Abstract
Although l-serine proceeds in just three steps from the glycolytic intermediate 3-phosphoglycerate, and as much as 8% of the carbon assimilated from glucose is directed via l-serine formation, previous attempts to obtain a strain producing l-serine from glucose have not been successful. We functionally identified the genes serC and serB from Corynebacterium glutamicum, coding for phosphoserine aminotransferase and phosphoserine phosphatase, respectively. The overexpression of these genes, together with the third biosynthetic serA gene, serAΔ197, encoding an l-serine-insensitive 3-phosphoglycerate dehydrogenase, yielded only traces of l-serine, as did the overexpression of these genes in a strain with the l-serine dehydratase gene sdaA deleted. However, reduced expression of the serine hydroxymethyltransferase gene glyA, in combination with the overexpression of serAΔ197, serC, and serB, resulted in a transient accumulation of up to 16 mM l-serine in the culture medium. When sdaA was also deleted, the resulting strain, C. glutamicum ΔsdaA::pK18mobglyA′(pEC-T18mob2serAΔ197CB), accumulated up to 86 mM l-serine with a maximal specific productivity of 1.2 mmol h−1 g (dry weight)−1. This illustrates a high rate of l-serine formation and also utilization in the C. glutamicum wild type. Therefore, metabolic engineering of l-serine production from glucose can be achieved only by addressing the apparent key position of this amino acid in the central metabolism.
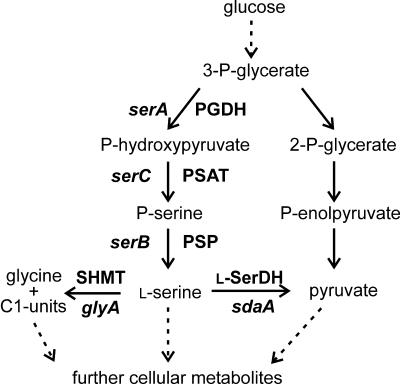
The demand of l-serine is about 300 tons per year, and this amino acid is required for the pharmaceutical and the cosmetic industries, in addition to being a building block for chemical and biochemical purposes (6). The current production relies mainly on its enzymatic or cellular conversion from the precursor glycine plus a C1 compound. Utilizing the condensing activity of serine hydroxymethyltransferase, an enzymatic system has been elaborated to convert glycine plus formaldehyde to l-serine (15). The cellular systems employed, among others, resting cells of methanol-utilizing bacteria such as Hyphomicrobium methylovorum where l-serine accumulation from glycine plus methanol was achieved (16). Also, a fermentative production of l-serine from glycine alone by Corynebacterium glycinophilum was described (19). However, there is not much information on the direct fermentative production of l-serine from glucose. Attempts to isolate l-serine-producing strains using different bacteria by applying undirected mutagenesis yielded mutants accumulating only traces of l-serine (38). Apparently, the direct conversion of glucose is a demanding challenge, probably due to the role of l-serine as a central intermediate for a number of cellular reactions (Fig. 1).
FIG. 1.
Scheme of l-serine biosynthesis and its metabolism in C. glutamicum during growth on glucose. Dotted arrows represent pathways consisting of more than one reaction. Genes are given in italics.
We are interested in the amino acid-synthesizing capabilities of Corynebacterium glutamicum, which is traditionally used for the large-scale production of l-glutamate and l-lysine (9). In general, the efforts to engineer producing strains were focused on the enzymes of the biosynthesis pathways. For instance, considerable formation of l-lysine resulted in the deregulation of the key enzyme aspartate kinase (4). Using similar approaches, C. glutamicum strains were developed by overproducing l-isoleucine, l-valine, l-threonine, or d-pantothenate (8, 13). Besides the supply of precursors (29, 31) or reducing power (23), export of amino acids was also found to be relevant (7). Another focus of strain development was degradation. For instance, production of l-threonine with C. glutamicum could be increased by decreasing its intracellular degradation (33) and in fact, one of the reasons for the success of l-lysine formation with C. glutamicum was its inability to degrade the product l-lysine. Thus, for the production of specific amino acids, a number of cellular reactions have to be considered in the ensemble and not just the biosynthesis pathway alone.
Since there is not yet a convincing strain for l-serine production from glucose, we met the challenge to engineer C. glutamicum for this purpose. In C. glutamicum as in other bacteria, l-serine is synthesized via phosphorylated intermediates starting with the glycolytic intermediate 3-phosphoglycerate, which is oxidized to phosphohydroxypyruvate. Subsequent transamination leads to the formation of phosphoserine, which is dephosphorylated to yield l-serine (Fig. 1). We have previously studied 3-phosphoglycerate dehydrogenase (PGDH; serA) from C. glutamicum catalyzing the initial reaction of the three-step pathway of l-serine biosynthesis (30). As a result of deleting the 197 carboxy-terminal amino acids of the SerA polypeptide, PGDH activity is no longer inhibited by l-serine (30). C. glutamicum possesses a high capacity to degrade l-serine in the presence of glucose, and we could demonstrate that sdaA-encoded l-serine dehydratase is involved in l-serine degradation (24). Based on these studies, we describe here the construction of an l-serine-producing strain from C. glutamicum by metabolic engineering.
