Abstract
Soil contaminated with polychlorinated biphenyls (PCB) was used as an inoculum to grow a complex biofilm community on PCB oil (Aroclor 1242) on a substratum (Permanox). The biofilm was monitored for 31 days by confocal laser scanning microscopy, community fingerprinting using single-strand conformational polymorphism (SSCP), amplicons of the 16S rRNA genes, and chemical analyses of the PCB congeners. SSCP analysis of the young biofilm revealed a rather diverse microbial community with species of the genera Herbaspirillum and Bradyrhizobium as dominant members. The biofilm developing on the PCB droplets displayed pronounced stages of PCB degradation and biofilm development not described before from pure-culture experiments. The first step was the colonization of the substratum while the PCB oil was hardly populated. When a certain density of bacteria was reached on the Permanox, the PCB was colonized, but soon the degradation of the congeners was markedly reduced and many cells were damaged, as seen by LIVE/DEAD staining. Finally, the biofilm formed aggregates and invaded the PCB oil, showing lower numbers of damaged cells than before and a dramatic increase in PCB degradation. This sequence of biofilm formation is understood as a maturation process prior to PCB oil colonization. This is followed by a thin biofilm on the PCB droplet, an aggregation process forming pockets in the PCB, and finally an invasion of the biofilm into the PCB oil. Only the mature biofilm showed degradation of pentachlorinated PCB congeners, which may be reductively dechlorinated and the resulting trichlorobiphenyls then aerobically metabolized.
Microbial communities organized in biofilms show a multitude of interactions, including carbon sharing (26), interspecies communication (33), and steep physicochemical gradients (10), and are very well protected against environmental stress factors such as radiation (32), water stress, or grazing (23). These characteristics make biofilms the preferred lifestyle of microorganisms in extreme habitats, and we speculated that complex biofilms might be able to degrade polychlorinated biphenyls (PCBs), which have been used in a number of industrial applications due to their excellent physical properties and extreme chemical stability but for various reasons finally leaked into the environment. The industrial chlorination of biphenyl produces a complex mixture of compounds, called congeners, differing in the number and position of chlorine atoms at the biphenyl rings. All congeners are poorly soluble in water but highly soluble in organic solvents, including fat. These properties make PCBs a major environmental pollutant with negative impacts on many biological systems (6) due to their enrichment in food chains (20).
Despite the fact that a number of PCB-degrading bacteria are known, this pollutant is recalcitrant in the environment, and efforts at bioremediation with optimized strains were often not very successful (11). To improve intrinsic bioremediation, a passive remedial approach that depends on microbial processes to degrade and dissipate pollutants in soil and groundwater, one has to understand the influence of this highly hydrophobic substrate on the autochthonous microbial community. We studied intensively a highly contaminated site near Wittenberg in Germany and developed a broad knowledge base of the bacterial community of the soil samples (29). The sandy soil is low in total organic carbon (TOC) and acidic, with a pH of 4.0 to 4.5. High PCB concentrations together with an overall low TOC content at the site make this pollutant the main carbon source here, promoting microbial communities which have been reported to degrade several congeners of PCB (1, 36).
To study the PCB-influenced microbial community, an approach to growing complex bacterial biofilms in microcosms has been developed (19). An inert substratum (Permanox) dotted with the industrial PCB mixture Aroclor 1242 was exposed to PCB-polluted soil samples. In a microcosm filled with a soil slurry from the site under study, a biofilm developed on the lower surface of the floating plastic device used as a substratum, separated from the soil by a water column. These biofilms were characterized by a polyphasic approach comprising single-strand conformational polymorphism (SSCP) community fingerprinting, PCB chemical analysis, and confocal laser scanning microscopy (CLSM). We report here on the spatial and compositional structure and complex dynamics of the developing biofilm and its PCB-degrading activity, leading to the identification of three stages of biofilm development.
MATERIALS AND METHODS
Characterization of the site.
The site, situated north of Wittenberg, Germany, had been polluted by the Russian army with PCB oil more than a decade ago (29). The soil used in our experiments contained mainly sand with some clay and had a pH of 4.54, a low TOC content of 0.38% ± 0.07%, a low total nitrogen (TN) content of 200 ± 40 ppm (giving a TOC/TN ratio of 19.0), and a PCB concentration of 813 ppm (451 ppm C). The PCB mixture was found to consist mostly of low-chlorinated congeners (29) with 2.9% dichlorobiphenyls, 44.7% trichlorobiphenyls, 51.6% tetrachlorobiphenyls, and 0.8% pentachlorobiphenyls, a composition similar to that of Aroclor 1242; therefore, this commercial PCB mixture was used in our experiments (15).
Soil samples.
Soil samples (approximately 500 g) were taken from the PCB-polluted site and stored at 4°C until use (12). Soil samples were air dried, and 2 to 10 mg of soil was exactly weighed and submitted to analysis in an EA 1108 elemental analyzer (Fisons, Italy) with CHN packing for analysis of TN and TOC after a previous reaction with hydrochloric acid. Results are averages of five analyses.
PCB analysis.
