. The swine fecal samples were collected from six different swine farms located in North Carolina and Ohio. Samples were also collected from the manure treatment systems associated with three of these farms. These systems included a conventional flush and lagoon storage system in North Carolina, an Ekokan upflow biofilter system in North Carolina (41), and a high-rise hog house system in Ohio (20). For the conventional and high-rise hog systems, samples were collected from two different herds. The treatment system samples collected included flush water and lagoon storage liquid on two separate occasions. The bacterial biomass was harvested by centrifugation at 16,000 × g at 4°C. Samples of compost were collected after 1, 2.5, and 3 months of composting. Representative sampling was conducted by collecting composites of multiple samples from various locations, depths, and cross sections from the compost. All samples were frozen immediately after sampling and stored at −20°C prior to analysis.
Total microbiome DNA was extracted from beef cattle manure samples using the RBB+C method (45), and a QIAamp DNA stool mini kit (QIAGEN, Inc., Valencia, CA) was used for all other samples. Genomic DNAs and plasmid DNAs from pure bacterial cultures were extracted using standard protocols (6). After visual assessment of the DNA quality by agarose gel electrophoresis, the resultant community DNA was quantified spectrophotometrically.
Phylogenetic analysis of tet genes, primer design, and specificity tests.
The primers used for this study are described in Table 1. All of the tet gene sequences comprising Tet classes A to E, G, and H currently available in GenBank were retrieved and aligned using ClustalX (36). The cmlA5 gene, which encodes an efflux protein for chloramphenicol resistance in E. coli, was used as an outgroup, and a neighbor-joining tree was inferred as described previously (44). The tet gene sequences of each cluster within the neighbor-joining tree were then separated and realigned, and the most conserved regions were used for primer design. The candidate primer sequences were then used to query all GenBank DNA sequences using BLAST to ensure that there were no nonspecific matches outside of the targeted tet gene groups. The candidate primers that matched exclusively with the desired groups of tet genes were then analyzed using PRIMER DESIGNER (version 2; Scientific & Educational Software, Durham, NC). Wherever necessary, degenerate bases were introduced into the primers to match all the sequences in the alignments. Using these methods, three primer pairs were designed to target two groups (three classes) of efflux tet genes (Table 1). The RPP tet primers Ribo2-FW and Ribo2-RV designed by Aminov et al. (5) were used to amplify seven classes of RPP tet genes.
TABLE 1.
PCR primer sequences, targets, annealing temperatures, and amplicon lengths
| Primer | Class targeted | Primer sequence (5′→3′) | Primer annealing temp (°C)c | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| tetAC-150fa | Tet A, C | GCT RTA TGC GTT GRT GCA AT | 58 | 567 | This study |
| tetAC-716ra | Tet A, C | TCC TCG CCG AAA ATG ACC | |||
| tetG-247fb | Tet G | GTC GAT TAC ACG ATT ATG GC | 57 | 432 | This study |
| tetG-678rb | Tet G | CAC TTG GCC GAT CAG TTG A | |||
| Ribo2-FW | Tet M, O, P, Q, S, T, W | GGM CAY RTG GAT TTY WTI GC | 52 | 1,315 | 5 |
| Ribo2-RV | Tet M, O, P, Q, S, T, W | TCI GMI GGI GTR CTI RCI GGR C |
“Regular” PCR was done on a PTC-100 thermocycler (MJ Research, Waltham, MA) in a 50-μl volume containing 1× PCR buffer (20 mM Tris-HCl [pH 8.4] and 50 mM KCl), a 200 μM concentration of each deoxynucleoside triphosphate, a 500 nM concentration of each primer, 1.75 mM MgCl2, 670 ng/μl bovine serum albumin, 1.25 U Platinum Taq DNA polymerase (Invitrogen Corporation, Carlsbad, CA), which allows hot-start PCR, and 1.0 μl DNA. After an initial denaturation step at 94°C for 4 min, 5 cycles of touchdown PCR (denaturation at 94°C for 30 s, annealing for 30 s with a 1°C-per-cycle decrement from 5°C above the annealing temperature to the final annealing temperature indicated in Table 1, and extension at 72°C for 1 min) were performed, followed by 30 regular cycles of PCR (94°C for 30 s, 30 s at the respective annealing temperature, and 72°C for 30 s) and a final extension for 7 min at 72°C. With primers Ribo2-FW and Ribo2-RV, an annealing temperature of 52°C and a longer extension time (1 min longer) were used. The optimal annealing temperatures were predetermined by gradient PCR using a RoboCycler (Stratagene). No-template controls were included in parallel.