MATERIALS AND METHODS
Bacteria and plasmids.
Bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5αMCR | endA1 supE44 recA1 gyrA96 relA1 deoR U169 φ80dlacZΔM15 mcrA Δ(mrr-hsdRMS-mcrBC) | 11 |
| GM2929 | dam-13::Tn9 dcm-6 hsdR2 recF143 mcrA mcrB | 27 |
| C. glutamicum strains | ||
| WT | Wild type, ATCC 13032 | ATCCb |
| ATCC 14752 | Requires biotin | ATCC |
| WTΔsdaA | WT carrying a deletion in the sdaA gene | 24 |
| WT::pK18mobglyA′ | WT with glyA under control of tac promoter | 32 |
| WTΔsdaA::pK18mobglyA′ | WTΔsdaA with glyA under control of tac promoter | This work |
| Plasmids | ||
| pCGL0040 | Donor of Tn5531 (IS1207, Kmr), AproriVEc | U53587c |
| pUC18 | Cloning vector; Apr | 26 |
| pUC18serC | pUC18 with 1.8-kb PCR product containing serC | This work |
| pUC18serB | pUC18 with 1.8-kb PCR product containing serB | This work |
| pUC18serCB | Ligation of 3.5-kb NotI-ScaI fragment from pUC18serC with 2.8-kb NotI-ScaI fragment from pUC18serB | This work |
| pUC18serAΔ197 | pUC18 with 1.25-kb PCR fragment containing serA with a deletion of 197 aa at the C terminus | 30 |
| pZ1serA | pZ1 with 1.9-kb EcoRI-BamHI fragment from pUC18serA containing serA | 30 |
| pZ1serAΔ197 | pZ1 with 1.3-kb EcoRI-BamHI fragment from pUC18serAΔ197 containing serAΔ197 | 30 |
| pEC-T18mob2 | E. coli-C. glutamicum shuttle vector, Tetr | 35 |
| pEC-T18mob2serCB | pEC-T18mob2 with 3.6-kb EcoRI-XbaI fragment containing serC and serB from pUC18serCB | This work |
| pEC-T18mob2serAΔ197CB | pEC-T18mob2serCB with 3.6-kb EcoRI-XbaI fragment containing serC and serB from pUC18serCB | This work |
| pK18mobglyA′ | Mobilizable vector, nonreplicative in C. glutamicum, Kmr, containing lacIq and Ptac fused to 5′-terminal fragment of glyA | 32 |
Kmr, kanamycin resistance; Apr, ampicillin resistance; Cmr, chloramphenicol resistance.
ATCC, American Type Culture Collection.
GenBank accession number.
Growth conditions.
Luria-Bertani medium (LB) was used as the standard medium for Escherichia coli, while brain heart infusion medium (BHI; Difco) was used as complex medium for C. glutamicum. As minimal medium, CGXII was used (17), but including 30 mg protocatechuic acid l−1 and 220 mM glucose as a carbon source. When appropriate, E. coli strains were cultured with carbenicillin (50 μg ml−1), kanamycin (50 μg ml−1), chloramphenicol (20 μg ml−1), or tetracycline (5 μg ml−1) and C. glutamicum strains with kanamycin (25 μg ml−1) or tetracycline (5 μg ml−1). A reduced concentration of kanamycin (15 μg ml−1) or tetracycline (4 μg ml−1), respectively, was used to obtain transformants of C. glutamicum. E. coli was grown at 37°C and C. glutamicum at 30°C in 50 ml medium in 500 ml baffled shake flasks and 120 rpm agitation. Strains harboring the chromosomally integrated plasmid pK18mobglyA′ were cultivated in the presence of 100 μM isopropyl-β-thiogalactoside (IPTG). For l-serine production experiments, these strains were cultivated without IPTG.
Isolation of serine-auxotrophic mutants and localization of transposon insertion sites.
The transposon (Tn) delivery vector pCGL0040 was isolated from E. coli GM2929 grown in the presence of 50 μg of kanamycin and 20 μg of chloramphenicol ml−1 (1). The plasmid was used to transform C. glutamicum ATCC 14752 to kanamycin resistance by using LBHIS plates (20) with 15 μg of kanamycin ml−1. The resulting Tn mutants were transferred to CGXII plates containing 25 μg of kanamycin ml−1 and either no peptide or a 3 mM concentration of the dipeptide Ser-Ala. Four clones that exhibited only growth in the presence of Ser-Ala were isolated. These clones were retrieved from the LBHIS master plate and tested on CGXII plates containing 25 μg of kanamycin ml−1, 2 mM Ser-Ala, 1 mM l-serine, 1 mM l-alanine, or no supplement. Three mutants required Ser-Ala or l-serine for growth. The cloning and sequencing of the Tn insertion site in these mutants was performed as described previously (32).
Construction of plasmids and strains.
