Abstract
Salinispora is the first obligate marine genus within the order Actinomycetales and a productive source of biologically active secondary metabolites. Despite a worldwide, tropical or subtropical distribution in marine sediments, only two Salinispora species have thus far been cultivated, suggesting limited species-level diversity. To further explore Salinispora diversity and distributions, the phylogenetic diversity of more than 350 strains isolated from sediments collected around the Bahamas was examined, including strains cultured using new enrichment methods. A culture-independent method, using a Salinispora-specific seminested PCR technique, was used to detect Salinispora from environmental DNA and estimate diversity. Overall, the 16S rRNA gene sequence diversity of cultured strains agreed well with that detected in the environmental clone libraries. Despite extensive effort, no new species level diversity was detected, and 97% of the 105 strains examined by restriction fragment length polymorphism belonged to one phylotype (S. arenicola). New intraspecific diversity was detected in the libraries, including an abundant new phylotype that has yet to be cultured, and a new depth record of 1,100 m was established for the genus. PCR-introduced error, primarily from Taq polymerase, significantly increased clone library sequence diversity and, if not masked from the analyses, would have led to an overestimation of total diversity. An environmental DNA extraction method specific for vegetative cells provided evidence for active actinomycete growth in marine sediments while indicating that a majority of sediment samples contained predominantly Salinispora spores at concentrations that could not be detected in environmental clone libraries. Challenges involved with the direct sequence-based detection of spore-forming microorganisms in environmental samples are discussed.
Bacteria belonging to the order Actinomycetales, commonly referred to as actinomycetes, perform significant biogeochemical roles in terrestrial soils and are highly valued for their unparalleled ability to produce biologically active secondary metabolites. These bacteria account for ca. 45% of the bioactive microbial metabolites discovered (3) and have played a central role in the development of the modern pharmaceutical industry. The immense biotechnological utility of actinomycetes has led to exhaustive surveys of cultivars from terrestrial habitats and an associated increase in the numbers of known compounds being rediscovered due to a high rate of redundancy in the strains isolated. Given recent advances in our understanding of marine actinomycetes (7, 13, 24; see also a recent issue of Antonie van Leeuwenhoek, vol. 87, 2005, devoted to the subject), strains isolated from the marine environment represent a relatively unexplored frontier for the discovery of new actinomycete biodiversity and a resource for novel secondary metabolites.
Recently, we reported the discovery of the first obligate marine actinomycete genus for which the name “Salinospora” was originally proposed (19) and subsequently revised to Salinispora (17). This new taxon belongs to the family Micromonosporaceae and is the source of novel secondary metabolites including salinosporamide A, a potent anticancer agent specifically targeting the 20S subunit of the mammalian proteasome (10). The discovery of this taxon provides clear evidence for the existence of autochthonous populations of marine actinomycetes, and the compounds being discovered from them indicate that cultured strains are an important resource for novel secondary metabolites.
Cultivation-based surveys have shown that Salinispora spp. occur at abundances of up to 104 CFU/ml of sediment and can be isolated from worldwide locations, including the Caribbean Sea, the Sea of Cortez, the Red Sea, the tropical Atlantic Ocean off The Bahamas, the tropical Pacific Ocean off Guam (13, 19), and from a sponge collected from the Great Barrier reef in Australia (15). Despite significant effort, we have yet to cultivate Salinispora strains from temperate Pacific or Arctic sediments, suggesting that their distribution may be restricted to tropical and subtropical latitudes. Polyphasic taxonomic studies of isolates obtained from various locations indicate that the cultivated diversity to date is restricted to the two species S. tropica and S. arenicola (17), while a third phylotype cultured from Palau is being examined to determine its taxonomic status. This low level of species diversity, as well as the historical difficulties associated with the cultivation of marine bacteria, raises the possibility that the full extent of Salinispora species diversity has yet to be realized.
The present study was designed to further explore Salinispora distributions and species diversity in marine sediments using selective cultivation methods together with cultivation-independent techniques. Our results revealed limited species-level diversity in sediments collected around the Bahamas, the cultivation of Salinispora from a record depth (1,100 m), and evidence that these bacteria are actively growing in some sediment samples while existing predominantly as spores in others. Special challenges involving cultivation-independent studies of spore-forming microorganisms are discussed.