PCR products were cloned into the TOPO-TA cloning vector (Invitrogen). Randomly selected clones were sequenced by the Plant and Microbe Genome Facility at The Ohio State University. Both strands of the cloned efflux tet genes were completely sequenced, while the cloned RPP tet genes, which are about 1.3 kb long, were sequenced from both ends. Following visual examination for base calling, all of these newly obtained sequences were first compared among themselves with BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and those sharing >99% identity were regarded as the same tet gene sequences and are listed as one phylotype. Putative tet sequences were identified by BLASTn searches (2). The BLASTn search output alignments were also examined for the presence of breakage, which can result from chimeric sequences.
Real-time PCR.
The regular PCR described above was used to generate sample-derived DNA standards for each real-time PCR assay. Two sets of such DNA standards were prepared from the two sets of microbiome DNA, i.e., the DNAs extracted from bovine manures and the DNAs derived from swine manures, swine lagoons, swine manure composts, and the Ekokan upflow biofilter system. The PCR products derived with each primer pair from each DNA set were pooled and purified using a QIAquick PCR purification kit (QIAGEN). The resultant DNA concentrations were quantified fluorimetrically using a PicoGreen dsDNA quantitation kit (Molecular Probes, Inc., Eugene, OR). The copy number of each DNA standard was calculated based on the mass concentration and the average molecular weight of the respective tet amplicons. Tenfold serial dilutions in Tris-EDTA of each DNA standard were prepared prior to real-time PCR assays. In total, six real-time PCR standards were prepared from the two sets of microbiome DNA samples for the three real-time PCR assays. Each of these standards was used in real-time PCR assays.
The conditions for real-time PCR were the same as those described above, with the following exceptions: a decreased primer concentration (250 nM each) was used, and 0.133× SYBR green I (Molecular Probes) and a 30 nM reference dye (Stratagene) were included. The thermal profiles consisted of the following four segments: (i) initial denaturation at 95°C for 4 min; (ii) 5 touchdown cycles of 94°C for 30 s, the respective annealing temperature (Table 1) for 30 s, with a 1°C decrement per cycle, and 72°C for 40 s (90 s for RPP tet genes); (iii) 45 cycles of 94°C for 30 s, the respective annealing temperature for 30 s, 72°C for 30 s (75 s for RPP tet genes), with a 1-s increment per cycle, and 86°C for 18 s; and (iv) 95°C for 2 min, 55°C for 30 s, and 95°C for 30 s. Fluorescence data were collected at the 72°C and 86°C steps (end points) of the third segment and during ramping from 55°C to 95°C (all points) of the last segment. All real-time PCR assays were performed using an Mx3000p machine (Stratagene). Baseline and threshold calculations were performed with Mx3000p software, using the fluorescence signals acquired at 86°C, at which primer dimers completely denature and will not affect quantification. Following real-time PCR, all products were analyzed by agarose gel electrophoresis and melting curve analysis. All real-time PCRs were done in triplicate for both the standards and the microbiome DNA samples.
To assess the precision and accuracy of each real-time PCR assay as well as to evaluate whether the microbiome DNA extracts contained a factor(s) that was inhibitory to PCR, the sample-derived real-time PCR standards were serially diluted to give 1 to 108 copies per μl, and a 1.0-μl aliquot of each dilution was used to “spike” 100-ng amounts of microbiome DNA. A parallel series of samples containing 100 ng microbiome DNA mixed with 1.0 μl Tris-EDTA buffer were also prepared. The tet gene copies in both series of samples were quantified as described above against respective sample-derived real-time PCR standards. The spiked samples were then corrected for the background copies of tet genes—derived from the microbiome DNA itself—allowing the actual copy number of tet genes measured from each standard addition to be plotted against its theoretical amount, and by doing so, allowing the linear range of the PCR assay to be determined. The detection limit of each real-time PCR assay was determined from serial dilutions of the sample-derived standard templates. Following these validation experiments, the abundance of each tet gene group present in each microbiome DNA sample was quantified against its respective sample-derived standard, using the real-time PCR conditions described above. The abundance (copies g−1, or copies ml−1 in the case of liquid samples) of each tet gene group was calculated by multiplying the copy number value per real-time PCR by the number of reactions that could be done with the DNA derived from 1 g or ml of each sample.
Statistical analysis.
The data were analyzed using the mixed procedure of SAS 9.1 (SAS Institute, Cary, NC). Least-square means (LSM) were generated for all data. Mean separation was conducted by using Fisher's protected least significant difference test, with significance declared at P values of ≤0.05.
