Abstract
Lactobacillus sobrius sp. nov., which was recently isolated from the intestine of weaning piglets, has potential probiotic properties. To follow the fate of L. sobrius strain 001T in dietary interventions, a novel and strain-specific quantitative detection procedure was developed. This procedure was based on the isolation of specific genomic fragments from the type strain by representational difference analysis and their detection by real-time PCR. The described strain-specific quantification approach may be used in studies aimed at tracking bacterial strains added to specific environments.
Lactobacilli are widely used for the production of fermented foods and feeds and are marketed as probiotics for humans and farm animals (3, 13, 14). Recently, Lactobacillus sobrius sp. nov. has been identified as an abundant member of the porcine intestinal microbiota (4, 5). Moreover, in vitro experiments demonstrated that L. sobrius 001T exerted a significant protective effect against intestinal damages caused by enterotoxigenic Escherichia coli K88 (4, 10). The need for in vivo studies prompted the development of a strain-specific quantitative approach to monitor L. sobrius 001T given via diet to weaning piglets separately from the indigenous populations present prior to the intervention. Discrimination between different strains of L. sobrius solely on the basis of the phenotypic characteristics or 16S rRNA gene-targeted phylogenetic analysis, however, is difficult. Such microdiversity has often been reported for many intestinal lactobacilli (9, 11). Hence, the identification by representational difference analysis (RDA) (6, 8) of different nucleic acid stretches among two otherwise highly similar genomes may well be a valuable approach toward the identification of closely related bacterial species and strains. RDA has been adapted recently to study the genomic diversity of bacterial strains belonging to a novel lineage of predominant soil Bacillus spp. (2). The potential of RDA to unravel genomic microdiversity of lactobacilli, however, has not been examined.
Here, we used L. sobrius sp. nov. strain 001T, a newly isolated abundant member of the porcine gastrointestinal tract community, for the development of a novel strain detection system. L. sobrius 001T strain-specific genomic fragments were first identified by RDA and were further detected by real- time PCR.
RDA was adapted to study the microdiversity between highly similar genomes of four strains of L. sobrius sp. nov. (L. sobrius 001T, L. sobrius 003, L. sobrius 004, and L. sobrius 005) (4). Lactobacillus acidophilus DSMZ20079T, Lactobacillus amylovorus DSMZ 20531T, Lactobacillus crispatus DSMZ 20584T, Lactobacillus gallinarum DSMZ 10532T and Lactobacillus helveticus DSMZ 20075T were used as reference strains. All Lactobacillus strains were propagated at 37°C, anaerobically, in deMan-Rogosa-Sharpe broth (Difco, Le Point de Claix, France). After 24 h of growth, the cells were harvested at 5,000 × g for 10 min and washed with filtered (0.2 μm pore size) phosphate-buffered saline (8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, and 0.24 g KH2PO4 [pH 7.2] per liter). The isolation of genomic DNA was done using a Fast DNA spin kit (Qbiogene, Inc., Carlsbad, CA).
