Abstract
A metagenomic library was constructed from the anaerobic sediments of a mesophilic sulfur spring. Thirty-five bacterial 16S rRNA gene-containing clones were identified in this library. Analysis of a genomic fragment belonging to candidate division OD1 provided useful insights into the physiology and biochemistry of this novel, yet-uncultured candidate division.
16S rRNA gene-based surveys clearly demonstrate that the scope of microbial diversity is much broader than implied by culture-dependent studies (12, 14). One of the most important challenges to microbial ecologists is to elucidate the physiological properties, energy conservation pathways, and ecological significance of recently discovered, yet-uncultured microorganisms. Although creative isolation strategies are bringing someof these “unculturable” microorganisms to pure cultures (3, 9, 18) or stable enrichments (7), the majority of novel bacterial lineages still evade isolation. An interesting alternative to isolation involves cloning and sequencing DNA directly from various ecosystems. This sequence-based analysis (metagenomics) allows an in silico investigation of various metabolic pathways utilized by novel microbial groups (8, 10, 15).
We are currently investigating the microbial diversity in Zodletone Spring, an anaerobic sulfide- and sulfur-rich spring in Oklahoma (4, 5, 11), with the goal of determining physiological features of uncultivated members of the microbial community. In this study, we report on the construction and screening of a metagenomic library from Zodletone Spring sediments and analysis of a 35.7-kb DNA fragment that belongs to candidate division OD1.
The spring location, geochemical characteristics, and microbial diversity have been previously documented (5, 17). DNA extracted from the spring source sediments was separated on 1% low-melting-point agarose using a field inversion gel electrophoresis box (MJ Research Inc, Watertown, MA). The DNA fraction greater than 30 kb was excised from the gel, and the DNA obtained was ligated into a CopyControl cloning vector, pCC1FOS (Epicenter Corp., Madison, WI), and transfected into Escherichia coli strain EPI300 according to the manufacturer's instructions.
Library screening for fosmids containing 16S rRNA genes was performed on pooled DNA from 384 clones, which had been treated with Plasmid-Safe ATP-dependent DNase (Epicenter) to minimize E. coli chromosomal DNA interference. Each pool was screened using the Bacteria-specific primer pairs 8F/805R (5) and 1054-16SF/21-23S R (6). The latter pair amplifies the intergenic spacer region along with approximately the last 500 bp of the 16S rRNA. 16S rRNA gene-containing fosmids within pools were located at the intersection of pooled plate rows and columns using additional sets of primers created to identify hypervariable regions within the target 16S rRNA gene. The detailed procedures for cloning, shotgun library construction, fluorescent-based DNA sequencing, and subsequent analysis were as described previously (1, 2, 13, 16).
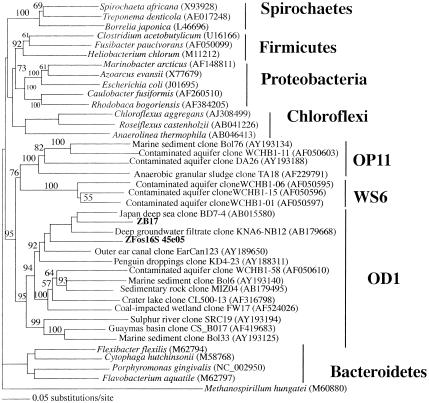
Screening of a total of 19,200 clones resulted in the identification of 35 16S rRNA gene-containing fosmids (Table 1). The phylogenetic affiliations of these clones were in accordance with our previous studies regarding the importance of sulfur-transforming and anaerobic fermentative processes in Zodletone Spring source sediments (5). Fosmids with 16S rRNA gene sequences monophyletic with members of the chemolithotrophic sulfide-oxidizing genus Acidithiobacillus and the sulfur-respiring genus Sulfurospirillum were detected, as well as fosmids belonging to the order Chromatiales and to the δ-Proteobacteria that could putatively be involved in anoxygenic photosynthesis and sulfur (or sulfate) reduction, respectively. Clones belonging to anaerobic fermentative groups included members of the genus Syntrophus, class Bacteroides, and order Clostridiales. Finally, several fosmids were closely related to 16S rRNA sequences previously encountered in Zodletone Spring (5), including ZFos45e05, related to the Zodletone 16S rRNA clone ZB17, both of which are members of candidate division OD1 (Fig. 1).