PCBs were extracted by washing the slides with n-hexane using a modified method described previously (7). The extracts were concentrated under a gentle stream of N2 to about 200 μl and filled up to 1,000 μl with hexane. Aliquots (1 μl) were analyzed by capillary gas chromatography performed on a Hewlett-Packard 5890 Series II gas chromatograph equipped with an HP Ultra 2 capillary column (50 m by 0.2 mm; film thickness, 0.11 mm) and a flame ionization detector. Hydrogen served as the carrier gas. The injector temperature was set to 250°C, and the detector temperature was 300°C. The oven program was as follows: 80°C for 3 min, 90°C to 288°C at 6°C min−1, and an isothermal period of 20 min. The PCB congeners were identified by gas chromatography-mass spectrometry (7) and by comparison with authentic standards. Because previous experiments with PCB microdroplets possessing a much larger surface-to-volume ratio than the PCB droplets used in this study have shown that only small amounts of PCB congeners were leached by water in sterile microcosms (36), 2,2′,5,5′-tetrachloro-PCB (PCB 52) (4), present in Aroclor 1242, was used as an internal standard (8).
Microcosm experiments.
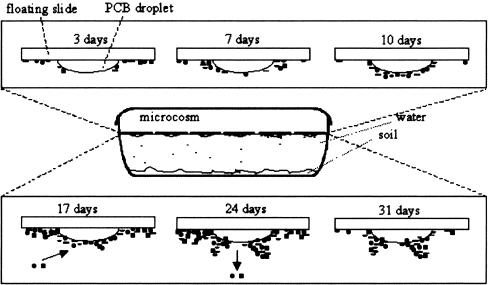
Droplets (0.7 μl) of Aroclor 1242 (Promochem, GB) were placed on a sterile slide (Permanox [Nunc], 100 by 20 mm, sterilely packed by the manufacturer) (17). Permanox is a solvent-resistant polymer which is inert against PCB oil, although solvents such as xylene or toluene cause Permanox plastic slides to warp. Because autofluorescence at commonly used wavelengths is very low, Permanox products are recommended for fluorescent applications. The slides, bearing 10 droplets of PCB, were placed with the PCB droplets downwards on the water surface of a reservoir filled with 1 liter sterile tap water and 100 g of PCB-contaminated soil, which were thoroughly mixed before the reservoir was filled. The microcosm was kept at room temperature without agitation. After 7, 14, 21, 24, and 28 days, new slides with 10 PCB droplets each were added, and all were harvested 3 days later. This resulted in incubation times of 3, 7, 10, 17, 24, and 31 days. Pieces with 2 PCB droplets per slide were cut off and immediately examined by CLSM. Biofilms were harvested with a sterile spatula, and DNA was extracted from material representing 3 or 4 PCB droplets by using the Fast-DNA-Spin kit for soil (Bio 101, La Jolla, CA) according to the manufacturer's instructions (31). Four to 5 different droplets of PCB from the same slide were extracted, and for each droplet the PCB composition was analyzed as described above.
SSCP fingerprint analysis.
The primers chosen for amplification of bacterial 16S rRNA genes were the forward primer Com1 and the reverse primer Com2-Ph as published by Schwieger and Tebbe (35). The phosphorylated strand of the PCR products was digested by lambda exonuclease (New England Biolabs, Schwalbach, Germany), proteins were removed by the Mini-elute kit (QIAGEN, Hilden, Germany) as recommended by the manufacturer, and the remaining single-stranded DNA was dried under a vacuum. The DNA was then resuspended in denaturing SSCP loading buffer (47.5% formamide, 5 mM sodium hydroxide, 0.12% bromophenol blue, and 0.12% xylene cyanol) and subjected to electrophoresis (35). Gels were run at 400 V for 17 h at 20°C in a Macrophor electrophoresis unit (LKB, Bromma, Sweden) and subsequently silver stained (5).
Sequence determination of SSCP bands.
Bands were excised from the gel, eluted in buffer (10 mM Tris buffer, 5 mM KCl, 1.5 mM MgCl2 · 6H2O, 0.1% Triton X-100, pH 9.0), and extracted at 95°C for 15 min. Extracts were centrifuged, and the supernatant was used as a DNA template in the PCR with the primers described above. The PCR product was purified (Mini-elute kit; QIAGEN, Hilden, Germany) and sequenced with a sequencing kit (DYEnamic ET Terminator cycle sequencing kit; Amersham Biosciences, Freiburg, Germany) and both primers. The product was cleaned with the Dye Ex Spin kit (QIAGEN, Hilden, Germany), and the sequence was analyzed on an ABI PRISM 337 DNA sequencer and an ABI PRISM 3100 genetic analyzer. The sequences were compared using the N-FASTA program and the EMBL and GenBank databases (Table 1) (2).
TABLE 1.
Identification of the main bands of the SSCP gels by excision, sequencing, and comparison with 16S and RNA gene sequences from public databases
| Band no. | Accession no. | Cultured closest matcha | Size (bp) | Identity (%) |
|---|---|---|---|---|
| 1 | AJ884727 | Bradyrhizobium sp. | 371 | 99 |
| 2 | AJ884728 | Herbaspirillum sp. | 371 | 97 |
| 3 | AJ884729 | Desulfovibrio sp. | 371 | 82 |
| 4 | AJ884730 | Oscillatoria sp. | 355 | 83 |
| 5 | AJ884731 | Bradyrhizobium sp. | 371 | 100 |
| 6 | AJ884732 | Bdellovibrio sp.* | 158 | 80 |
| 7 | AJ884733 | Herbaspirillum sp. | 371 | 98 |
| 8 | AJ884734 | Acidovorax sp. or Variovorax sp. | 257 | 98 |
| 9 | AJ884735 | Pelotomaculum sp. | 370 | 85 |
| 10 | AJ884736 | Sphingomonas sp. or Paracoccus sp. | 370 | 81 |
| 11 | AJ884737 | Rhodovibrio sp. | 370 | 87 |
| 12 | AJ884738 | Trichlorobacter sp. | 304 | 84 |
| 13 | AJ884739 | Pectinatus sp. | 371 | 82 |
| 14 | AJ884740 | Bradyrhizobium sp. | 371 | 98 |
| 15 | AJ884741 | Bdellovibrio sp.* | 170 | 89 |
| 16 | AJ884742 | Acidithiobacillus sp.* | 171 | 91 |
| 17 | AJ884743 | Beggiatoa sp.* | 206 | 86 |
| 18 | AJ884744 | Polynucleobacter sp.* | 165 | 87 |
| 19 | AJ884745 | Herbaspirillum sp. | 371 | 98 |
| 20 | AJ884746 | Acanthamoeba sp. mitochondrion | 371 | 99 |
*, sequence obtained for only one primer.