Nucleotide sequence accession numbers.
The tet gene sequences produced by this study have been deposited in GenBank under the accession numbers listed in Table 2.
TABLE 2.
Affiliations of sequenced tet genes, as determined by comparison to GenBank sequences
| tet class | tet clone; prevalence (accession no.) | Most similar match (accession no.) | Identity (%) |
|---|---|---|---|
| tet(A) | BC-AC-1; 8/10 (AY171576) | Gram-negative bacteria, Tn1721 (X61367) | 100 |
| BC-AC-2; 1/10 (AY171577) | Gram-negative bacteria, Tn1721 (X61367) | 98.2 | |
| tet(C) | BC-AC-3; 1/10 (AY171578) | Aeromonas salmonicida, pRAS3.2 (AY043299) | 99.8 |
| tet(G) | BC-G-1; 3/14 (AY171579) | Salmonella enterica serovar Typhimurium (AF261825) | 94.2 |
| BC-G-2; 1/14 (AY171580) | Salmonella enterica serovar Typhimurium (AF261825) | 94.4 | |
| BC-G-3; 2/14 (AY171581) | Mannheimia haemolytica (AJ276217) | 96.0 | |
| BC-G-4; 3/14 (AY171582) | Pseudomonas sp./pPSTG1 (AF133139) | 100 | |
| BC-G-5; 2/14 (AY171583) | Pseudomonas sp./pPSTG1 (AF133139) | 98.3 | |
| BC-G-6; 3/14 (AY171584) | Pseudomonas sp./pPSTG1 (AF133139) | 95.6 | |
| tet(O) | BC-RPP-1; 2/31 (AY171585) | Campylobacter jejuni (M18896) | 99.4 |
| BC-RPP-2; 3/31 (AY171586) | Streptococcus mutans (M20925) | 99.0 | |
| BC-RPP-3; 2/31 (AY171587) | Campylobacter jejuni (M18896) | 94.8 | |
| tet(M) | BC-RPP-4; 1/31 (AY171588) | Clostridium septicum (AB054984) | 98.9 |
| tet(Q) | BC-RPP-5; 3/31 (AY171589) | Bacteroides fragilis tet(Q)3 (Y08615) | 96.5 |
| BC-RPP-6; 11/31 (AY171590) | Bacteroides fragilis tet(Q)3 (Y08615) | 100 | |
| BC-RPP-7; 1/31 (AY171591) | Bacteroides fragilis tet(Q)3 (Y08615) | 96.7 | |
| BC-RPP-8; 1/31 (AY171592) | Bacteroides fragilis tet(Q)3 (Y08615) | 96.0 | |
| BC-RPP-9; 2/31 (AY171593) | Bacteroides fragilis tet(Q)3 (Y08615) | 97.8 | |
| tet(W) | BC-RPP-10; 1/31 (AY171594) | Butyrivibrio fibrisolvens (AJ427421) | 99.9 |
| BC-RPP-11; 2/31 (AY171595) | Bifidobacterium sp. strain ISO3519 (AF202986) | 96.8 | |
| BC-RPP-12; 2/31 (AY171596) | Arcanobacterium pyogenes (AY049983) | 94.9 |
RESULTS
Primer specificity and tet gene diversity.
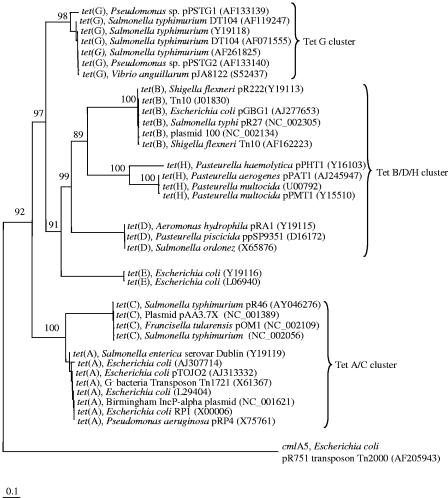
It was not possible to design “universal” primers for all seven prevalent classes (A to E, G, and H) of efflux tet genes because of the degree of sequence divergence among them (Fig. 1). However, our in silico analysis suggested that it was possible to design at least three primer pairs that would target six classes of efflux tet genes (Table 1). The tetAC-150f/716r, tetG-247f/678r, and Ribo2-FW/RV primer pairs all generated single bands of the expected size from DNA preparations from both pure cultures and microbiomes, suggesting that these primers are specific (data not shown). The tetBDH-55f/1029r primer pair (not shown) produced the expected PCR product from DNA prepared from the E. coli strain carrying tet(H), whereas multiple bands resulted from microbiome DNA, even at an increased stringency (data not shown). This primer pair was not used in further analyses. The Ribo2-FW/RV primer set was reported to produce nonspecific bands from microbiome DNA (5), but the modifications made here (the use of a hot start and an elevated annealing temperature) improved the primer specificity, enabling specific amplification of the intended RRP tet gene group from microbiome DNA.
