Abstract
Random PCR mutagenesis is a powerful tool for structure-function analysis of targeted proteins, especially when coupled with DNA integration through natural transformation followed by selection for loss of function. The technique has been applied successfully to structure-function analysis of transcriptional regulators, enzymes, and transporters in Acinetobacter sp. strain ADP1. However, the mismatch repair system prevents the full spectrum of nucleotide substitutions that may be selected at the level of protein function from being recovered. This barrier may be overcome by introducing PCR-mutagenized genes into strains in which the corresponding genes have been deleted.
Random PCR mutagenesis provides an array of mutations, many of which cause amino acid substitutions that may alter the functional properties of proteins. When coupled to natural transformation in Acinetobacter sp. strain ADP1, the procedure can be streamlined, as PCR amplicons of targeted genes are provided directly to the host and integrated into the chromosome by homologous recombination (15). When followed by a suitable screen or selection, the process can provide an overview of the amino acid side chains that contribute to function. A merit of the procedure is its objectivity because mutations are identified on the basis of phenotypic variation. No presumptions are brought to bear on the identification of targets for mutagenesis, so amino acid contributions that might not have been predicted can be characterized (7, 10, 24). Nevertheless, the procedure must be regarded with caution because not all nucleotide substitutions are made with equal probability during PCR amplification (34). Furthermore, as described here, mismatch repair (MMR) also contributes to making the process less than fully random because it reduces the frequencies of transition mutations that are introduced into the chromosome.
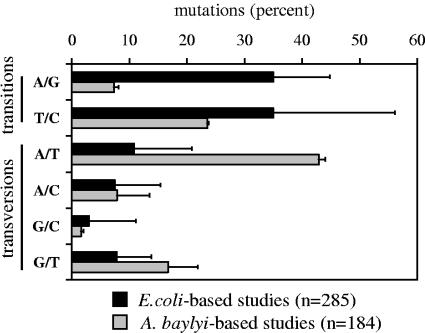
Transformation-facilitated random PCR mutagenesis has been successfully applied to proteins involved in the catabolism and regulation of phytochemicals in the nutritionally versatile Acinetobacter sp. strain ADP1. These investigations have given valuable insight into structure-function relationships in these proteins (7, 15, 16, 17, 24, 29). However, when we applied the random PCR mutagenesis approach to pcaK, a gene encoding a protocatechuate transporter belonging to the ubiquitous major facilitator superfamily (28), evidence of bias in the mutant collection was observed. A library of 117 pcaK mutants was analyzed, and in 20 cases, identical base pair substitutions were recovered in independently derived mutants (A. Buchan and L. N. Ornston, unpublished data). While saturation was being approached with respect to the field of pcaK variants that could be recovered using this technique, absent from the collection were several anticipated mutations, including those abolishing conserved charged residues in transmembrane-spanning regions. Furthermore, the nucleotide substitutions were heavily biased towards transversions (71%) relative to transitions (29%). This pattern contrasts the distribution of nucleotide substitutions recovered from random PCR mutagenesis approaches coupled with cloning and expression of a target gene in Escherichia coli (Fig. 1). As generation of PcaK mutants in Acinetobacter sp. strain ADP1 depends upon replacement of the wild-type pcaK allele with mutated variants via homologous recombination, a transformation-associated bias caused by MMR was considered.
FIG. 1.
Distribution of single-nucleotide substitutions generated by a general random PCR mutagenesis approach (41). Natural transformation through which heteroduplex DNA was formed (25, 26) was used to obtain Acinetobacter mutants used in this analysis (15 [n = 76]; A. Buchan and L. N. Ornston, unpublished data [n = 108]). Mutants derived from E. coli-based approaches in which heteroduplex DNA is not formed were obtained from the following references: references 18 (n = 48), 30 (n = 14), 1 (n = 10), 36 (n = 35), 2 (n = 8), 33 (n = 9), 35 (n = 6), 42 (n = 8), 9 (n = 5), 19 (n = 18), 37 (n = 11), 40 (n = 13), 27 (n = 19), 4 (n = 5), and 20 (n = 76). If nucleotide substitutions are not explicitly described in the references, they were deduced from protein substitutions, and ambiguous substitutions were omitted. Error bars represent variations in frequencies among studies conducted with the two organisms. A. baylyi, Acinetobacter baylyi.