Microscopy analysis—biofilm staining.
Samples were first stained for hydrophobic compounds (PCBs) and then for nucleic acids (bacteria). PCB droplets on the slides were stained with Nile Red (Sigma, St. Louis, MO). For this purpose a stock solution of 2 mg Nile Red in 1 ml acetone-water (1:1, vol/vol) was diluted 1:1,000 in demineralized water. After staining for 15 min, the sample was carefully rinsed twice and counterstained using a nucleic acid-specific stain. Bacterial cell distribution was determined with SYBR green (Molecular Probes, Eugene, OR) (27). The nucleic acid-specific SYBR green stain was used as supplied and diluted 1:1,000 in demineralized water. Samples were incubated for 5 min at room temperature. Live and damaged cells in biofilms were stained with the BacLight kit (Molecular Probes, Eugene, OR) applied as described by the manufacturer. All samples were examined immediately after staining using CLSM (25).
CLSM.
Laser scanning microscopy was performed using the model TCS SP (Leica, Heidelberg, Germany) attached to an upright microscope. The instrument was controlled by Leica Confocal software, version 2.5, Build 1347d. The system was equipped with three visible lasers: an Ar laser (458, 476, 488, and 514 nm), a laser diode (561 nm), and a He-Ne laser (633 nm). The spectrophotometer feature allowed flexible and optimal adjustment of sliders on the detector side. The following settings were used for excitation and recording of emission signals, respectively: SYBR green, 488 and 500 to 530 nm; Nile Red, 488 and 550 to 700 nm; Syto9, 488 and 500 to 540 nm; propidium iodide, 561 and 590 to 650 nm. Biofilm samples were observed with 10× 0.3-numerical aperture (NA), 20× 0.5-NA, and 63× 0.9-NA water-immersible lenses (24).
Digital image analysis.
Images were visualized by using the microscope software (Leica) for maximum-intensity projections and Imaris, version 4.06 (Bitplane, Zürich, Switzerland), for XYZ projections and isosurface rendering. Live/Dead staining was quantified using the ConAn program (written especially for digital image analysis of microbiological CLSM data) after object definition with an intensity threshold of 51 pixels. Images were mounted in Photoshop CS (Adobe, San Jose, Calif.) without any image adjustments.
Statistical analysis.
The data were analyzed using Statistica 5.0 software (Statsoft, Tulsa, Okla.). For cluster analysis of PCB degradation, we used the relative amount of the remaining congeners, the unweighted-pair group method using average linkages, and 1 − Pearson r as a distance measure.
RESULTS
Structural development of the biofilm.
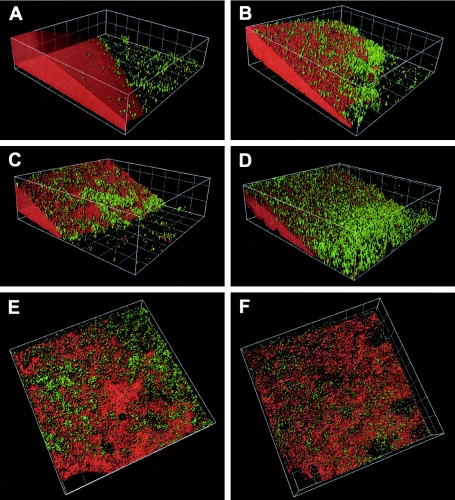
The biofilm was monitored by CLSM over 31 days using Nile Red to stain PCBs and SYBR green for the bacteria. Figure 1 depicts the formation and development of the biofilm on the PCB droplet, showing the spatial progress of the biofilm on the PCB oil droplet. After 3 days of incubation, a biofilm was detected on the Permanox slide close to the PCB, but only a few cells were observed on the PCB droplet (Fig. 1A). It is noteworthy that Nile Red stained not only PCB but also some groups of bacteria, as shown in Fig. 2B, indicating a hydrophobic cell surface. Staining with the BacLight kit revealed a number of damaged cells staining red. A quantitative analysis gave a living/damaged cell ratio of 1.0:0.76, which means 24% more living than defective cells. Subsequently, after 7 days, substantial biofilm accumulation was observed on the margins of the droplet as well as directly on the PCB droplet (Fig. 1B). Furthermore, after 7 days, the number of live cells was higher than that of defective cells, with a ratio of 1.0:0.68 (Fig. 3G).
FIG. 1.
Three-dimensional surface rendering of PCB biofilm development. (A) Three-day-old biofilm; (B) 7-day-old biofilm; (C) 10-day-old biofilm; (D) 17-day-old biofilm; (E) 24-day-old biofilm; (F) 31-day-old biofilm. Green, SYBR green; red, Nile Red. Grid size, 20 μm.