FIG. 1.
Neighbor-joining tree of six classes of tet genes encoding efflux pump proteins. The tree was inferred from DNA sequences, and it was arbitrarily rooted with the cmlA5 gene, which encodes an efflux pump protein rendering resistance to chloramphenicol in E. coli. Bootstrap values were calculated from 100 trees, and the number at each node indicates the number of times that the node was supported in the bootstrap analysis. The bar represents a 0.1 estimated change per nucleotide. Each primer pair listed in Table 1 targets a corresponding cluster.
Cloning and sequencing of the PCR products derived from the primer specificity tests confirmed the specificity of these primers. Based on BLASTn comparisons, all of the sequenced clones matched known tet genes in GenBank, with sequence identities ranging from 94.2% to 100% (Table 2). None of our sequences was broken into two segments in the BLASTn search alignments, suggesting a very low probability of chimeric sequences among our tet sequences. The clone BC-AC-1 represents 8 of the 10 clones from the Tet A/C library and is 100% identical to the tet(A) gene present in the transposon Tn1721. The Tet G clone library was shown to contain a more diverse set of clones. The clone BC-G-4 is 100% identical to the tet(G) gene present in the plasmid pPSTG1 from a Pseudomonas sp. The clones BC-G-1 and BC-G-2 are 92.6% identical to each other and share relatively low identities (≤94.2% and ≤94.4%, respectively) to any tet(G) genes currently available in GenBank. Clones belonging to classes Tet M, Tet O, Tet Q, and Tet W were obtained from the RPP clone library. The most abundant type of clones matched class Tet Q, and clones related to class Tet M were the least abundant (Table 2). Collectively, these sequencing results confirmed the specificity of the tetAC-150f/716r, tetG-247f/678r, and Ribo2-FW/RV primers and their utility with microbiome DNA samples. In addition to known tet genes, these primer pairs also amplified heretofore unidentified members of the respective tet gene classes present in bovine manure microbiomes.
Validation of real-time PCR assays and quantification of tet genes.
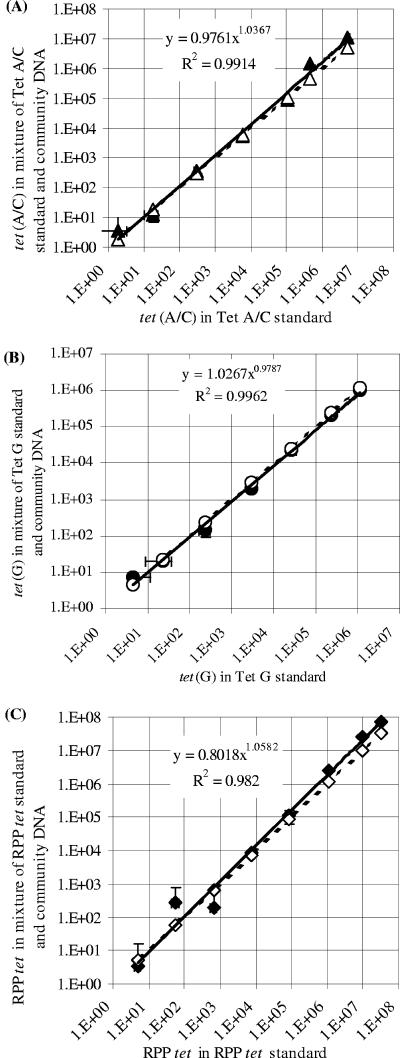
The accuracy of each real-time PCR assay was validated by quantifying known numbers of tet gene templates mixed into microbiome DNA samples. The resultant slopes of the standard curves for the real-time PCR assays for tet(A/C), tet(G), and RPP tet genes were −2.957, −3.361, and −3.633, respectively, and the R2 values were 0.991, 0.970, and 0.997, respectively. When the copy numbers of tet genes spiked into the samples were plotted against the corresponding copy numbers of tet genes quantified in the validation experiments, after correcting for the background numbers of tet genes present in the microbiome DNA itself, high R2 values over at least 6 orders of magnitude were obtained, and both the slope and the exponent values were close to 1.0 (Fig. 2). Each of these plots also nearly superimposed its corresponding theoretical plot, assuming 100% accuracy. Collectively, these results not only show that the assays are precise and accurate but also indicate that the microbiome DNA samples did not have significant inhibition in each of the real-time PCR assays. The limits of detection for all of the real-time PCR assays were <10 tet gene copies per real-time PCR.