Prior studies of MMR systems of divergent bacterial lineages provide a framework for how cellular machinery might block integration of transition-containing PCR products in Acinetobacter. MMR plays a well-recognized role in repair of replication-based errors, but it is also active in preventing recombination between divergent sequences (31, 32, 38, 39). Heteroduplex DNA containing even single-nucleotide mismatches formed during homologous recombination can be targets of MMR-mediated heteroduplex rejection, also known as antirecombination (5, 23). MutS, the mismatch binding protein and an essential component of MMR, preferentially binds mismatches that arise from transition mutations over those arising from transversion substitutions (3), thereby affecting the frequency with which mutant alleles are integrated into the chromosome during recombination (11). Furthermore, recent studies of E. coli have demonstrated that MMR-mediated sequence-specific biases in genetic engineering approaches employing bacteriophage λ are relieved with MutS-defective cells (6). Therefore, an interpretation of the conflicting substitution profiles presented in Fig. 1 is that the Acinetobacter MMR system reduces the integration efficiency of transition mutation-bearing PCR products during the transformation step. To test this hypothesis, PCR mutagenesis of pcaK was conducted with strains containing or lacking functional MutS, and the resulting nucleotide substitutions in the mutant genes were compared.
Acinetobacter strain ADP8502 contains the ΔvanK7602 and ΔpcaBD mutations that facilitate selection of PcaK-deficient strains on medium containing protocatechuate and a nonselecting growth substrate (7). The basis of the selection is twofold. First, as both VanK and PcaK transport protocatechuate (8), selection for strains defective in PcaK activity is carried out in a ΔvanK background. Second, PcaB-deficient strains that take up and metabolize protocatechuate accumulate a toxic metabolite that inhibits growth on the selective medium (12). Inactivation of MutS in strain ADP8504 was achieved by an Ω(SpR/StR) insertion by the following procedure: strain ADP8502 was transformed with the linearized plasmid pZR7008 (38) followed by selection for resistance to streptomycin and spectinomycin at respective concentrations of 10 and 40 μg/ml. PCR mutagenesis was performed using crude cell lysates of Acinetobacter sp. strain ADP1 with the primers PK1 and PK2 (7) in 100-μl volumes. Twenty-five microliters from each reaction mixture was added in parallel to 500-μl competent cultures of ADP8502 and ADP8504. Following 3 h of incubation at 37°C with shaking, the transformation reaction mixtures were plated on minimal medium (31) containing 5 mM fumarate and 1 mM protocatechuate and incubated in the dark at 22°C for 28 h. The ΔpcaBD deletion was corrected by transformation of purified strains with a plasmid carrying wild-type pcaBD (pZR8550). The pcaK genes of mutant strains were sequenced in their entirety by using PK1, PK2, and an internal primer, PK3 (5′ CCGATTTCTAACAGGTATCGGATTG 3′). pcaK was sequenced from representative mutants derived from ADP8502 (n = 13) and ADP8504 (n = 18) transformed with Taq-amplified pcaK DNA. Substitutions are presented relative to the pcaK coding strand and as “nucleotide/nucleotide” (e.g., T/C). The arrangement of the nucleotide designations does not necessarily reflect the original substitution (i.e., T/C represents either a T-to-C or C-to-T substitution).
Comparison of PCR-generated mutations in strains containing and lacking functional MutS.