Plasmids were constructed in E. coli DH5αMCR from PCR-generated fragments (Expand High Fidelity PCR kit; Roche Diagnostics) by using C. glutamicum ATCC 13032 DNA as a template prepared according to a method described elsewhere (10). E. coli was transformed by the RbCl2 method (12) and C. glutamicum via electroporation (36). All transformants were analyzed by plasmid analysis and/or PCR with appropriate primers, respectively.
In order to construct pEC-T18mob2serCB, serC and serB were amplified by PCR using the upstream primers serC-upper (5′-GACCACCCACAGCCACCGTAATC-3′; the nucleotide (nt) corresponding to nt 877628 of NC003450 is underlined) and serB-upper (5′-GCGGCCGCGTTGATGATCCTTGGGGTTACG-3′; the nucleotide corresponding to nt 2671294 of NC003450 is underlined), respectively, and the respective reverse primers serC-lower (5′-GCGGCCGCTTTCCCGCATGTTGACTCCTTCTA-3′; the nucleotide corresponding to nt 875874 of NC003450 is underlined) and serB-lower (5′-GAAGGATCCTCGCTATGTGG-3′; the nucleotide corresponding to nt 2669483 of NC003450 is underlined). Boldfaced nucleotides correspond to the introduction of a NotI restriction site. The PCR fragments were blunted and cloned into the SmaI site of pUC18. The obtained plasmids, pUC18serC and pUC18serB, were digested with ScaI and NotI, and the inserts containing serC and serB were isolated and ligated together, resulting in plasmid pUC18serCB. The plasmid was digested with EcoRI and XbaI, and the serCB-containing insert obtained was ligated into EcoRI- and XbaI-treated pEC-T18mob2, resulting in plasmid pEC-T18mob2serCB.
To construct pEC-T18mob2serAΔ197CB, plasmid pUC18serAΔ197 (30) was digested with EcoRI and BamHI and the serAΔ197-containing insert obtained was blunted and ligated in EcoRI-linearized and blunted plasmid pEC-T18serCB.
In order to place the glyA gene in the chromosome of C. glutamicum under the control of the IPTG-inducible tac promoter, the respective strains were transformed via electroporation with the nonreplicative plasmid pK18mobglyA′ to kanamycin resistance. Selection for kanamycin resistance was performed in the presence of 100 μM IPTG. The correct integration into the chromosome via homologous glyA sequences was verified by PCR with appropriate primer pairs and controls. The resulting mutants carried one intact copy of glyA under the control of the inducible tac promoter and one incomplete copy under its own promoter.
Enzyme assays.
Phosphoserine phosphatase activity was analyzed as previously described (3) by the determination of inorganic phosphate (Pi) released from phosphoserine. Assays were performed discontinuously in mixtures (100 μl) containing 20 mM Tris-HCl (pH 7.5), 1 mM MgCl2, and 5 mM phosphoserine. The reaction was stopped after 5 and 10 min by adding 10 μl of 0.2 M EDTA and placing it on ice. The amount of Pi released was determined with an EnzChek phosphate assay kit (Molecular Probes) as described previously.
Serine hydroxymethyltransferase was assayed discontinuously by the quantification of glycine formed from serine and 5,10-methylene tetrahydrofolate via high-performance liquid chromatrography as previously described (33).
3-Phosphoglycerate dehydrogenase activity was determined spectrophotometrically by the formation of NADH as described elsewhere (30).
RESULTS
Identification of the genes coding for phosphoserine aminotransferase and phosphoserine phosphatase.
While the serA gene was known (30), the aim was to identify the complete l-serine biosynthetic pathway in C. glutamicum. Therefore, we used a recently established transposon mutant bank of C. glutamicum ATCC 14752 with Tn5531 to screen for l-serine auxotrophs (18). Three clones unable to grow on minimal medium CGXII unless supplemented with 1 mM l-serine were identified. Sequencing of the transposon flanking regions revealed that in two mutants, the insertions were at different sites within the open reading frame NCgl2436, whereas in the remaining mutant, the insertion was apparently in the promoter region of NCgl0794, 35 nucleotides upstream of the deduced open reading frame. The latter open reading frame encodes a polypeptide of 376 amino acids which exhibits identities of 20% over its entire length to the phosphoserine aminotransferase (PSAT; serC) of E. coli (14). NCgl2436 encodes a deduced polypeptide of 433 amino acids and its C-terminal half shares 41% identical amino acids with the phosphoserine phosphatase (PSP, serB) from E. coli (25). Interestingly, the N-terminal half of the C. glutamicum polypeptide, which exhibits an ACT-like domain (2), is absent from the E. coli PSP, which is 93 amino acids shorter. The sequence similarities to E. coli and the serine auxotrophy of the transposon mutants identified the genes as serC and serB from C. glutamicum. Additionally, we constructed plasmid pEC-T18mob2serCB carrying both genes. With this moderate-copy-number plasmid, the PSP activity was increased threefold, from 110 nmol min−1 mg (protein)−1 in the wild type (WT) to 320 nmol min−1 mg (protein)−1 in strain WT(pEC-T18mob2serCB). Furthermore, we demonstrated that this plasmid complemented the serC and serB transposon mutants as expected (not shown).