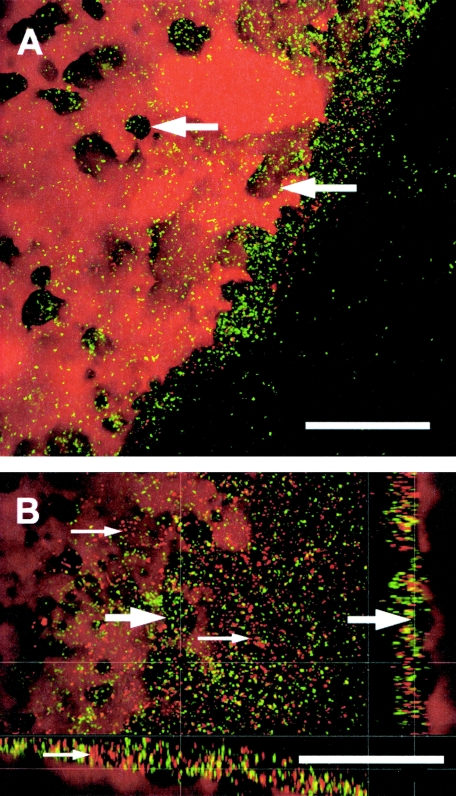
FIG. 2.
Top and side views of a 31-day-old biofilm after staining with Nile Red and SYBR green. (A) PCB surface as maximum-intensity projections showing local depressions in the PCB droplet (arrows). (B) PCB surface as top view (large image) and side views (bottom, XZ; right, YZ), demonstrating the local depressions from the top and from the side (large arrows). Note that some bacteria could be stained with Nile Red, which was used for PCB staining (small arrows indicate red cells). Bars, 50 μm.
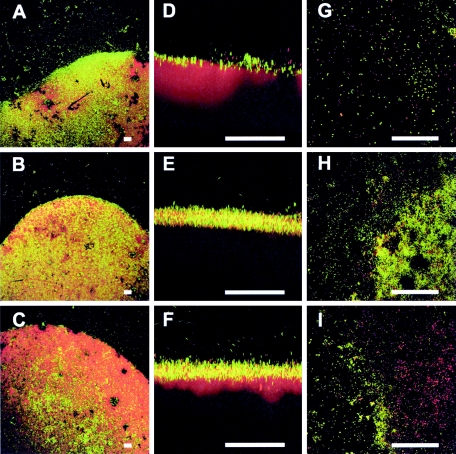
FIG. 3.
Biofilm development after 7 days (A, D, and G), after 10 days (B, E, and H), and after 31 days (C, F, and I). The PCB biofilm stained with SYBR green (bacteria) and Nile Red (PCB droplet) is shown as XY maximum-intensity projections (A, B, and C) and as XZ projections (D, E, and F). The PCB biofilm after staining with LIVE (green)/DEAD (red) is presented as maximum-intensity projections (G, H, and I).
The monitoring detected an interesting phenomenon in the 10-day-old biofilm: on the one hand, the biofilm previously on the Permanox surface detached to a large extent—leaving the slide surface around the droplet almost without cells—but on the other hand, the biofilm on the PCB oil grew significantly and started to form some aggregates (Fig. 3D and E). The densest population was observed in the 10-day-old biofilm. At this point the accumulation of cells was not only at the droplet boundary but widespread on the PCB oil. The numbers of live and damaged cells also changed after 10 days and became equally distributed, with a 1.0:0.97 ratio (Fig. 3H). After 17 days the PCB biofilm showed the highest number of species (see Fig. 5), but at this time the bacterial population on the Permanox substratum was lower than that for the 7-day-old biofilm (Fig. 1B and D). Twenty-four days after incubation, large microbial aggregates could be noticed on the PCB surface, as schematically depicted in Fig. 4, exposing some areas of the PCB surface. For the first time, biofilm invasion into the PCB droplet was observed. The 31-day-old biofilm revealed more detachment from the PCB surface, and the microbial aggregates became larger (Fig. 3C and F). In addition, larger “holes” in the PCB droplet were observed (Fig. 1E, 1F, 2A, and 2B). As at 10 days, the Permanox surface around the droplet was almost empty at this time. After 17, 24, and 31 days, damaged cells were dominant, with a ratio of 1.0:1.7 (Fig. 3I).
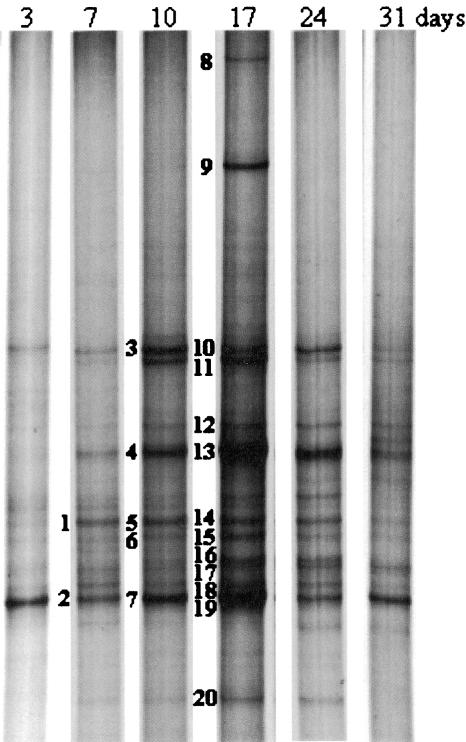
FIG. 5.
Dynamics of biofilm development as revealed by SSCP fingerprinting. Marked bands were excised, sequenced, and compared with sequences of described species (Table 1).
FIG. 4.
Scheme of PCB biofilm development in the microcosm.