Consistent with the mutator phenotype typical of MutS-deficient strains (5), the frequency of spontaneous mutations giving rise to protocatechuate-resistant colonies was sevenfold higher in strain ADP8504 than that in strain ADP8502. Transformation of ADP8504 with Taq-amplified pcaK DNA resulted in 30-fold more protocatechuate-resistant colonies than did that of the same strain receiving no PCR product (see Fig. S1 in the supplemental material). This is similar to the 24-fold increase seen when amplified products are provided to the wild-type mutS strain ADP8502 and suggests that more than 95% of the protocatechuate-resistant colonies derived from either strain transformed with pcaK PCR products possess Taq-introduced mutations.
The spectrum of PCR-generated mutations identified in the MutS-deficient strain (ADP8504) receiving pcaK PCR products was distinct from that found with the MutS-positive strain (ADP8502). Transitions accounted for 84% of all pcaK nucleotide substitutions derived from the MutS-deficient strain compared to 42% of substitutions obtained from ADP8502 provided with the same PCR product pools (see Table S2 in the supplemental material). The substitution profiles for the 12 pcaK variants obtained in the functional MutS background parallel those found in a previous study of PCR-generated mutations in pcaK (A. Buchan and L. N. Ornston, unpublished data) and were folded into the larger data set for comparisons. This substitution pattern was also remarkably consistent with that of another large mutant library generated in Acinetobacter sp. strain ADP1 (PobR) (see Table S1 in the supplemental material) (15). Relative to substitutions recovered in functional MutS strains, the frequencies of both A/G and T/C transitions were elevated about three- and twofold, respectively, in the MutS-deficient strain. A second feature characteristic of the PcaK mutants derived from the MutS-deficient strain was that most (73%) contained multiple (ranging from two to five) mutations within pcaK (see Table S2 in the supplemental material). In contrast, only 15% of the mutants derived from the functional MutS strain receiving the same PCR product pools had more than one mutation in pcaK, a finding consistent with previous studies of PcaK and PobR (see Table S1 in the supplemental material). However, significantly higher frequencies of multiple mutations at a target locus have been reported using this approach (24). As MMR systems are subject to saturation (13, 21), the prevalence of multiple mutations in a target gene is likely a reflection of the capacity of the MMR system and the composition of the PCR pool provided as donor DNA.
Circumventing the transformation-associated bias.
Application of the transformation-facilitated random PCR mutagenesis approach to a heterologous gene yielded a different pattern of nucleotide substitutions. Kok et al. (15) designed a docking site in the Acinetobacter sp. strain ADP1 chromosome that would allow integration and expression of foreign genes and PCR-mutagenized variants of that gene. In this system, the recipient strain does not have DNA corresponding to the gene targeted for PCR mutagenesis. Of the 25 nucleotide substitutions identified in 17 mutants, 92% were transitions. This pattern is analogous to that of E. coli-based libraries (see Table S1 in the supplemental material) and consistent with our interpretation of the role of MMR. If the gene targeted for PCR mutagenesis is not present in the chromosome of the Acinetobacter recipient strain, heteroduplexes are avoided and the MMR-mediated biases associated with the transformation- facilitated random PCR mutagenesis approach are circumvented. Therefore, through chromosomal deletion of the gene of interest and the use of a counterselectable marker, such as sacB (14, 22), to facilitate integration of mutagenized variants, it is likely that a different spectrum would be recovered. It would be beneficial to take this approach in the presence of a functional MutS because, as noted above, deficiencies in MutS give rise to a high frequency of strains with multiple mutations in the target gene.
Supplementary Material
Acknowledgments
We are grateful to David Young and Graham Walker for valuable discussions.