FIG. 2.
Validation curves plotting actual tet gene copies versus quantified tet gene copies by real-time PCR assays. (A) tet(A); (B) tet(G); (C) RPP tet genes. The actual numbers of tet gene copies (x axis) were plotted against the quantification values (y axis) for the tet genes (solid lines). Theoretical plots assume 100% accuracy (dashed lines). Error bars (both x and y) indicate standard deviations (n = 3).
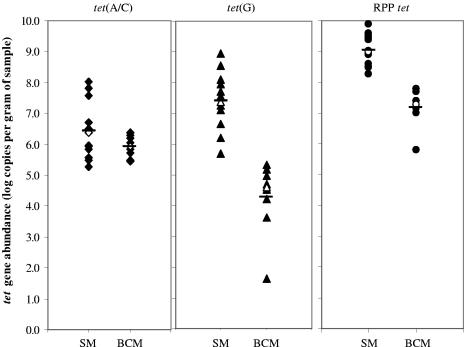
All of the tet gene groups targeted in this study were detected in all of the bovine and swine manure microbiome DNA samples analyzed. The RPP tet genes were found to be the most abundant in both types of manures, and although there were large variations in total tet gene abundance in each type of sample, the LSM values (expressed as log copies g−1 manure) were significantly higher for swine manures (7.63 ± 0.126) than for bovine manures (5.79 ± 0.164). In relation to specific tet gene groups, these differences were also statistically significant for the tet(G) and RPP tet genes (Fig. 3).
FIG. 3.
Abundance of tet genes present in fresh beef cow manures (BCM) and swine manures (SM). Each data point represents one manure sample. The horizontal bars indicate LSMs, and the open symbols represent the median values.
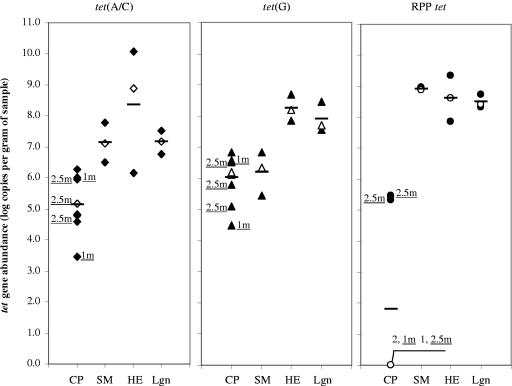
All of the samples collected throughout the Ekokan upflow biofilter system and its lagoon had similar tet gene abundances to those determined for the swine manure and hog house effluent entering the system, with the exception of the RPP tet gene abundance measured in the separated liquid sample (data not shown). Similar observations were made with the other lagoon samples (Fig. 4): the LSM of total tet gene abundance (7.848 ± 0.124 log g−1) in these samples was comparable to that determined for untreated swine manures (7.409 ± 0.183) as well as that for hog house effluent (8.405 ± 0.422). There were also no significant differences within the respective tet gene groups in terms of their abundance in all of these types of samples (Fig. 4). Conversely, the composted swine manure samples all had substantially lower tet gene abundances, with the LSM of total tet gene abundance g−1 of sample being 4.32 (±0.327) and 7.409 (±0.183) log for the composted and untreated swine manures, respectively. Both the tet(A/C) and RPP tet genes were significantly reduced in the composted manures, and while the tet(G) abundance was also reduced, the differences were not found to be statistically significant (Fig. 4).
FIG. 4.
Abundance of tet genes present in treated and untreated swine manures. CP, compost of swine manures; SM, swine manures taken from hog houses; HE, house effluent from hog houses; Lgn, lagoons receiving hog house effluent. Each data point represents one sample. The horizontal bars indicate LSMs, and the open symbols represent the median values. 1m, 1-month compost; 2.5m, 2.5-month compost. The composting time for the remaining compost samples was 3 months.
DISCUSSION
Quantitative measurements of AR in entire microbiomes are required for monitoring of the dynamics of AR in agricultural and other environmental scenarios. The three real-time PCR assays developed and tested for this study permit the quantification of 10 major tet gene classes in entire microbiomes from samples such as animal manures and manure treatment facilities, including wastewater lagoons, biofilter systems, and composts. All of these samples are complex, in terms of both their physicochemical composition and bacterial diversity. All of the clones selected for sequencing carried the selected classes of tet genes and included both known and heretofore unidentified tet sequences (Table 2). For these reasons, we believe the primer sets possess the desired characteristics suitable for application to animal, human, and environmental DNA samples.