The genomic DNA of all four L. sobrius isolates (approximately 1 μg) was digested with CfoI (Promega, Madison, WI) for 1.5 h at 37°C in the respective buffer provided by the manufacturer and in the presence of bovine serum albumin (BSA) (5 ng/μl). The total digestion reaction had a volume of 20 μl. The restriction enzyme recognizes a 4-bp sequence motif of high GC content (GCGC), which is present at a relatively low frequency in the DNA of low-GC gram-positive bacteria (2). The restriction reaction yielded the highest frequency of DNA fragments in the range of 0.7 to 3 kb (data not shown), which were further used to produce DRIVER and TESTER using the procedure previously described by Felske (2), with some modification. The ligation of DRIVER adapters was carried out by the following reaction: 8 μl of the digested DNA was mixed with 8 μl of DRIVER adapter (50 μM; 1:1 mixture of T7-long [THEBEN] with Short [KORINTH]) (Table 1), 1 μl (400 U) of T4 DNA ligase, 2 μl 10× ligase buffer, and 1 μl (25 μg ml−1) BSA to a final volume of 20 μl. The ligation mixture was incubated at 4°C for 16 h. Adapter residues were removed from the ligation reaction using a DNA Clean & Concentrator-5 kit (Zimo Research, Orange, CA). The DNA was eluted from the filter with 20 μl mQ water. The DRIVER amplification was performed with a Taq DNA polymerase kit from Life Technologies (Gaithersburg, MD). PCRs (50 μl) contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 300 μM concentrations of each deoxynucleoside triphosphate, a 0.8 μM concentration of primer T7, 2.5 units of recombinant Taq DNA polymerase, and 5 μl of template (ligation reaction). The samples were amplified in a thermocycler T1 Whatman Biometra starting with a predenaturation of 94°C for 60 s and then using 40 cycles of 94°C for 15 s, 44°C for 30 s, and 68°C for 90 s and a final elongation step of 4.5 min at 68°C. This PCR product was purified with a QIAquick PCR purification kit (Westburg, Leusden, The Netherlands), eluted with 50 μl of TE buffer and further used as the DRIVER. For TESTER production, 1 μl of DRIVER was digested with 10 U of CfoI for 1.5 h at 37°C in the respective buffer plus BSA (5 ng/μl) in a total volume of 20 μl. The digested DNA was purified using a DNA Clean & Concentrator-5 kit (Zimo Research) and eluted with mQ in a final volume of 20 μl. Ten microliters of this was amended with 7.5 μl TESTER adapter Sp6-long (SPARTA) and Short (KORINTH) (1:1 mixture of 50 μM each), and the ligation and removal of the adapter residues were achieved as described above for the DRIVER. The DNA was eluted from the filter with 50 μl mQ water, yielding the TESTER preparation. These concentrations resulted in a 100-fold DRIVER excess when the same volumes of DRIVER and TESTER were applied in the RDA reaction. Subsequently, the subtractive hybridization was performed by mixing 5 μl (each) of DRIVER and TESTER, and 10 μl of 5× EE buffer [50 mM N-(2-hydroxyethyl)piperazine-N′-(3-propanesulfonic acid) (EPPS) and 5 mM EDTA, pH 8.0] (12). The reaction was covered by a drop of mineral oil and denatured for 5 min at 99°C. Without a cooling step, 5 μl of a 5 M NaCl solution was added, and then the sample was hybridized at 67°C for 3 h.
TABLE 1.
DNA oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ to 3′) | Target | Reference |
|---|---|---|---|
| T7-long (THEBEN) | TTT CTA ATA CGA CTC ACT ATA GGC CGC CAG CG | RDA | BACREXa |
| SP6-long (SPARTA) | TTT ATT TAG GTG ACA TAG ATT AGG CCG CCA | RDA | BACREX |
| Short (KORINT) | CTG GCG GCC TAC CA | RDA | BACREX |
| T7-RDA primers | AAT ACG ACT CAC TAT AG | RDA | 2 |
| SP6-RDA primers | ATT TAG GTG ACA CTA TAG A | RDA | 2 |
| S-G-Lab-0159-a-A-2 | CGG TAT TAG CAC CTG TTT C | 16S rRNA | This study |
| L-*-OTU171-0077-a-S-2 | ACT TCG GTA ATG ACG TTG | 16S rRNA | This study |
| OTU171_RDA_F | TTC TGC CTT TTT GGG ATC AA | RDA fragment A | This study |
| OTU171_RDA_R | CCT TGT TTA TTC AAG TGG GTG A | RDA fragment A | This study |
| T7 | TAA TAC GAC TCA CTA TAG G | Promega | |
| Sp6 | GAT TTA GGT GAC ACT ATA G | Promega |
BACREX homepage, http://mik.gbf.de/bacrex. European Communities project QLK3-2000-01678.