This research was supported by NIH grant GM063628. A.B. was supported by NSF Postdoctoral Research Fellowship DBI-0200164.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barnard, T. J., M. E. Watson, Jr., and M. A. McIntosh. 2001. Mutations in the Escherichia coli receptor FepA reveal residues involved in ligand binding and transport. Mol. Microbiol. 41:527-536. [DOI] [PubMed] [Google Scholar]
- 2.Blaesing, F., C. Weigel, M. Welzeck, and W. Messer. 2000. Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol. Microbiol. 36:557-569. [DOI] [PubMed] [Google Scholar]
- 3.Brown, J., T. Brown, and K. R. Fox. 2001. Affinity of mismatch-binding protein MutS for heteroduplexes containing different mismatches. Biochem. J. 354:627-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cicero, M. P., M. M. Sharp, C. A. Gross, and K. N. Kreuzer. 2001. Substitutions in bacteriophage T4 AsiA and Escherichia coli σ70 that suppress T4 motA activation mutations. J. Bacteriol. 183:2289-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claverys, J. P., and S. A. Lacks. 1986. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol. Rev. 50:133-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costantino, N., and D. L. Court. 2003. Enhanced levels of λ Red-mediated recombinants in mismatch repair mutants. Proc. Natl. Acad. Sci. USA 100:15748-15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Argenio, D. A., A. Segura, W. M. Coco, P. V. Bunz, and L. N. Ornston. 1999. The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK. J. Bacteriol. 181:3505-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Argenio, D. A., M. W. Vetting, D. H. Ohlendorf, and L. N. Ornston. 1999. Substitution, insertion, deletion, suppression, and altered substrate specificity in functional protocatechuate 3,4-dioxygenases. J. Bacteriol. 181: 6478-6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deckert, J., A. M. Rodriguez Torres, J. T. Simon, and R. S. Zitomer. 1995. Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6109-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ermakova-Gerdes, S., Z. Yu, and W. Vermaas. 2001. Targeted random mutagenesis to identify functionally important residues in the D2 protein of photosystem II in Synechocystis sp. strain PCC 6803. J. Bacteriol. 183: 145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34:359-399. [DOI] [PubMed] [Google Scholar]
- 12.Hartnett, G. B., B. Averhoff, and L. N. Ornston. 1990. Selection of Acinetobacter calcoaceticus mutants deficient in the p-hydroxybenzoate hydroxylase gene (pobA), a member of a supraoperonic cluster. J. Bacteriol. 172: 6160-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humbert, O., M. Prudhomme, R. Hakenbeck, C. G. Dowson, and J. P. Claverys. 1995. Homeologous recombination and mismatch repair during transformation in Streptococcus pneumoniae: saturation of the Hex mismatch repair system. Proc. Natl. Acad. Sci. USA 92:9052-9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, R. M., and P. A. Williams. 2003. Mutational analysis of the critical bases involved in activation of the AreR-regulated σ54-dependent promoter in Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol. 69:5627-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kok, R. G., D. A. D'Argenio, and L. N. Ornston. 1997. Combining localized PCR mutagenesis and natural transformation in direct genetic analysis of a transcriptional regulator gene, pobR. J. Bacteriol. 179:4270-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kok, R. G., D. A. D'Argenio, and L. N. Ornston. 1998. Mutation analysis of PobR and PcaU, closely related transcriptional activators in Acinetobacter. J. Bacteriol. 180:5058-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kok, R. G., D. M. Young, and L. N. Ornston. 1999. Phenotypic expression of PCR-generated random mutations in a Pseudomonas putida gene after its introduction into an Acinetobacter chromosome by natural transformation. Appl. Environ. Microbiol. 65:1675-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, J., L. Passaglia, I. Rombel, D. Yan, and S. Kustu. 1999. Mutations affecting motifs of unknown function in the central domain of nitrogen regulatory protein C. J. Bacteriol. 181:5443-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobocka, M., and M. Yarmolinsky. 1996. P1 plasmid partition: a mutational analysis of ParB. J. Mol. Biol. 259:366-382. [DOI] [PubMed] [Google Scholar]