Influence of the overexpression of the l-serine biosynthesis genes on l-serine accumulation.
We previously showed that truncation of serA from C. glutamicum by 197 amino acids at its C terminus (encoded by the serAΔ197 allele) provided a 3-phosphoglycerate dehydrogenase devoid of feedback inhibition by l-serine (30). Here, we studied whether overexpression of serA or serAΔ197 is sufficient to increase l-serine accumulation in the WT. Therefore, the strains WT(pZ1serA)(pEC-T18mob2) and WT(pZ1serAΔ197)(pEC-T18mob2) were grown in minimal medium with 220 mM glucose as the carbon source and the l-serine concentration in the culture medium was determined (Table 2). However, neither the overexpression of mutant serAΔ197 nor the WT allele yielded significant l-serine concentrations. Enzyme assays confirmed, for WT(pZ1serA) (pEC-T18mob2), a specific PGDH activity of 700 nmol (min mg)−1 and for WT(pZ1serAΔ197) (pEC-T18mob2), a specific activity of 690 nmol (min mg)−1 equivalent to an 8- to 10-fold overexpression compared to the WT (30).
TABLE 2.
l-Serine accumulation of various C. glutamicum strains
| Strain |
l-Serine concn (mM)a at:
|
|
|---|---|---|
| 28 h | 54 h | |
| WT | <0.01 | <0.01 |
| WT(pZ1)(pEC-T18mob2) | <0.01 | <0.01 |
| WT(pZ1serA)(pEC-T18mob2) | <0.01 | 0.12 ± 0.10 |
| WT(pZ1serAΔ197)(pEC-T18mob2) | <0.01 | 0.08 ± 0.05 |
| WT(pZ1)(pEC-T18mob2serCB) | <0.01 | 0.04 ± 0.03 |
| WT(pZ1serA)(pEC-T18mob2serCB) | <0.01 | 0.11 ± 0.03 |
| WT(pZ1serAΔ197)(pEC-T18mob2serCB) | <0.01 | 0.05 ± 0.02 |
| WT ΔsdaA(pZ1)(pEC-T18mob2) | 0.08 ± 0.03 | 0.05 ± 0.01 |
| WT ΔsdaA(pZ1serA) (pEC-T18mob2serCB) | 0.09 ± 0.01 | 0.15 ± 0.01 |
| WT ΔsdaA(pZ1serAΔ197) (pEC-T18mob2serCB) | 0.44 ± 0.28 | 0.14 ± 0.01 |
l-Serine accumulation was determined on minimal medium CGXII with 220mM glucose in two or three independent experiments.
In order to test whether the additional expression of serC and serB or even their expression alone resulted in l-serine accumulation, the respective strains WT(pZ1)(pEC-T18mob2serCB), WT(pZ1serA)(pEC-T18mob2serCB), and WT(pZ1serAΔ197)(pEC-T18mob2serCB) were constructed. Surprisingly, also with these strains, no substantial l-serine accumulation occurred (Table 2).
Influence of deletion of sdaA on l-serine accumulation.
Based on the result that the overexpression of the serine biosynthetic genes is not sufficient for l-serine production and our previous observation of a significant contribution of sdaA-encoded l-serine dehydratase (l-SerDH) to l-serine degradation in C. glutamicum (24), we used the wild-type derivative containing the sdaA deletion (WTΔsdaA) to assay for the influence of the overexpression of the l-serine biosynthetic genes serA, serAΔ197, serC, and serB on l-serine accumulation in this background (Table 2). The control strain WTΔsdaA(pZ1)(pEC-T18mob2) accumulated 0.08 mM l-serine after 28 h of cultivation and <0.05 mM after 54 h, showing that at the early time point, sdaA deletion alone resulted in traces of l-serine. C. glutamicum WTΔsdaA(pZ1serA)(pEC-T18mob2serCB) accumulated 0.09 mM l-serine after 24 h and up to 0.15 mM after 54 h, showing a slight increase in l-serine accumulation compared to the control. However, with strain WTΔsdaA(pZ1serAΔ197)(pEC-T18mob2serCB), 0.44 mM (28 h) and 0.14 mM (54 h) l-serine concentrations were determined. This comparison shows an advantage of the serAΔ197 allele over serA but that, despite the deletion of sdaA, degradation of l-serine is still occurring.
Influence of reduced serine hydroxymethyltransferase activity on l-serine accumulation.