The PCRs contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 300 μM concentrations of each deoxynucleoside triphosphate, and 2.5 units of recombinant Taq DNA polymerase and were prewarmed to 68°C in a thermocycler T1 Whatman Biometra. Five microliters of the hybridization reaction was then directly transferred from the 67°C heating block into the 40-μl prewarmed PCR mix. The RDA amplification was performed with a preincubation of 1 min at 68°C; the addition of 5 μl (1 μM) primer SP6 (during a pause of 3 min at 4°C); 45 cycles of 94°C for 15 s, 44°C for 30 s, and 68°C for 90 s; and a final incubation for 4.5 min at 68°C.
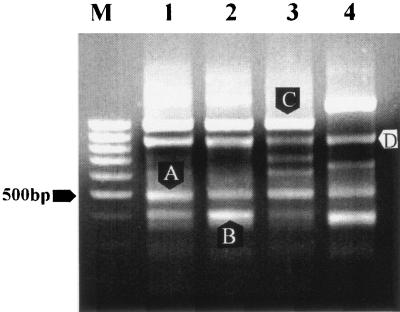
In this study, L. sobrius strain 001T was used as the TESTER and was mixed separately with 100-fold excess DNA from each of the related strains used as the DRIVERs (L. sobrius strains 003, 004, and 005). The resulting subtractive hybridization reactions between the TESTER and DRIVERs aimed to eliminate DNA fragments shared between the strains. The SP6 primer complements required for PCR amplification are present on TESTER homoduplexes only after subtractive hybridization, allowing for the amplification of fragments unique to the TESTER. Figure 1 displays the RDA fragments obtained after PCR with SP6 primer (0.4 to 3 kb), comparing L. sobrius strain 001T with three highly related strains. Identical DRIVERs were found to yield the same RDA patterns (lane 1 and 2), suggesting minor tube-to-tube variation. Subsequently, the PCR fragments were isolated from gel, reamplified, and cloned, and four of the fragments were sequenced as previously described and compared to sequences available in public databases by using BLASTx analysis (5).
FIG. 1.
RDA analysis of L. sobrius strain 001T and other L. sobrius strains. Subtractive hybridization between L. sobrius strain 001T (TESTER) and the following strains: lanes 1 and 2, L. sobrius strain 003 (DRIVER); lane 3, L. sobrius strain 004 (DRIVER); lane 4, L. sobrius strain 005 (DRIVER). RDA fragments identified after sequencing analysis and BLASTx were as follows: A, a 500-bp fragment predicted to code for a polypeptide with highest similarity (29% at the AA level) to LBA0036, a hypothetical protein from L. acidophilus (1); B, a 400-bp fragment predicted to code for a polypeptide with highest similarity (70% at the AA level) to LBA0860, a hypothetical protein from L. acidophilus (1); C, a 950-bp fragment predicted to code for a polypeptide with highest similarity (41% at the AA level) to E. coli putative cytoplasmic protein (gi 32470041); and D, a 850-bp fragment predicted to code for a polypeptide with high similarity (95% at the AA level) to A. camphorata manganese superoxide dismutase (gi 33186704). M, standard 100-bp DNA ladder.
The predicted translation products of two of the sequences (Fig. 1, A and B) were found to show similarity with the hypothetical proteins LBA0036 (29% at the amino acid [AA] level) and LBA0860 (70% at the AA level) of L. acidophilus NCFM (1). The other two RDA fragments (Fig. 1, C and D) were predicted to code for proteins most closely related to the E. coli putative cytoplasmic protein (41% at the AA level; gi 32470041) and Antrodia camphorata manganese superoxide dismutase (95% at the AA level; gi 33186704), respectively. The genomic fragments, identified by RDA, were further used as a target for the strain-specific PCR detection and quantification of L. sobrius 001T. Primers were developed for each RDA fragment using the Primer3 program (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). First, we tested the strain-specific primers with L. sobrius strain 001T genomic DNA as the template. Efficient amplification of genomic DNA of L. sobrius strain 001T resulting in unique amplicons with the expected size was obtained only with theprimer set targeting RDA fragment A (data not shown). Therefore, this primer set was further used for strain-specific L. sobrius 001T PCR detection (Table 1). In addition, species-specific primers targeting the 16S rRNA gene of L. sobrius sp. nov. were designed using the ARB software package (7).