Previous studies have shown that the use of 16S rRNA genes from a single bacterial strain as real-time PCR standards can lead to inaccuracies in quantifying the total bacteria present in a complex sample (26), suggesting that differences in the sequence diversity of targeted genes between the real-time PCR standards and the samples to be quantified can lead to inaccuracies. When a group of related genes present in microbiome DNA samples are to be quantified by real-time PCR, the sequence diversity of the targeted gene in the sample is unknown and may be quite variable. With these points in mind, we prepared sample-derived real-time PCR standards by pooling the tet gene amplicons produced from all the microbiome DNA samples to be quantified rather than by selecting one or a few bacterial strains carrying a tet gene. We think that the preparation of sample-derived standards is a practical way to produce real-time PCR standards for accurate and simultaneous quantification of a group of related genes in microbiomes.
When the abundance of one bacterial species is to be quantified, real-time PCR standards can be validated using either known numbers of bacterial cells (17) or a known number of target gene templates (29). In this study, we validated each of the real-time PCR assays by using its respective sample-derived standard (Fig. 2), for a number of reasons. First, the efficiency of cell lysis and DNA recovery does not influence the results. Second, and more importantly, the real-time PCR assay could be validated against potentially all target tet genes present in the samples analyzed rather than against a few selected tetracycline-resistant laboratory strains, which may or may not be present in the samples. Third, the tet gene copy number per bacterial cell may also vary among different bacterial strains, e.g., due to the plasmid copy number. Therefore, the choice of strain used as a standard might confound the results. Fourth, as opposed to cultivation-based analyses that determine bacterial abundance as CFU (or most probable number) per unit mass of sample, real-time PCR assays directly quantify gene copies. As such, even if resistant bacterial strains were used for validation, it would be difficult to convert the tet gene abundance quantified by the real-time PCR assays into numbers of tetracycline-resistant bacteria because of the reason mentioned above. The results of our validation experiments demonstrate that all three real-time PCR assays are accurate for quantification of the tet genes over at least 6 orders of magnitude, as indicated by the R2, slope, and exponent values calculated from the validation plots (Fig. 2).
Real-time PCR assays using SYBR green I are versatile, but their accuracy can be confounded by primer dimer formation during amplification (40). Even with a reduced primer concentration, melting curve analysis and gel electrophoresis indicated the formation of primer dimers, especially in reactions where the target was present at a low abundance and in the no-template controls. All primer dimers in the three real- time PCR assays denatured completely at 86°C, while the three types of tet amplicons remained undenatured. Therefore, we used the fluorescence measurements acquired at 86°C in our real-time PCR assays to eliminate the fluorescence induced by primer dimers, for improved accuracy. This approach can be used for improved accuracy and reproducibility of SYBR green-based real-time PCR assays quantifying a group of related genes for which a sequence-specific, fluorescently labeled probe cannot be designed. Of course, appropriate fluorescence acquisition temperatures need to be determined for different real-time PCR assays.
Although our sample sets were limited, the real-time PCR assays revealed some interesting differences among the animal manures and the means used to either store or treat these manures prior to their reintroduction into the environment. All of the swine manures had a significantly greater total tet gene abundance than the bovine manures (Fig. 3). However, whether these differences can be attributed to the use of tetracyclines in swine feeds, species differences in the fecal microflora, or both requires more study, perhaps through the examination of conventional and organic swine farms. The results presented here also suggest that the treatment of hog house effluents by an upflow biofilter system and/or lagoon storage did not appreciably reduce the tet gene abundance (data not shown). These findings are similar to a previous report describing the abundance of resistant E. coli and Salmonella in lagoon samples (12). A limited reduction in tet gene abundance during lagoon storage is also consistent with the detection of tet genes in groundwater downstream of a swine wastewater lagoon (4, 9), and more in-depth studies of the persistence and dissemination of AR genes surrounding lagoon facilities seem warranted. Conversely, all of the composted swine manure samples had a substantially reduced tet gene abundance, especially the abundance of RPP tet genes (Fig. 4), which were no longer detectable in two-thirds of the compost samples. These results suggest that effective tet gene reduction may be achieved during the composting process. The physicochemical conditions created during composting are known to reduce the pathogen load (11, 37, 38), and this may have contributed, at least partially, to the reductions in tet gene abundances, but the differences observed among the samples also suggest that more systematic and comparative studies are required to confirm this observation.