TABLE 1.
Fosmids containing 16S rRNA genes identified in the library
| Phylum | Fosmid name | Closest match (accession no.) | % Similarity | Affiliation |
|---|---|---|---|---|
| Proteobacteria | ||||
| α-Proteobacteria | ZFos25g11 | Azospirillum sp. strain 5C (AF413109) | 99 | Genus Azospirillum |
| γ-Proteobacteria | ZFos3b04 | Acidithiobacillus albertensis (AJ459804) | 97 | Genus Acidithiobacillus |
| ZFos29d02 | Heavy metal impacted soil clone KCM-B-83 (AJ581596) | 94 | Order Chromatiales | |
| ZFos32e11 | Acinetobacter sp. strain ATCC 31012 (AF542963) | 95 | Genus Acinetobacter | |
| ZFos33d04 | Stenotrophomonas maltophilia (DQ41193) | 96 | Genus Stenotrophomonas | |
| ZFos44D | Psychrobacter frigidicola (AJ609556) | 92 | Genus Psychrobacter | |
| ZFos39g11 | Acinetobacter sp. strain Wuba16 (AF336348) | 97 | Genus Acinetobacter | |
| δ-Proteobacteria | ZFos33i05 | Desulfobacterium cetonicum (AJ237603) | 87 | δ-Proteobacteria |
| ZFos14e13 | Syntrophus gentianae (X85132) | 95 | Genus Syntrophus | |
| ZFos52C | Syntrophus gentianae (X85132) | 97 | Genus Syntrophus | |
| ɛ-Proteobacteria | ZFos2f11 | Sulfurospirillum sp. strain EK7 (AJ535704) | 96 | Genus Sulfurospirillum |
| ZFos9d02 | Sulfurospirillum sp. strain EK7 (AJ535704) | 99 | Genus Sulfurospirillum | |
| Chloroflexi | ZFos9b11 | Coastal marine sediment clone TIHP368-10 (AB031632) | 93 | Group V sediment Chloroflexi |
| ZFos48f09 | Coastal marine sediment clone TIHP368-10 (AB031632) | 91 | Group V sediment Chloroflexi | |
| ZFos40d07 | Uranium waste pile clone Sh765B-TzT-6 (AJ519642) | 91 | Class Dehalococcoidetes | |
| Bacteroidetes | ZFos25h14 | Protein-degrading consortium clone BSA1B-13 | 97 | Class Bacteroides |
| ZFos44a23 | Clinical isolate strain 47077 (AF227830) | 94 | Order Sphingobacteriales | |
| ZFos45e01 | Bovine rumen clone p2b04ct-1 (AY578446) | 99 | Genus Prevotella | |
| ZFos45c09 | Bovine rumen clone p1g07ct-1 (AY578417) | 91 | Class Bacteroides | |
| Planctomyces | ZFos39a01 | Salt marsh clone SIMO-1913 (AY711279) | 89 | Class Planctomycetaceae |
| Candidate division OD1 | ZFos45e05 | Japan trench clone BD7-4 (AB015580) | 82 | Candidate division OD1 |
| Actinobacteria | ZFos12D | Dichloropropane dechloronating clone SHA-34 (AJ306762) | 96 | Family Streptomycetaceae |
| ZFos29a01 | Arthrobacter gandensis (AJ491108) | 97 | Genus Arthrobacter | |
| ZFos33B | Oil storage cavity clone KB20 (AB074931) | 98 | Family Coriobacteriaceae | |
| ZFos35D | Oil storage cavity clone KB20 (AB074931) | 98 | Family Coriobacteriaceae | |
| Firmicutes | ZFos1A | Trichlorobenzene consortium clone SJA-143 (AJ009494) | 92 | Family Acidaminococcaceae |
| ZFos19h03 | Anaerospora hongkongensis (AY372051) | 97 | Family Acidaminococcaceae | |
| ZFos28B | Sporomusa aerivorans (AJ506192) | 97 | Genus Sporomusa | |