Real-time PCR was performed with an iCycler IQ real-time detection system associated with the iCycler optical system interface software version 2.3 (Bio-Rad, Veenendaal, The Netherlands). A reaction mixture (25 μl) consisted of 12.5 μl of IQ SYBR Green Supermix (Bio-Rad), 0.2 μM concentrations of each primer set, and 5 μl of the template DNA. The PCR conditions were an initial DNA denaturation step at 95°C for 3 min, 40 cycles of denaturation at 95°C for 15 s, and primer annealing and extension at 53.9 to 63.9°C for 45 s. After optimization as described below, annealing and extension temperatures were 60.3°C and 62.5°C for the strain- and species-specific primers, respectively. Conditions for the two primer sets were first optimized using temperature gradient PCR, with amplification efficiency, linearity, and specificity as criteria. The efficiencies of amplification, calculated from the formula Eff(n) = [10(−1/slope) − 1], were between 95 and 99% for the 16S rRNA gene-targeting primers and between 96 and 103% for the primers amplifying the RDA fragment. The linear range of amplification for both primer sets comprised DNA aliquots equivalent to dilutions of from 107 to 10 cells per PCR (data not shown). In addition, the real-time PCR detection level was tested using genomic DNA isolated from a 10-fold dilution series of a L. sobrius strain 001T cell suspension. No amplification was observed when the concentration of the DNA was below 100 cell equivalents per PCR. The 10-fold difference between the two approaches suggested a significant effect of the DNA isolation on the PCR detection limit. The data concur with a previous report that DNA isolation protocols affect the detection limit of PCR approaches (15). The specificity of the primer sets was further optimized with closely related Lactobacillus sp. reference strains and, in the case of RDA fragment A targeting primers, also with the other L. sobrius strains.
PCR primers targeting RDA fragment A were found to confer strain-specific L. sobrius 001T detection, as demonstrated by real-time PCR using genomic DNA of L. sobrius strain 001T and other nontarget L. sobrius strains and Lactobacillus spp. The specificity was also not influenced by the size of the ratio between the target and nontarget DNA, as confirmed in a detection experiment using different ratios, varying from 1 to 1,000, between the L. sobrius 001T and L. sobrius 003 genomic DNA (data not shown).
Species-specific primers targeting the 16S rRNA gene of L. sobrius sp. nov. did not amplify the 16S rRNA gene of other Lactobacillus reference strains, except for L. amylovorus (DSMZ 20531T), as was expected due to identical target sequences. Melt curve analysis was used to detect the formation of the correct amplification product. For the L. sobrius 16S rRNA targeting primer set, primer dimers with lower Tm values were observed when small amounts of diluted DNA (up to 10 cell equivalents) were used in PCRs. The design of improved 16S rRNA-targeted primer pairs that would allow for the discrimination between L. sobrius sp. nov. and L. amylovorus, however, was hindered by the overall sequence identity, which was above 97%.
As the PCR detection of a specific RDA fragment provided a high degree of resolution between related Lactobacillus species and strains, we focused on this approach rather than on using 16S rRNA as a target, and we tested the strain-specific detection in vivo. Four porcine ileal lumen samples, obtained from piglets after a large feeding trial (4), were spiked with different concentrations of L. sobrius strain 001T (107 to 103 cells/ml). The isolation of genomic DNA was done using a Fast DNA spin kit (Qbiogene). In samples spiked with L. sobrius strain 001T in concentrations of 1 × 103, 1 × 105, and 1 × 107 cells/ml, real-time PCR detection yielded 1.1 × 103 ± 0.4, 1.2 × 105 ± 0.5, and 1.1 × 107 ± 0.2 (mean count ± standard deviation for four samples), respectively, confirming that accurate enumeration of L. sobrius in animal samples can be achieved.