In conclusion, this study has validated real-time PCR assays that can be used to accurately quantify the abundance of three different tet gene groups present in manure, compost, lagoon, and bioreactor samples. We also developed approaches to generate sample-derived standards that can be used for gene quantification and to eliminate fluorescence signals derived from primer dimers. Such approaches should be useful for other applications of real-time PCR to accommodate the effects of sequence divergence on accurate quantification. Based on our preliminary results, there also appears to be a great deal of variation in the efficacy of manure treatment methods to reduce tet gene abundance, and these should be evaluated in greater detail.
Acknowledgments
This research was supported by a Board of Regents award from The Ohio State University and a USDA-CSREES award (M.M. and Z.Y.) as well as by a USDA-IFAFS award (F.M.).
We thank John Sylvester for his assistance with statistical analysis, Srinand Sreevatsan for providing the DNAs extracted from the swine manure samples, and Mike Williams and Brian Sheldon of North Carolina State University for providing samples collected from the Ekokan upflow biofilter system and the lagoons. We also thank Roderick Mackie for helpful discussions during the preparation of the manuscript.
REFERENCES
- 1.Aarestrup, F. M., F. Bager, M. F. Jensen, M. Madsen, A. Meyling, and H. C. Wegener. 1998. Resistance to antimicrobial agents used for animal therapy in pathogenic, zoonotic and indicator bacteria isolated from different food animals in Denmark: a baseline study for the Danish Integrated Antimicrobial Resistance Monitoring Programme (DANMAP). APMIS 106:745-770. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.American Society for Microbiology. 1999. Antimicrobial resistance: an ecological perspective. Report from the American Academy of Microbiology Colloquium. [Online.] http://www.asm.org/Academy/index.asp?bid=2167. [PubMed]
- 4.Aminov, R. I., J. C. Chee-Sanford, N. Garrigues, B. Teferedegne, I. J. Krapac, B. A. White, and R. I. Mackie. 2002. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 68:1786-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology. Greene Publishing Associates and John Wiley & Sons, New York, N.Y.
- 7.Bach, H.-J., J. Tomanova, M. Schloter, and J. C. Munch. 2002. Enumeration of total bacteria and bacteria with genes for proteolytic activity in pure cultures and in environmental samples by quantitative PCR mediated amplification. J. Microbiol. Methods 49:235-245. [DOI] [PubMed] [Google Scholar]
- 8.Barton, M. D. 2000. Antibiotic use in animal feed and its impact on human health. Nutr. Res. Rev. 13:279-299. [DOI] [PubMed] [Google Scholar]
- 9.Chee-Sanford, J. C., R. I. Aminov, I. J. Krapac, N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Commission on Antimicrobial Feed Additives. 1997. Antimicrobial feed additives. Ministry of Agriculture, Stockholm, Sweden.
- 11.Elorrieta, M. A., F. Suarez-Estrella, M. J. Lopez, M. C. Vargas-Garcia, and J. Moreno. 2003. Survival of phytopathogenic bacteria during waste composting. Agric. Ecosyst. Environ. 96:141-146. [Google Scholar]
- 12.FAAIR Scientific Advisory Panel. 2002. Policy recommendations. Clin. Infect. Dis. 34:S76-S77. [DOI] [PubMed] [Google Scholar]
- 13.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 14.General Accounting Office. 1999. Food safety: the agricultural use of antibiotics and its implications for human health. General Accounting Office, Washington, D.C.
- 15.Guillaume, G., D. Verbrugge, M.-L. Chasseur-Libotte, W. Moens, and J.-M. Collard. 2000. PCR typing of tetracycline resistance determinants (TetA-E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS Microbiol. Ecol. 32:77-85. [DOI] [PubMed] [Google Scholar]
- 16.Heid, C., J. Stevens, K. Livak, and P. Williams. 1996. Real-time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 17.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (New York) 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 19.Johnston, L. M., and L.-A. Jaykus. 2004. Antimicrobial resistance of Enterococcus species isolated from produce. Appl. Environ. Microbiol. 70:3133-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keener, H. M., D. L. Elwell, T. A. Menke, and R. R. Stowell. 2001. Design and performance of a high-rise hog facility manure drying bed. Appl. Eng. Agric. 17:703-709. [Google Scholar]
- 21.Kim, D. 2001. Real-time quantitative PCR. Exp. Mol. Med. 33:101-109. [PubMed] [Google Scholar]
- 22.LiPuma, J. J. 2003. Burkholderia cepacia complex as human pathogens. J. Nematol. 35:212-217. [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda, H., C. Fujimoto, Y. Haruki, T. Maeda, S. Kokeguchi, M. Petelin, H. Arai, I. Tanimoto, F. Nishimura, and S. Takashiba. 2003. Quantitative real-time PCR using TaqMan and SYBR green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med. Microbiol. 39:81-86. [DOI] [PubMed] [Google Scholar]
- 24.Manch-Citron, J. N., G. H. Lopez, A. Dey, J. W. Rapley, S. R. MacNeill, and C. M. Cobb. 2000. PCR monitoring for tetracycline resistance genes in subgingival plaque following site-specific periodontal therapy. J. Clin. Periodontol. 27:437-446. [DOI] [PubMed] [Google Scholar]
- 25.Morrison, T. B., Y. Ma, J. H. Weis, and J. J. Weis. 1999. Rapid and sensitive quantification of Borrelia burgdorferi in infected mouse tissue by continuous fluorescent monitoring of PCR. J. Clin. Microbiol. 37:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primer set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 27.National Research Council Institute of Medicine. 1998. The use of drugs in food animals: benefits and risks. Committee on Drug Use in Food Animals report. National Academy Press, Washington, D.C.