In growing C. glutamicum, only 16% of the l-serine synthesized is incorporated into protein (21) whereas the remainder is cleaved by serine hydroxymethyltransferase (SHMT; glyA) to provide 5,10-methylene tetrahydrofolate and glycine. A reduced activity of SHMT was already shown to be favorable for l-threonine production with C. glutamicum due to an l-threonine-degrading side activity of the enzyme. Since the glyA gene could not be deleted or disrupted in C. glutamicum, even when supplemented with glycine (33), plasmid pK18mobglyA′ was employed to reduce the SHMT activity by replacing the native glyA promoter with the IPTG-inducible tac promoter (33). We used strain WT::pK18mobglyA′ to analyze the influence of a reduced SHMT activity on l-serine production. In the first experiments, this strain already accumulated up to 1 mM l-serine (not shown), illustrating the principal importance of SHMT reduction for l-serine accumulation. In order to overexpress serAΔ197 together with serC and serB in the kanamycin-resistant strain WT::pK18mobglyA′, all three genes were cloned into vector pEC-T18mob2 (35) to generate pEC-T18serAΔ197CB (see Materials and Methods). Using this tetracycline resistance-conferring plasmid, strain WT::pK18mobglyA′(pEC-T18serAΔ197CB) was generated.
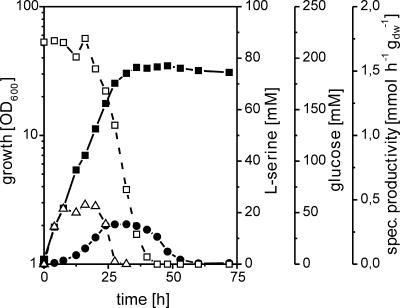
This strain was cultivated with or without 100 μM IPTG in minimal medium CGXII containing 220 mM glucose as the carbon source. Whereas in the presence of IPTG, the SHMT activity was 40 nmol min−1 mg (protein)−1, it was 10 nmol min−1 mg (protein)−1 without IPTG, confirming the successful application of pK18mobglyA′. With IPTG, l-serine accumulation was below 1 mM (not shown), but in the absence of the inducer, up to 16 mM l-serine accumulated (Fig. 2). Rate calculations showed that constant specific productivities of about 0.4 mmol h−1 g (dry weight)−1 occurred within 8 to 20 h of the cultivation. However, almost all l-serine was degraded again, which is consistent with our prior finding of the strong utilization of externally added l-serine by C. glutamicum unless l-SerDH (sdaA) was deleted (24).
FIG. 2.
Growth (▪), glucose (□) and l-serine (•) concentrations in the medium, and specific l-serine productivity (▵) of strain WT::pK18mobglyA′(pEC-T18mob2serAΔ197CB) on minimal medium with 220 mM glucose. OD600, optical density at 600 nm; dw, dry weight.
Influence of combining the deletion of sdaA with reduced SHMT activity on growth and l-serine accumulation.
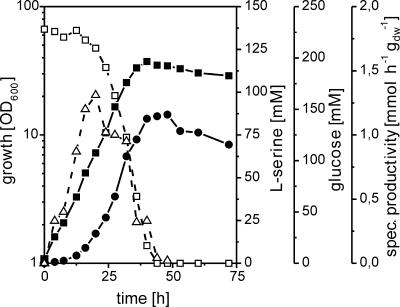
We used plasmid pK18mobglyA′ to exchange the native glyA promoter with the tac promoter in strain WTΔsdaA as well as plasmid pEC-T18mob2serAΔ197CB to overexpress the l-serine biosynthesis genes. The resulting strain, WTΔsdaA::pK18mobglyA′(pEC-T18mob2serAΔ197CB), was cultivated on CGXII medium with 220 mM glucose as the carbon source. In order to reduce glyA expression, IPTG was omitted. Enzyme activity determinations confirmed the expected low SHMT activity (not shown). A typical cultivation profile is shown in Fig. 3. l-Serine accumulated up to about 86 mM in the culture medium, with a maximum specific productivity of 1.2 mmol h−1 g (dry weight)−1 at about 20 h and a molar yield (YP/S) of 0.64 mol/mol. This confirmed the positive effect of sdaA deletion, which was also visible, although at a drastically reduced level when just the l-serine biosynthesis genes were overexpressed, without reducing SHMT activity (see above). Despite the efficient l-serine production with strain WTΔsdaA::pK18mobglyA′(pEC-T18mob2serAΔ197CB), there was still significant degradation of l-serine at later time points (Fig. 3). A comparison of the growth rates of the different genetically modified strains with that of the wild type revealed that overexpression of the genes serAΔ197, serC, and serB alone or in combination with a deletion of the sdaA gene resulted in a decreased growth rate (Table 3). The largest reduction of growth rate was observed as a consequence of reduced glyA expression. The strains WT::pK18mobglyA′(pEC-T18mob2serAΔ197CB) and WTΔsdaA::pK18mobglyA′(pEC-T18mob2serAΔ197CB) exhibited three- to fourfold-decreased growth rates under production conditions compared to the wild type and a twofold rate compared to the respective strains without reduced glyA expression (Table 3). This corroborates our previous finding that a reduced SHMT activity correlates with a reduced growth rate (33).
FIG. 3.
Growth (▪), glucose (□) and l-serine (•) concentrations in the medium, and specific l-serine productivity (▵) of strain WTΔsdaA::pK18mobglyA′(pEC-T18mob2serAΔ197CB) on minimal medium with 220 mM glucose. OD600, optical density at 600 nm; dw, dry weight.
TABLE 3.