The RDA real-time PCR method reported here can be applied in further studies aiming to monitor the addition of specific bacterial strains to various environments. Together with other molecular approaches, including various genetic fingerprinting techniques, genetic marking, monoclonal antibodies assay, and antibiotic resistance markers, the RDA may also be employed for species identification or subspecies discrimination. The method might be used in the future as a cultivation-independent method for the quantification of microdiversity in vivo. Specifically, we anticipate that the RDA real-time PCR approach will provide reliable strain detection in the mammalian gastrointestinal tract.
Nucleotide sequence accession numbers.
The sequences reported in this study were deposited in GenBank under accession numbers AY899195 to AY899197.
Acknowledgments
This work has been carried out with financial support from the European Community specific RTD program “Quality of Life and Management of Living Resources,” research project HEALTHYPIGUT (QLK5-LT2000-00522).
Andreas D. Felske is acknowledged for providing excellent help regarding the application of RDA protocol.
REFERENCES
- 1.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felske, A. 2002. Streamlined representational differences analysis for comprehensive studies of numerous genomes. J. Microbiol. Methods 50:305-311. [DOI] [PubMed] [Google Scholar]
- 3.Klaenhammer, T. R., R. Barrangou, B. L. Buck, M. A. Azcarate-Peril, and E. Altermann. 2005. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 29:393-409. [DOI] [PubMed] [Google Scholar]
- 4.Konstantinov, R. S. 2005. Lactobacilli in the porcine intestine: from composition to functionality. Ph.D. thesis. Wageningen University, Wageningen, The Netherlands.
- 5.Konstantinov, S. R., A. Awati, H. Smidt, B. A. Williams, A. D. L. Akkermans, and W. M. de Vos. 2004. Specific response of a novel and abundant Lactobacillus amylovorus-like phylotype to dietary prebiotics in the guts of weaning piglets. Appl. Environ. Microbiol. 70:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lisitsyn, N. A., N. Lisitsyn, and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 7.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Middendorf, B., and R. Gross. 1999. Representational difference analysis identifies a strain-specific LPS biosynthesis locus in Bordetella spp. Mol. Gen. Genet. 262:189-198. [DOI] [PubMed] [Google Scholar]
- 9.Mukai, T., K. Arihara, A. Ikeda, K. Nomura, F. Suzuki, and H. Ohori. 2003. Lactobacillus kitasatonis sp. nov., from chicken intestine. Int. J. Syst. Evol. Microbiol. 53:2055-2059. [DOI] [PubMed] [Google Scholar]
- 10.Roselli, M., A. Finamore, S. R. Konstantinov, M. S. Britti, W. M. de Vos, H. Smidt, and E. Mengheri. 2005. A Lactobacillus amylovorus-like strain from the resident porcine gastrointestinal microbiota protects against intestinal damages promoted by enterotoxigenic Escherichia coli strain K88 in vitro, abstr. 69C. Abstr. Am. Soc. Microbiol. Conf. Beneficial Microbes, Lake Tahoe, Nev.
- 11.Satokari, R. M., E. E. Vaughan, H. Smidt, M. Saarela, J. Matto, and W. M. de Vos. 2003. Molecular approaches for the detection and identification of bifidobacteria and lactobacilli in the human gastrointestinal tract. Syst. Appl. Microbiol. 26:572-584. [DOI] [PubMed] [Google Scholar]
- 12.Straus, D., and F. M. Ausubel. 1990. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc. Natl. Acad. Sci. USA 87:1889-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tannock, G. W. 2004. A special fondness for lactobacilli. Appl. Environ. Microbiol. 70:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaughan, E. E., M. C. de Vries, E. G. Zoetendal, K. Ben-Amor, A. D. Akkermans, and W. M. de Vos. 2002. The intestinal LABs. Antonie Leeuwenhoek 82:341-352. [PubMed] [Google Scholar]
- 15.Zoetendal, E. G., K. Ben-Amor, A. D. Akkermans, T. Abee, and W. M. de Vos. 2001. DNA isolation protocols affect the detection limit of PCR approaches of bacteria in samples from the human gastrointestinal tract. Syst. Appl. Microbiol. 24:405-410. [DOI] [PubMed] [Google Scholar]