- 28.Nogva, H. K., K. Rudi, K. Naterstad, A. Holck, and D. Lillehaug. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl. Environ. Microbiol. 66:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Mahony, J., and C. Hill. 2004. Rapid real-time PCR assay for detection and quantitation of Mycobacterium avium subsp. paratuberculosis DNA in artificially contaminated milk. Appl. Environ. Microbiol. 70:4561-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pusterla, N., J. Huder, C. Leutenegger, U. Braun, J. Meteghan, and H. Lutz. 1999. Quantitative real-time PCR for detection of members of the Erhlichia phagocytophila genogroup in host animals and Ixodes ricinus ticks. J. Clin. Microbiol. 37:1329-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlegelová, J., V. Babák, E. Klímová, J. Lukášová, P. Navrátilová, A. Šustácková, I. Šedivá, and D. Ryšánek. 2002. Prevalence of and resistance to anti-microbial drugs in selected microbial species isolated from bulk milk samples. J. Vet. Med. Ser. B 49:216-225. [DOI] [PubMed] [Google Scholar]
- 32.Smith, D. L., A. D. Harris, J. A. Johnson, E. K. Silbergeld, and J. G. Morris, Jr. 2002. Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proc. Natl. Acad. Sci. USA 99:6434-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stubner, S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen detection. J. Microbiol. Methods 50:155-164. [DOI] [PubMed] [Google Scholar]
- 34.Tauxe, R. V. 2002. Emerging foodborne pathogens. Int. J. Food Microbiol. 78:31-41. [DOI] [PubMed] [Google Scholar]
- 35.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Leeuwenhoek 76:115-137. [PubMed] [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiquia, S. M., N. F. Y. Tam, and I. J. Hodgkiss. 1998. Salmonella elimination during composting of spent pig litter. Bioresour. Technol. 63:193-196. [Google Scholar]
- 38.Turner, C. 2002. The thermal inactivation of E. coli in straw and pig manure. Bioresour. Technol. 84:57-61. [PubMed] [Google Scholar]
- 39.van den Bogaard, A., and E. Stobberingh. 1999. Antibiotic usage in animals: impact on bacterial resistance and public health. Drugs 58:589-607. [DOI] [PubMed] [Google Scholar]
- 40.Vandesompele, J., A. De Paepe, and F. Speleman. 2002. Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal. Biochem. 303:95-98. [DOI] [PubMed] [Google Scholar]
- 41.William, C. M. 26. July 2004, posting date. Development of environmentally superior technologies: phase 1 report for technology determination per agreements between the attorney general of North Carolina and Smithfield Foods, Premium Standard farms and frontline farmers. [Online.] http://www.cals.ncsu.edu/waste_mgt/smithfield_projects/phase1report04/front.pdf.
- 42.Witte, W. 2000. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents 16:S19-S24. [DOI] [PubMed] [Google Scholar]
- 43.World Health Organization. 1997. The medical impact of the use of antimicrobials in food animals. World Health Organization, Geneva, Switzerland.
- 44.Yu, Z., V. J. J. Martin, and W. W. Mohn. 1999. Occurrence of two resin acid-degrading bacteria and a gene encoding resin acid degradation in pulp and paper mill effluent biotreatment systems assayed by PCR. Microbiol. Ecol. 38:114-125. [DOI] [PubMed] [Google Scholar]
- 45.Yu, Z., and M. Morrison. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 36:808-812. [DOI] [PubMed] [Google Scholar]