Microbial community structure development.
Qualitatively the PCB biofilm community did not show dramatic changes over the whole biofilm development (Fig. 5). SSCP analysis of the young biofilm revealed a rather diverse microbial community after 3 days. Analysis of the sequence of the main SSCP band resulted in its identification as a close relative of the genus Herbaspirillum (band 2). Band 2 (as well as bands 7 and 19) stayed prominent over the whole observation period until the final sampling after 31 days. A second major band, band 1 (as well as bands 5 and 14), showed high identity to a Bradyrhizobium sp. On the 7-day-old biofilm two other bands, showing mobilities on the SSCP gel equal to those of bands 10 and 13, were identified as a Sphingomonas or Paracoccus sp. and a Pectinatus sp., respectively. The 10-day-old PCB biofilm revealed the same bands found after 7 days and two additional intensive bands, bands 11 and 6, corresponding to Rhodovibrio sodomensis and a Bdellovibrio sp., respectively. Microbial diversity increased further over the following days and reached an apparent maximum at day 17, when bands 8 and 9, which were not seen on the other days, could be observed. The new band 9 was found to be a Pelotomaculum sp., and band 8 an Acidovorax or Variovorax sp. Bands 8 and 9 were present only in this biofilm and disappeared afterwards. Furthermore, bands 16, 17, and 18 appeared and were identified as an Acidothiobacillus sp., a Beggiatoa sp., and a Polynucleobacter sp., respectively. On days 17 and 24, band 20, which was identified as the mitochondrion of an Acanthamoeba species (Table 1), was more prominent. The biofilm remained relatively constant on days 24 and 31, and no new significant bands appeared.
Dynamics of PCB degradation.
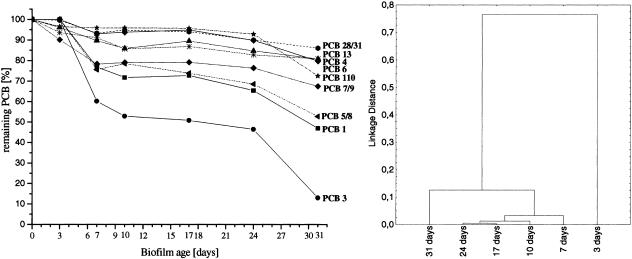
The PCB degradation profile of the biofilm at each time point revealed a differentiated decrease of several PCB congeners. Some of these congeners had a significantly delayed decrease, e.g., 2-chlorobiphenyl (PCB 1), 2,3- and 2,4′-dichlorobiphenyl (PCB 5 and PCB 8, respectively), and especially 4-chlorobiphenyl (PCB 3). The latter congener did not show any degradation after 3 days but declined rapidly after day 7. Between days 10 and 24, degradation remained relatively stable for all congeners. After that time an additional decrease took place for many PCB congeners, reaching 87% degradation for 4-chlorobiphenyl. A cluster analysis based on the degradation of all congeners of the PCB oil at the various time points gives a much better comparison of the metabolic activities of the biofilm community than a comparison based only on the degradation of single congeners. Such an analysis showed three different stages of degradation (Fig. 6, right panel). While the PCB oil was hardly degraded after 3 days, some degradation was apparent after 7 days. The degradation patterns of the next three time points clustered together, showing very similar compositions of the PCB oil. The 31-day-old biofilm was separated from this group because of a second round of PCB consumption (also clearly seen in Fig. 6, left panel).
FIG. 6.
(Left) Time course of the degradation of individual PCB congeners; (right) dendrogram of the PCB composition of the biofilms. Congeners are as follows: PCB 1, 2-chlorobiphenyl; PCB 3, 4-chlorobiphenyl; PCB 4, 2,2′-dichlorobiphenyl; PCB 5, 2,3-dichlorobiphenyl; PCB 6, 2,3′-dichlorobiphenyl; PCB 7, 2,4-dichlorobiphenyl; PCB 8, 2,4′-dichlorobiphenyl; PCB 9, 2,5-dichlorobiphenyl; PCB 13, 3,4′-dichlorobiphenyl; PCB 28, 2,4,4′-trichlorobiphenyl; PCB 31, 2,4′,5-trichlorobiphenyl; PCB 110, 2,3,3′,4′,6-pentachlorobiphenyl.
DISCUSSION
While the community analysis showed no fundamental changes in the biodiversity of the biofilm, confocal laser scanning microscopy revealed a clear sequence of biofilm colonization of the PCB oil. The bacteria settled first on the Permanox substratum, and only a very few bacteria were found on the PCB droplet at this stage. Often something like a “zone of avoidance” could be seen between the colonized Permanox and the PCB droplet, leaving a small zone free of bacteria on the Permanox substratum close to the PCB. Interestingly, no “clay hutches,” i.e., bacterial clay aggregates, which have been reported previously (18), could be detected on the substratum. One explanation could be that in the clay hutches the clay was loaded with PCBs from the contaminated soil, which served as a substrate for the bacteria within the hutches, while in the experiments discussed here, PCB was presented on the substratum to the microbial community, which could use the substrate directly without any recruitment of clay. Another reason could be that the clay aggregates forming microcosms were gently shaken, but slides and reservoirs were not shaken in the current study. The next step in biofilm development was a concentration of bacteria around the droplet, which approached the PCB oil and started to colonize it. The number of bacteria found on the Permanox became more and more depleted, and after 17 days the biofilm was found mainly on the PCB droplet. Here it formed aggregates, and in the late stage of biofilm formation, depressions in the PCB droplet could regularly be seen, where bacterial aggregates started to grow into the droplet. A similar behavior has been described for a biofilm with Acinetobacter venetianus, using diesel fuel droplets, where the bacteria adhere to the oil surface (3, 30), and it can be speculated that this is a common behavior of biofilm communities colonizing hydrophobic substances.