Comparison of l-serine fluxesa in recombinant C. glutamicum strains
| Characteristics | WT | WT (pserA)(pserCB)b | WTΔsdaA (pserA)(pserCB)b | WT::pK18mobglyA′ (pserACB)c | WTΔsdaA::pK18mobglyA′ (pserACB)c |
|---|---|---|---|---|---|
| μmaxd (h−1) | 0.38 | 0.26 | 0.21 | 0.11 | 0.11 |
| Serine flux for cellular demand | 9.50 | 6.5 | 5.25 | 2.75 | 2.75 |
| Serine excretion flux | 0 | 0 | 0 | 10 | 21 |
| Sum of serine fluxes | 9.5 | 6.5 | 5.25 | 12.75 | 23.75 |
Fluxes are the maximal fluxes observed in nmol min−1 mg (dry weight)−1.
(pserA)(pserCB) stands for (pZ1serAΔ197)(pEC-T18mob2serCB).
pserACB stands for (pEC-T18mob2serAΔ197CB).
μmax, maximum growth rate.
DISCUSSION
Our functional studies identified the PSAT (serC) and PSP (serB) of Corynebacterium glutamicum. Although there are two further open reading frames (NCgl0400, NCgl0294) annotated as PSP in the genome of C. glutamicum, only NCgl2436 encodes a functional PSP. PSP of C. glutamicum contains 433 amino acyl residues, and its N terminus is extended by 93, 198, and 190 residues compared to the PSP proteins from E. coli, Methanococcus jannaschii, and humans, respectively (25, 28, 37). Notably, the additional N-terminal segment of C. glutamicum PSP includes a domain with similarity to an “ACT domain” that has been found in a number of proteins, including PGDH of E. coli, Mycobacterium tuberculosis, and C. glutamicum (2, 5, 30). This domain is proposed to represent a conserved regulatory ligand binding fold, but experimental evidence for PSP is absent. The N-terminal extension as characteristic for the C. glutamicum PSP is also present in that of M. tuberculosis (Rv3042c) and in PSPs of other Actinomycetales. On the other hand, PSAT of C. glutamicum has high identity (61%) to the protein of M. tuberculosis (Rv0884c) but only reduced identity (20%) to that of E. coli (14). The structural differences between PSAT from C. glutamicum and E. coli might be due to a second enzymatic function which is present only in the E. coli protein and which is involved in pyridoxal-5′-phosphate synthesis. These differences are also apparent from the fact that only the N-terminal part of the C. glutamicum PSAT protein, but the entire E. coli PSAT protein, is classified as an aminotransferase class V (pfam00266).
The wild-type serA derivative and the serAΔ197 allele were overexpressed either alone or in combination with the genes serB and serC, but l-serine accumulated only in traces not exceeding 0.1 mM. This distinguishes l-serine accumulation from, for instance, l-lysine formation by C. glutamicum, where deregulation of the initial enzyme of the pathway already results in substantial l-lysine accumulation of around 40 mM (4), indicating that intracellular l-serine turnover precludes its production. This is corroborated by our findings that l-serine production was high only when glyA expression was reduced and l-serine dehydratase activity absent. The corresponding strain WTΔsdaA::pK18mobglyA′(pEC-T18mob2serAΔ197CB) accumulated l-serine up to 86 mM, which is in the same range as that observed for l-threonine accumulation with a respective threonine-producing C. glutamicum strain (33). Moreover, this strain produced a yield of 0.64 mol l-serine per mol glucose, which is about two- to sixfold higher than that obtained with processes where glycine or glycine plus methanol were used as substrates (16, 19). However, the reduced glyA expression in the l-serine-producing strain led to a slower growth rate and a lower final optical density compared to a strain with native glyA expression likely due to perturbation of the C-1 metabolism. Moreover, since the deletion of sdaA alone or in combination with overexpression of the l-serine biosynthetic genes did not result in an appreciable l-serine accumulation, it is inconclusive whether a significant intracellular flux increase due to the overexpression of the serA alleles together with serB and serC is present. These results denote that the l-serine pathway is a rather unusual amino acid biosynthesis pathway. This idea is supported by the atypical PGDH (serA) inhibition of C. glutamicum (30) and the facts that the equilibrium of the PGDH-catalyzed reaction is on the substrate side (34) and an ACT domain is present in PSP (serB).