| ZFos31b07 | Artesian basin clone R82 (AF407695) | 94 | Family Clostridiaceae | |
| ZFos39g02 | Oil reservoir clone PL-35B11 (AY570624) | 96 | Family Clostridiaceae | |
| ZFos37b08 | Uranium-contaminated sediment clone ph5Lac302-37 (AY527741) | 94 | Family Acidaminococcaceae | |
| ZFos40d08 | Sporomusa aerivorans (AJ506192) | 96 | Genus Sporomusa | |
| ZFos44a01 | Butyrivibrio fibrisolvens (X89980) | 93 | Family Lachnospiraceae | |
| ZFos46b02 | Anaerospora hongkongensis (AY372051) | 97 | Family Acidaminococcaceae | |
| ZFos50a03 | Bovine rumen clone p3a03ct-1 (AY578513) | 93 | Family Lachnospiraceae |
FIG. 1.
Distance dendrogram representing the phylogenetic affiliation of selected members of the novel candidate division OD1, including the 16S rRNA gene from fosmid ZFos45e05 and 16S rRNA clone ZB17, previously encountered during a 16S rRNA gene survey of Zodletone Spring (5). The tree was constructed as previously described (4). A more detailed phylogenetic analysis, as well as a list of partial and complete OD1 16S rRNA clones identified in the GenBank database, is available in the supplemental materials (Fig. S1; Table S1).
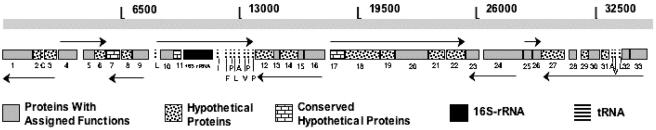
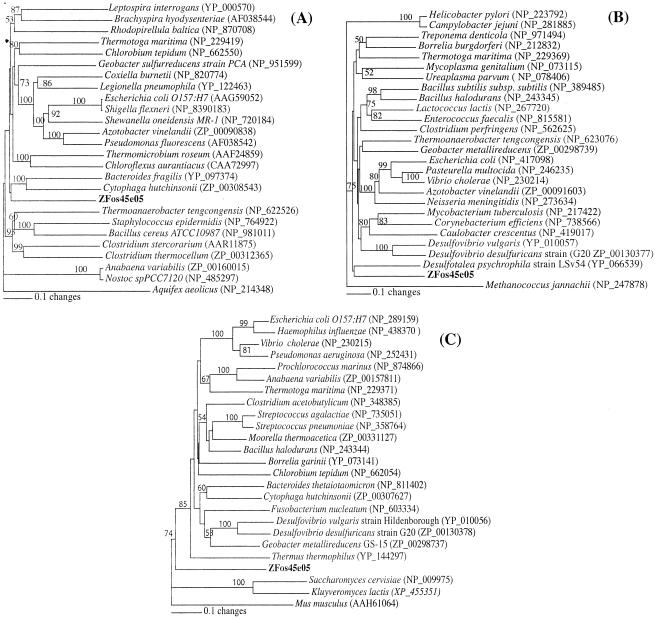
Novel candidate division OD1 members are globally distributed in marine and terrestrial habitats and appear to be mainly present in anoxic environments (Fig. 1; see also Table S1 in the supplemental materials). Fosmid ZFos45e05, belonging to candidate division OD1, was sequenced and open reading frames characterized (Fig. 2 and Table 2; see also Table S2). The rRNA operon organization within ZFos45e05 was different from that observed within most bacterial genomes. The 16S rRNA gene appears to be separate from both the 23S and the 5S rRNA genes. Thirteen tRNA genes were identified around the 16S rRNA gene. Fosmid ZFos45e05 has an overall low G+C content (34.9%). The consistently low G+C content in all genes argues that this value is a true reflection of the G+C content of the entire genome of the microorganism. Out of 33 protein-coding open reading frames (ORFs), 13 had no orthologs in the database (Table 2). Apparent phylogenetic affiliation of the remaining 20 ORFs indicated that their most closely related orthologs were dispersed among the bacterial, archaeal, and eukaryotic phyla. Phylogenetic analysis of three putative housekeeping genes in ZFos45e05 [DNA polymerase 1 (ORF 1), tRNA (guanine-N1)-methyltransferase (ORF 10), and large ribosomal subunit protein L19 (ORF 32)] (Fig. 3) supported the hypothesis that candidate division OD1 is not closely related to any bacterial division with genome-sequenced representatives. The large sequence divergence which resulted in deep branching points to the potential genomic novelty of this yet-uncultured phylum of Bacteria.