This sequence of PCB biofilm formation was accompanied by a marked increase in the number of damaged cells after 10 days, which has also been observed in other mature biofilms (37) and appeared, therefore, not to be caused only by the hydrophobicity of PCB. Live-dead staining revealed the presence of damaged cells from the very beginning, but their number increased dramatically after 2 weeks. Similar results have been described for mature toluene-degrading biofilms, which grew in a bioreactor on polypropylene rings using toluene as a carbon source (37). Recently, this increase in the number of damaged cells has been also described as an aging process of biofilms (38). If conditions are becoming unfavorable, some bacteria possess the ability to destroy the biofilm matrix, setting the cells free as planktonic cells which can then move to more favorable sites (21). Because of the small changes in the composition of the microbial community over the entire experiment, it can be assumed that the bacteria that formed the PCB biofilm were probably the same that accumulated at the margins of the droplets. A Herbaspirillum sp. was the first to appear significantly in the PCB biofilm community, and it stayed present over the whole experiment; for this reason, it can be assumed that the Herbaspirillum sp. was very important for PCB degradation. Herbaspirillum sp. strain K1 (22) and the recently described Herbaspirillum chlorophenolicum (14) have been shown to degrade 2,3,4,6-tetrachlorophenol and 4-chlorophenol, but no other involvement in biofilm has been reported until now. Interestingly, no Burkholderia species could be detected, although they are reported to be abundant in the soil (29), among the isolates grown on biphenyl (1) and involved in the degradation of PCB congeners (36). A second major band in the 7-day biofilm (band 2) showed 100% identity with a Bradyrhizobium sp. that has previously been identified as a main degrader of 4-chlorobenzoate (9), and a putative 4-chlorobenzoate coenzyme A ligase has been found in the Bradyrhizobium japonicum USDA110 genome (16). Since a Bradyrhizobium sp. has also been reported from a PCB biofilm from the same soil (36), it can be speculated that Bradyrhizobium species may have their function in the lower degradation pathway of PCBs. Bradyrhizobium and/or Herbaspirillum species seem to be good candidates for taking over the role of the missing Burkholderia species in PCB degradation, but further studies are needed for confirmation. Furthermore, the observation of two Bradyrhizobium spp. in the PCB biofilm correlated well with the report of a number of Bradyrhizobium clones from soil based both on DNAs and on rRNAs of this site (28, 29). Pectinatus spp. are well known as beer spoilage bacteria and have been isolated from very different places, even from a mixture of oil with beer (13), which demonstrates at least their ability to survive in relatively extreme environments. The occurrence of a Pectinatus sp. (band 9), an anaerobic species (34), in the biofilm with an SSCP band intensity similar to those of the Herbaspirillum and Bradyrhizobium species points at least to anaerobic niches within the biofilm, which could also explain the degradation of the 2,3,3′,4′,6-pentachlorobiphenyl (PCB 110), probably degraded via reductive dechlorination. These three bacteria were significantly present in the SSCP analysis and showed strong bands in all biofilms (Fig. 5). It can be assumed that these bacteria played a crucial role in biofilm formation and activity.
The PCB biofilm showed three distinct stages of PCB degradation for most congeners. The first stage occurred before 10 days, when individual congeners were degraded in the range of 5 to 45%. Interestingly, within the period from 10 to 24 days, another stage occurred, in which the biofilm did not degrade much PCB. One reason could be metabolites inhibiting or damaging cells in the biofilm, because during this period (17 days) the highest ratio of damaged to living cells was observed, with about 70% damaged cells in the 17-day-old biofilm. This may also explain the attachment and detachment of the Pelotomaculum sp. and the Acidovorax or Variovorax sp. on the 17-day-old biofilm; these two bacteria possibly used compounds of the dead cells as a substrate. The detection of the Acanthamoeba sp. mitochondrion, which was most prominent in the 17-day biofilm, points additionally to some grazing pressure on the biofilm community, further contributing to the increase in damaged cells and the almost complete stop in PCB degradation.
The third degradation stage occurred in the period between 24 and 31 days. This stage showed a dramatic decrease in 4-chlorobiphenyl (PCB 3) to only 15% of its original amount, as well as in other congeners, with 2-chlorobiphenyl (PCB 1) and 2,3- and 2,4′-dichlorobiphenyl (PCB 5 and PCB 8, respectively), for example, decreasing by about 50%. A detailed analysis of the SSCP fingerprint revealed that a Pectinatus sp. (band 9) and a Sphingomonas or Paracoccus sp. (band 10), as well as a Herbaspirillum sp., were the most abundant species at this time. This is corroborated by the report of Nogales et al., who found Sphingomonas species to be one of the active groups in the same site (29). The reason for the second stage of degradation is unknown, but the presence of the anaerobic Pectinatus sp. and the invasion of the biofilm into the PCB oil point to anaerobic conditions, where reductive dechlorination is favored. This is corroborated by the beginning of the degradation of 2,3,3′,4,5′-pentachlorobiphenyl (PCB 110) after day 24, which is very likely initiated by a reductive dechlorination step. At this stage it is assumed that the biofilm showed an anaerobic section close to the PCB oil, where the higher-chlorinated congeners were dechlorinated, and an aerobic section where the ring cleavage occurred. However, the formation of an anaerobic zone in the biofilm obviously also caused some changes in the aerobic part, because here another round of degradation of mono- and dichlorobiphenyls was observed.