- 20.Lu, J., W. Zhao, and L. S. Frost. 2004. Mutational analysis of TraM correlates oligomerization and DNA binding with autoregulation and conjugative DNA transfer. J. Biol. Chem. 279:55324-55333. [DOI] [PubMed] [Google Scholar]
- 21.Maas, W. K., C. Wang, T. Lima, A. Hach, and D. Lim. 1996. Multicopy single-stranded DNA of Escherichia coli enhances mutation and recombination frequencies by titrating MutS protein. Mol. Microbiol. 19:505-509. [DOI] [PubMed] [Google Scholar]
- 22.Metzgar, D., J. M. Bacher, V. Pezo, J. Reader, V. Doring, et al. 2004. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 32:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modrich, P., and R. Lahue. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65: 101-133. [DOI] [PubMed] [Google Scholar]
- 24.Morawski, B., A. Segura, and L. N. Ornston. 2000. Substrate range and genetic analysis of Acinetobacter vanillate demethylase. J. Bacteriol. 182:1383-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmen, R., and K. J. Hellingwerf. 1997. Uptake and processing of DNA by Acinetobacter calcoaceticus—a review. Gene 192:179-190. [DOI] [PubMed] [Google Scholar]
- 26.Palmen, R., B. Vosman, P. Buijsman, C. K. Breek, and K. J. Hellingwerf. 1993. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J. Gen. Microbiol. 139:295-305. [DOI] [PubMed] [Google Scholar]
- 27.Panchal, R. G., and H. Bayley. 1995. Interactions between residues in staphylococcal alpha-hemolysin revealed by reversion mutagenesis. J. Biol. Chem. 270:23072-23076. [DOI] [PubMed] [Google Scholar]
- 28.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parke, D., and L. N. Ornston. 2004. Toxicity caused by hydroxycinnamoyl-coenzyme A thioester accumulation in mutants of Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol. 70:2974-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen, C., N. Orem, J. Trueheart, J. W. Thorner, and I. G. Macara. 2000. Random mutagenesis and functional analysis of the Ran-binding protein, RanBP1. J. Biol. Chem. 275:4081-4091. [DOI] [PubMed] [Google Scholar]
- 31.Rayssiguier, C., D. S. Thaler, and M. Radman. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396-401. [DOI] [PubMed] [Google Scholar]
- 32.Selva, E. M., L. New, G. F. Crouse, and R. S. Lahue. 1995. Mismatch correction acts as a barrier to homologous recombination in Saccharomyces cerevisiae. Genetics 139:1175-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tavakoli, N. P., and K. M. Derbyshire. 1999. IS903 transposase mutants that suppress defective inverted repeats. Mol. Microbiol. 31:1183-1195. [DOI] [PubMed] [Google Scholar]
- 34.Tindall, K. R., and T. A. Kunkel. 1988. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry 27:6008-6013. [DOI] [PubMed] [Google Scholar]
- 35.Tintut, Y., and J. D. Gralla. 1995. PCR mutagenesis identifies a polymerase-binding sequence of sigma 54 that includes a sigma 70 homology region. J. Bacteriol. 177:5818-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan, L., J. K. Kim, V. W. Pollard, and G. Dreyfuss. 2001. Mutational definition of RNA-binding and protein-protein interaction domains of heterogeneous nuclear RNP C1. J. Biol. Chem. 276:7681-7688. [DOI] [PubMed] [Google Scholar]
- 37.Williams, R. M., S. Rimsky, and H. Buc. 1996. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 178:4335-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young, D. M., and L. N. Ornston. 2001. Functions of the mismatch repair gene mutS from Acinetobacter sp. strain ADP1. J. Bacteriol. 183:6822-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zahrt, T. C., and S. Maloy. 1997. Barriers to recombination between closely related bacteria: MutS and RecBCD inhibit recombination between Salmonella typhimurium and Salmonella typhi. Proc. Natl. Acad. Sci. USA 94: 9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, X., and R. Schleif. 1998. Catabolite gene activator protein mutations affecting activity of the araBAD promoter. J. Bacteriol. 180:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, Y., X. Zhang, and R. H. Ebright. 1991. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 19:6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, Y., X. Zhang, and R. H. Ebright. 1993. Identification of the activating region of catabolite gene activator protein (CAP): isolation and characterization of mutants of CAP specifically defective in transcription activation. Proc. Natl. Acad. Sci. USA 90:6081-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.