FIG. 2.
Genomic map of OD1 fosmid ZFos45e05. ORFs are shaded according to their putative functions. More information regarding putative functions of ZFos45e05 ORFs is available in the supplemental materials (Table S2).
TABLE 2.
General characteristics of ZFos45e05 OD1 fosmid
| Characteristic | ZFos45e05 |
|---|---|
| Length (bp) | 35,743 |
| G+C% | 34.9 |
| G+C% (coding regions) | 40.4 |
| rRNA operon | 16S |
| No. of ORFs | 33 |
| No. of tRNA | 13 |
| Hypothetical proteins (unique) | 13 |
| Conserved hypothetical proteins (no. of hits to other hypothetical proteins) | 3 |
| Phylogenetic affiliations of most similar orthologsa | Proteobacteria (5), Firmicutes (3), Aquifex (1), Fusobacteria (1), Actinobacteria (1), Chloroflexi (1), Archaea (5), eukaryotes (3) |
Numbers in parentheses are numbers of ORFs within ZFos45e05 most closely similar to each lineage.
FIG. 3.
Distance dendrogram evaluating the phylogenetic position of three protein-encoding ORFs from candidate division OD1 fosmid ZFos45e05 compared to their orthologs from previously sequenced bacterial genomes: DNA polymerase 1 (ORF 1) (A), tRNA (guanine-N1)-methyltransferase (ORF 10) (B), and large ribosomal subunit protein L19 (ORF 32) (C).
Four genes in ZFos45e05 potentially encode enzymes involved in metabolic processes. These include two phosphoenolpyruvate synthase gene paralogs (ORFs 13 and 15), both of which have an archaeal affiliation based on BLASTp analysis. The other metabolism-related genes were pyruvate formate lyase-activating enzyme (ORF 23) and oxygen-sensitive ribonucleoside triphosphate reductase (ORF 24), both of which are present only in anaerobic or facultative anaerobic microorganisms (Table S2). The genomic organization in which the latter two genes are adjacent has also been observed in Archaea (e.g., fosmids belonging to anaerobic methane oxidizing microorganisms [8]), Pyrococcus abyssi, and Pyrolobus fumarii. This similar phylogeny and genomic organization might be indicative of ancient evolutionary traits or the occurrence of horizontal gene transfer between the aforementioned groups.
The small size of the sequenced OD1 fosmid coupled with our inability to detect more OD1 fosmids within the library renders information regarding this unique group of bacteria limited. However, this study provided several interesting observations, including an unusual rRNA operon organization, low G+C content, low sequence similarity of OD1 putative gene products to their orthologs, the presence of genes encoding oxygen-sensitive enzymes, and an apparent archaeal affiliation and archaeal genomic organization of OD1 genes involved in metabolic processes. We are currently evaluating different strategies to locate more OD1 fosmids based on the information generated by sequencing ZFos45e05. This could aid in elucidating some of the metabolic pathways utilized by this bacterial division as well as in understanding its ecological significance in anaerobic ecosystems.
Nucleotide sequence accession numbers.