Conclusions.
The biofilm developing from PCB-contaminated soil on PCB droplets displayed a pronounced pattern of maturation not reported before from pure-culture experiments. The first step was the colonization of the Permanox substratum, while the PCB oil was hardly populated. When a certain density of bacteria was reached on the Permanox, the PCB was colonized, but afterwards the degradation of the congeners was markedly reduced and many cells were damaged. Finally, the biofilm formed aggregates and invaded the PCB oil, showing fewer damaged cells and a dramatic increase in PCB degradation. The three stages of the biofilm development are as follows. (i) Up to 10 days, the PCB droplet is populated, degradation of low-chlorinated PCB congeners starts, and an increase in biodiversity is observed. (ii) Between 10 and 24 days, aggregates are formed in the mature biofilm on the PCB droplet, little PCB is degraded, and bacterial diversity reaches a maximum. (iii) Between 24 and 31 days, the aggregates increase, bacterial invasion of PCB oil starts, the biodiversity of the biofilm decreases, and a new round of PCB degradation, including that of the pentachlorobiphenyls, occurs.
This was reflected not only in the analysis of the CLSM data but also in the degradation pattern of the PCB congeners as shown in Fig. 6. This sequence of biofilm formation is understood as a maturation process (prior to PCB oil colonization) followed by the development of a thin biofilm on the PCB droplet, an aggregation process forming pockets in the PCB, and finally an invasion of the biofilm into the PCB oil. The mature biofilm was able to degrade higher-chlorinated PCB congeners, a process that may involve reductive dechlorination.
Acknowledgments
We are indebted to Jennifer Skerra for microbiological work and to Esther Surges for PCB and soil analysis.
A.J.M. acknowledges a Ph.D. stipend from the German Academic Exchange Service (DAAD).
REFERENCES
- 1.Abraham, W. R., H. Lünsdorf, C. Strömpl, B. Nogales, E. R. B. Moore, and K. N. Timmis. 2003. Microbial communities in composite biofilms participating in the degradation of PCB. Water Air Soil Pollut. Focus Bioremediation 3:57-64. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Baldi, F., N. Ivošević, A. Minacci, M. Pepi, R. Fani, V. Svetličić, and V. Žutić. 1999. Adhesion of Acinetobacter venetianus to diesel fuel droplets studied with in situ electrochemical and molecular probes. Appl. Environ. Microbiol. 65:2041-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballschmiter, K., and M. Zell. 1980. Analysis of polychlorinated biphenyls (PCB) by glass capillary gas chromatography. Fresenius J. Anal. Chem. 302:20-31. [Google Scholar]
- 5.Bassam, B. J., G. Caetano-Anolles, and P. M. Gresshoff. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196:80-83. [DOI] [PubMed] [Google Scholar]
- 6.Brouwer, A., M. P. Longnecker, L. S. Birnbaum, J. Cogliano, P. Kostyniak, J. Moore, S. Schantz, and G. Winneke. 1999. Characterization of potential endocrine-related health effects at low-dose levels of exposure to PCBs. Environ. Health Perspect. 107(Suppl. 4):639-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buthe, A., and E. Denker. 1995. Qualitative and quantitative determination of PCB congeners by using a HT-5 GC column and an efficient quadropole MS. Chemosphere 30:753-771. [Google Scholar]
- 8.Duinker, J. C., D. E. Schulz, and G. Petrick. 1988. Selection of chlorinated biphenyl congeners for analysis in environmental samples. Mar. Pollut. Bull. 19:19-25. [Google Scholar]
- 9.Gentry, T. J., G. Wang, C. Rensing, and I. L. Pepper. 2004. Chlorobenzoate-degrading bacteria in similar pristine soils exhibit different community structures and population dynamics in response to anthropogenic 2-, 3-, and 4-chlorobenzoate levels. Microb. Ecol. 48:90-102. (First published 19 April 2004; doi: 10.s00248-003-1048-1.) [DOI] [PubMed] [Google Scholar]
- 10.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 11.Harkness, M. R., J. B. McDermott, D. A. Abramowicz, J. J. Salvo, W. P. Flanagan, M. L. Stephens, F. J. Mondello, R. J. May, J. H. Lobos, and K. M. Carroll. 1993. In situ stimulation of aerobic PCB biodegradation in Hudson River sediments. Science 259:503-507. [DOI] [PubMed] [Google Scholar]
- 12.Harry, M., B. Gambier, and E. Garnier-Sillam. 2000. Soil conservation for DNA preservation for bacterial molecular studies. Eur. J. Soil Biol. 36:51-55. [Google Scholar]
- 13.Helander, I. M., A. Haikara, I. Sadovskaya, E. Vinogradov, and M. S. Salkinoja-Salonen. 2004. Lipopolysaccharides of anaerobic beer spoilage bacteria of the genus Pectinatus—lipopolysaccharides of a Gram-positive genus. FEMS Microbiol. Rev. 28:543-552. [DOI] [PubMed] [Google Scholar]
- 14.Im, W. T., H. S. Bae, A. Yokota, and S. T. Lee. 2004. Herbaspirillum chlorophenolicum sp. nov., a 4-chlorophenol-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54:851-855. [DOI] [PubMed] [Google Scholar]
- 15.Ivanov, V., and E. Sandell. 1992. Characterization of polychlorinated biphenyl isomers in Sovol and trichlorobiphenyl formulations by high-resolution gas chromatography with electron capture detection and high-resolution gas chromatography-mass spectrometry techniques. Environ. Sci. Technol. 26:2012-2017. [Google Scholar]
- 16.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 17.Lünsdorf, H. 2000. Anzucht und Charakterisierung von PCB-abbauenden Biofilmen, p. 35-38. In K. N. Timmis and W.-R. Abraham (ed.), Mikrobielle Ökologie von Altlasten und deren Bioremediation durch den Einsatz von Spezialkulturen. Final report, BEO Projekt FKZ 0319433C. GBF, Braunschweig, Germany.