Nevertheless, reduction of the glyA-encoded SHMT activity had clearly the major impact on l-serine accumulation. Already, the reduction of glyA expression alone resulted in an approximate 1 mM accumulation of l-serine (not shown), which was not the case upon sdaA deletion. The importance of reduced SHMT activity is also evident when comparing the maximal fluxes rates (Table 3). With SHMT reduction and overexpression of the biosynthesis pathway genes, the growth rate was 0.11 h−1. The calculated flux over the pathway to satisfy the need for cellular synthesis, like phospholipid synthesis and C1 generation under these conditions, is 2.75 nmol min−1 mg (dry weight)−1 (22). Taking a maximal l-serine excretion rate of 10 nmol min−1 mg (dry weight)−1 into account (Fig. 2), a total l-serine flux of 12.75 nmol min−1 mg (dry weight)−1 resulted (Table 3). Importantly, the comparison of this strain with the WT and WTDsdaA (pEC-T18mob2serAΔ197CB) illustrates that reducing the l-serine degradation to glycine and C1 units favors an increased total l-serine flux, indicative of a stimulation of the l-serine synthesis probably due to a reduced availability of glycine and C1 units (Table 3). The strong increase in l-serine flux by 11.02 to 23.75 nmol min−1 mg (dry weight)−1 due to the additional sdaA deletion in WTDsdaA::pK18mobglyA′ (pEC-T18mob2serAΔ197CB) is largely in agreement with the difference in the l-serine degradation rates observed for the wild type and its sdaA deletion mutant with externally added l-serine where the sdaA deletion caused a decrease in l-serine degradation by 7.7 nmol min−1 mg (dry weight)−1 (24). This work demonstrates that engineering l-serine production from glucose requires considering the position of l-serine in metabolism instead of considering l-serine as an end product of a biosynthetic pathway.
Acknowledgments
We thank A. Schmitz and S. Knebel for technical assistance and V. F. Wendisch for fruitful discussions and critical reading of the manuscript.
This work was supported in part by the Deutsche Bundesstiftung Umwelt (DBU AZ13037 and AZ13089).
REFERENCES
- 1.Ankri, S., I. Serebrijski, O. Reyes, and G. Leblon. 1996. Mutations in the Corynebacterium glutamicum proline biosynthetic pathway: a natural bypass of the proA step. J. Bacteriol. 178:4412-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chipman, D. M., and B. Shaanan. 2001. The ACT domain family. Curr. Opin. Struct. Biol. 11:694-700. [DOI] [PubMed] [Google Scholar]
- 3.Cho, H., W. Wang, R. Kim, H. Yokota, S. Damo, S. H. Kim, D. Wemmer, S. Kustu, and D. Yan. 2001. BeF3− acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF3− complex with phosphoserine phosphatase. Proc. Natl. Acad. Sci. USA 98:8525-8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cremer, J., L. Eggeling, and H. Sahm. 1991. Control of the lysine biosynthesis sequence in Corynebacterium glutamicum as analyzed by overexpression of the individual corresponding genes. Appl. Environ. Microbiol. 57:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dey, S., Z. Hu, X. L. Xu, J. C. Sacchettini, and G. A. Grant. 2005. d-3-Phosphoglycerate dehydrogenase from Mycobacterium tuberculosis is a link between the Escherichia coli and mammalian enzymes. J. Biol. Chem. 280:14892-14899. [DOI] [PubMed] [Google Scholar]
- 6.Drauz, K., B. Hoppe, A. Kleemann, H.-P. Krimmer, W. Leuchtenberger, and C. Weckbecker. 2003. Amino acids. In M. Bohnet, C. J. Brinker, and B. Cornils (ed.), Ullmann's encyclopedia of industrial chemistry. Wiley-VCH Verlag GmbH, Weinheim, Germany.
- 7.Eggeling, L., and H. Sahm. 2003. New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch. Microbiol. 180:155-160. [DOI] [PubMed] [Google Scholar]
- 8.Eggeling, L., S. Morbach, and H. Sahm. 1997. The fruits of molecular physiology: engineering the l-isoleucine biosynthesis pathway in Corynebacterium glutamicum. J. Biotechnol. 56:167-182. [Google Scholar]
- 9.Eggeling, L., W. Pfefferle, and H. Sahm. 2001. Amino acids, p. 281-303. In C. Ratledge and B. Kristiansen (ed.), Basic biotechnology. Cambridge University Press, Cambridge, England.