Sequences obtained in this study have been deposited in GenBank under accession numbers DQ227583 to DQ227617 and AC160099.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Science Foundation Microbial Observatories Program (grant MCB_0240683).
We thank Jennifer L. Groh, Anne M. Spain, and Kristen N. Savage for critical review of the manuscript.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bodenteich, A., S. L. Chissoe, Y. F. Wang, and B. A. Roe. 1993. Shotgun cloning as the strategy of choice to generate templates for high-throughput dideoxynucleotide sequencing, p. 42-50. In J. C. Venter (ed.), Automated DNA sequencing and analysis techniques. Academic Press, London, United Kingdom.
- 2.Chissoe, S. L., A. Bodenteich, Y. F. Wang, Y. P. Wang, S. W. Burian, C. Dennis, J. Crabtree, A. Freeman, K. Iyer, L. Jian, Y. Ma, H. J. McLaury, H. Q. Pan, O. Sharan, S. Toth, Z. Wong, G. Zhang, N. Heisterkamp, J. Groffen, and B. A. Roe. 1995. Sequence and analysis of the human ABL gene, the BCR gene, and regions involved in the Philadelphia chromosomal translocation. Genomics 27:67-82. [DOI] [PubMed] [Google Scholar]
- 3.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elshahed, M. S., F. Z. Najar, B. A. Roe, A. Oren, T. A. Dewers, and L. R. Krumholz. 2004. Survey of archaeal diversity reveals an abundance of halophilic Archaea in a low-salt, sulfide- and sulfur-rich spring. Appl. Environ. Microbiol. 70:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elshahed, M. S., J. M. Senko, F. Z. Najar, S. M. Kenton, B. A. Roe, T. A. Dewers, J. R. Spear, and L. R. Krumholz. 2003. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl. Environ. Microbiol. 69:5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Martinez, J., S. G. Acinas, A. I. Anton, and F. Rodriguez-Valera. 1999. Use of the 16S-23S ribosomal genes spacer region in studies of prokaryotic diversity. J. Microbiol. Methods 36:55-64. [DOI] [PubMed] [Google Scholar]
- 7.Girguis, P. R., V. J. Orphan, S. J. Hallam, and E. F. DeLong. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69:5472-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hallam, S. J., N. Putnam, C. M. Preston, J. C. Detter, D. Rokhsar, P. M. Richardson, and E. F. DeLong. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457-1462. [DOI] [PubMed] [Google Scholar]
- 9.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 269:1127-1129. [DOI] [PubMed] [Google Scholar]
- 10.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo, Q., L. R. Krumholz, F. Z. Najar, B. A. Roe, A. D. Peacock, D. C. White, and M. S. Elshahed. 2005. Diversity of the microeukaryotic community in sulfide-rich Zodletone Spring. Appl. Environ. Microbiol. 71:6175-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 13.Pan, H. Q., Y. P. Wang, S. L. Chissoe, A. Bodenteich, Z. Wang, K. Iyer, S. W. Clifton, J. S. Crabtree, and B. A. Roe. 1994. The complete nucleotide sequence of the SacBII domain of the P1 pAD10-SacBII cloning vector and three cosmid cloning vectors: pTCF, svPHEP, and LAWRIST16. Genet. Anal. Tech. Appl. 11:181-186. [DOI] [PubMed] [Google Scholar]
- 14.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 15.Riesenfeld, C. S., P. D. Schloss, and J. Handelsman. 2004. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genetics 38:525-552. [DOI] [PubMed] [Google Scholar]
- 16.Roe, B. A. 2004. Shotgun library construction for DNA sequencing. Methods Mol. Biol. 255:171-187. [DOI] [PubMed] [Google Scholar]
- 17.Senko, J. M., B. S. Campbell, J. R. Henricksen, M. S. Elshahed, T. A. Dewers, and L. R. Krumholz. 2004. Barite deposition mediated by phototrophic sulfide-oxidizing bacteria. Geochim. Cosmochim. Acta 68:773-780. [Google Scholar]
- 18.Zengler, K., G. Toledo, M. Rappe, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.