- 18.Lünsdorf, H., R. W. Erb, W. R. Abraham, and K. N. Timmis. 2000. ‘Clay hutches’: a novel interaction between bacteria and clay minerals. Environ. Microbiol. 2:161-168. [DOI] [PubMed] [Google Scholar]
- 19.Lünsdorf, H., C. Strömpl, A. M. Osborn, A. Bennasar, E. R. Moore, W. R. Abraham, and K. N. Timmis. 2001. Approach to analyze interactions of microorganisms, hydrophobic substrates, and soil colloids leading to formation of composite biofilms, and to study initial events in microbiogeological processes. Methods Enzymol. 336:317-331. [DOI] [PubMed] [Google Scholar]
- 20.MacDonald, R. W., L. A. Barrie, T. F. Bidleman, M. L. Diamond, D. J. Gregor, R. G. Semkin, W. M. J. Strachan, Y. F. Li, F. Wania, M. Alaee, L. B. Alexeeva, S. M. Backus, R. Bailey, J. M. Bewers, C. Gobeil, C. J. Halsall, T. Harner, J. T. Hoff, L. M. M. Jantunen, W. L. Lockhart, D. Mackay, D. C. G. Muir, J. Pudykiewicz, K. J. Reimer, J. N. Smith, G. A. Stern, W. H. Schroeder, R. Wagemann, and M. B. Yunker. 2000. Contaminants in the Canadian Arctic: 5 years of progress in understanding sources, occurrence and pathways. Sci. Total Environ. 254:93-234. [DOI] [PubMed] [Google Scholar]
- 21.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70:3232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mannisto, M. K., M. A. Tiirola, and J. A. Puhakka. 2001. Degradation of 2,3,4,6-tetrachlorophenol at low temperature and low dioxygen concentrations by phylogenetically different groundwater and bioreactor bacteria. Biodegradation 12:291-301. [DOI] [PubMed] [Google Scholar]
- 23.Matz, C., T. Bergfeld, S. A. Rice, and S. Kjelleberg. 2004. Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 6:218-226. [DOI] [PubMed] [Google Scholar]
- 24.Neu, T. R., U. Kuhlicke, and J. R. Lawrence. 2002. Assessment of fluorochromes for two-photon laser scanning microscopy of biofilms. Appl. Environ. Microbiol. 68:901-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neu, T. R., and J. R. Lawrence. 1997. Development and structure of microbial biofilms in river water studied by confocal laser scanning microscopy. FEMS Microbiol. Ecol. 24:11-25. [Google Scholar]
- 26.Nielsen, A. T., T. Tolker-Nielsen, K. B. Barken, and S. Molin. 2000. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ. Microbiol. 2:59-68. [DOI] [PubMed] [Google Scholar]
- 27.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 28.Nogales, B., E. R. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nogales, B., E. R. B. Moore, W. R. Abraham, and K. N. Timmis. 1999. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl polluted moorland soil. Environ. Microbiol. 1:199-212. [DOI] [PubMed] [Google Scholar]
- 30.Pepi, M., A. Minacci, F. Di Cello, F. Baldi, and R. Fani. 2003. Long-term analysis of diesel fuel consumption in a co-culture of Acinetobacter venetianus, Pseudomonas putida and Alcaligenes faecalis. Antonie Leeuwenhoek 83:3-9. [DOI] [PubMed] [Google Scholar]
- 31.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarro, M. I., D. A. Moreno, E. Chicote, P. I. Lorenzo, A. M. Garcia, and F. Montero. 2003. Biofouling on austenitic stainless steels in spent nuclear fuel pools. Materials Corrosion 54:535-540. [Google Scholar]
- 33.Schachter, B. 2003. Slimy business—the biotechnology of biofilms. Nat. Biotechnol. 21:361-365. [DOI] [PubMed] [Google Scholar]
- 34.Schleifer, K. H., M. Leuteritz, N. Weiss, W. Ludwig, G. Kirchhof, and H. Seidel-Rufer. 1990. Taxonomic study of anaerobic, gram-negative, rod-shaped bacteria from breweries: emended description of Pectinatus cerevisiiphilus and description of Pectinatus frisingensis sp. nov., Selenomonas lacticifex sp. nov., Zymophilus raffinosivorans gen. nov., sp. nov., and Zymophilus paucivorans sp. nov. Int. J. Syst. Bacteriol. 40:19-27. [DOI] [PubMed] [Google Scholar]
- 35.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tillmann, S., C. Strömpl, K. N. Timmis, and W. R. Abraham. 2005. Stable isotope probing reveals the dominant role of Burkholderia sp. in aerobic degradation of PCBs. FEMS Microbiol. Ecol. 52:207-217. [DOI] [PubMed] [Google Scholar]
- 37.Tresse, O., S. Lescob, and D. Rho. 2003. Dynamics of living and dead bacterial cells within a mixed-species biofilm during toluene degradation in a biotrickling filter. J. Appl. Microbiol. 94:849-854. [DOI] [PubMed] [Google Scholar]
- 38.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]