- 10.Eikmanns, B. J., N. Thum-Schmitz, L. Eggeling, K. Lüdtke, and H. Sahm. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817-1828. [DOI] [PubMed] [Google Scholar]
- 11.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1985. Techniques for transformation of Escherichia coli, p. 109-136. In D. M. Glover (ed.), DNA cloning, vol. 1. IRL Press, Oxford, United Kingdom. [Google Scholar]
- 13.Hermann, T. 2003. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 104:155-172. [DOI] [PubMed] [Google Scholar]
- 14.Hester, G., W. Stark, M. Moser, J. Kallen, Z. Markovic-Housley, and J. N. Jansonius. 1999. Crystal structure of phosphoserine aminotransferase from Escherichia coli at 2.3 Å resolution: comparison of the unligated enzyme and a complex with alpha-methyl-l-glutamate. J. Mol. Biol. 286:829-850. [DOI] [PubMed] [Google Scholar]
- 15.Hsiao, H.-Y., and T. Wei. 1985. Enzymatic production of l-serine with a feedback control system for formaldehyde addition. Biotechnol. Bioeng. 28:1510-1518. [DOI] [PubMed] [Google Scholar]
- 16.Izumi, Y., T. Yoshida, S. S. Miyazaki, T. Mitsunaga, T. Ohshiro, M. Shimao, A. Miyata, and T. Tanabe. 1993. l-Serine production by a methylotroph and its related enzymes. Appl. Microbiol. Biotechnol. 39:427-432. [DOI] [PubMed] [Google Scholar]
- 17.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennerknecht, N., H. Sahm, M. R. Yen, M. Patek, M. H. Saier, Jr., and L. Eggeling. 2002. Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J. Bacteriol. 184:3947-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubota, K., and K. Yokozeki. 1989. Production of l-serine from glycine by Corynebacterium glycinophilum and properties of serine hydroxymethyltransferase, a key enzyme in l-serine production. J. Ferment. Bioeng. 67:387-390. [Google Scholar]
- 20.Liebl, W., A. Bayerl, U. Stillner, and K. H. Schleifer. 1989. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol. Lett. 65:299-304. [DOI] [PubMed] [Google Scholar]
- 21.Marx, A., A. A. deGraaf, W. Wiechert, L. Eggeling, and H. Sahm. 1996. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol. Bioeng. 49:111-129. [DOI] [PubMed] [Google Scholar]
- 22.Marx, A., K. Striegel, A. A. deGraaf, H. Sahm, and L. Eggeling. 1997. Response of the central metabolism of Corynebacterium glutamicum to different flux burdens. Biotechnol. Bioeng. 56:168-180. [DOI] [PubMed] [Google Scholar]
- 23.Marx, A., S. Hans, B. Möckel, B. Bathe, A. A. de Graaf, A. C. McCormack, C. Stapleton, K. Burke, M. O'Donohue, and L. K. Dunican. 2003. Metabolic phenotype of phosphoglucose isomerase mutants of Corynebacterium glutamicum. J. Biotechnol. 104:185-197. [DOI] [PubMed] [Google Scholar]
- 24.Netzer, R., P. Peters-Wendisch, L. Eggeling, and H. Sahm. 2004. Cometabolism of a nongrowth substrate: l-serine utilization by Corynebacterium glutamicum. Appl. Environ. Microbiol. 70:7148-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuwald, A. F., and G. V. Stauffer. 1985. DNA sequence and characterization of the Escherichia coli serB gene. Nucleic Acids Res. 13:7025-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 27.Palmer, B. R., and M. G. Marinus. 1994. The dam and dcm strains of Escherichia coli: a review. Gene 143:1-12. [DOI] [PubMed] [Google Scholar]
- 28.Peeraer, Y., A. Rabijns, C. Verboven, J. F. Collet, E. Van Schaftingen, and C. De Ranter. 2003. High-resolution structure of human phosphoserine phosphatase in open conformation. Acta Crystallogr. Sect. D Biol. Crystallogr. 59:971-977. [DOI] [PubMed] [Google Scholar]
- 29.Peters-Wendisch, P., B. Schiel, V. F. Wendisch, E. Katsoulidis, B. Möckel, H. Sahm, and B. J. Eikmanns. 2001. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3:295-300. [PubMed] [Google Scholar]
- 30.Peters-Wendisch, P., R. Netzer, L. Eggeling, and H. Sahm. 2002. 3-Phosphoglycerate dehydrogenase from Corynebacterium glutamicum: the C-terminal domain is not essential for activity but is required for inhibition by l-serine. Appl. Microbiol. Biotechnol. 60:437-441. [DOI] [PubMed] [Google Scholar]
- 31.Riedel, C., D. Rittmann, P. Dangel, B. Möckel, S. Petersen, H. Sahm, and B. J. Eikmanns. 2001. Characterization of the phosphoenolpyruvate carboxykinase gene from Corynebacterium glutamicum and significance of the enzyme for growth and amino acid production. J. Mol. Microbiol. Biotechnol. 3:573-583. [PubMed] [Google Scholar]
- 32.Simic, P., H. Sahm, and L. Eggeling. 2001. l-Threonine export: use of peptides to identify a new translocator from Corynebacterium glutamicum. J. Bacteriol. 183:5317-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simic, P., J. Willuhn, H. Sahm, and L. Eggeling. 2002. Identification of glyA (encoding serine hydroxymethyltransferase) and its use together with the exporter ThrE to increase l-threonine accumulation by Corynebacterium glutamicum. Appl. Environ. Microbiol. 68:3321-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugimoto, E., and L. I. Pizer. 1968. The mechanism of end product inhibition of serine biosynthesis. I. Purification and kinetics of phosphoglycerate dehydrogenase. J. Biol. Chem. 243:2081-2089. [PubMed] [Google Scholar]
- 35.Tauch, A., O. Kirchner, B. Löffler, S. Gotker, A. Pühler, and J. Kalinowski. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45:362-367. [DOI] [PubMed] [Google Scholar]
- 36.van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 37.Wang, W., R. Kim, J. Jancarik, H. Yokota, and S. H. Kim. 2001. Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution. Structure 9:65-71. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida, H., and K. Nakayama. 1974. Production of l-serine analog-resistant mutants from various bacteria and the effect of l-threonine and l-homoserine on the production of l-serine. J. Agric. Chem. Soc. Jpn. 48:201-208. [Google Scholar]