MATERIALS AND METHODS
Sediment collection and processing.
During research expeditions in 2000, 2002, and 2003, a total of more than 200 marine sediment samples were collected around the islands of The Bahamas. Individual sediment samples were homogenized and portions used immediately for cultivation experiments with the remainder frozen at −20°C (−80°C upon return to the laboratory) for subsequent DNA extraction and enrichment cultivation. Sediment samples ranged from fine carbonate muds to coral rubble and were collected using SCUBA or a modified, surface deployed sampler (model #214WA110; Kahlsico, El Cajon, CA) to depths of 1,100 m. Sediment samples yielding 2 × 103 to 5 × 103 Salinispora CFU/ml, determined by using previously described methods (17), were considered for subsequent DNA extraction. From this sample pool, 13 sediment samples representing various geographical locations, depths, and sampling dates (Table 1) were chosen for additional cultivation-dependent and cultivation-independent experiments.
TABLE 1.
Sediment samples and Salinispora-specific PCR amplification results
| Samplea | Depth (m) | Location | Environmental DNAb,c | Enrichment culture DNAb,d |
|---|---|---|---|---|
| BA00-11 | 30 | Little San Salvador | + | NT |
| BA00-12 | 22 | Little San Salvador | − | NT |
| BA00-13 | 10 | Little San Salvador | − | NT |
| BA00-14 | 1 | Little San Salvador | − | NT |
| BA02-35 | 1,025 | Little San Salvador | − | + |
| BA02-36 | 1,100 | Little San Salvador | + | + |
| BA02-47 | 845 | Long Island | − | + |
| BA02-50 | 7 | Long Island | − | + |
| BA02-60 | 18 | Stirrup Cay | − | + |
| BA03-03 | 20 | Sweetings Cay | − | + |
| BA03-58 | 714 | Little San Salvador | − | + |
| BA03-63 | 2 | Little San Salvador | − | + |
| BA03-114 | 765 | Grand Bahama | − | + |
All samples were collected from the Bahamas (BA) in the years 2000 (BA00), 2002 (BA02), or 2003 (BA03).
In the nested PCR, all DNA samples yielded a primary amplification product using the primer set F270, RC1492. “+” and “−” symbols indicate secondary amplification results using the Salinispora-specific primer set F468, RC1492.
Identical amplification results were obtained using both mechanical and chemical lysis methods for DNA extraction. Clone libraries were prepared from the two samples that yielded positive, nested PCR amplification products.
Enrichment culture DNA was obtained using a chemical lysis method. NT, not tested. All enrichment cultures tested yielded a Salinispora-specific PCR product.
Selective actinomycete cultivation.
All sediment samples were processed in the field as soon as possible after collection by using desiccation and heat shock as selective cultivation methods (19). These methods were designed to reduce the numbers of gram-negative bacteria and to enrich for slow-growing, spore-forming actinomycetes. Treated samples were then inoculated onto medium M1 (1% starch, 0.4% yeast extract, 0.2% peptone, natural seawater, and 2% agar) or M5 (natural seawater and 2% agar) and incubated for 4 to 8 weeks at room temperature. Novobiocin (10 μg/ml) or rifampin (5 μg/ml) was added to reduce the number of unicellular bacteria. The antifungal agent cycloheximide (100 μg/ml) was added to all isolation media. Actinomycete colonies were recognized by the presence of branching, vegetative filaments and the formation of tough, leathery colonies that adhered to the agar surface. Hence, only mycelium-forming bacteria belonging to the order Actinomycetales were included in the present study. Morphologically diverse actinomycetes possessing Salinispora-like features (19) were repeatedly transferred on solid media until pure cultures were obtained. All pure strains were grown in M1 broth and cryopreserved at −80°C in 10% glycerol.
In addition to direct plating, enrichment methods were used in an attempt to cultivate new Salinispora diversity. These methods were designed based on Salinispora antibiotic resistances and the ability of actinomycetes to degrade recalcitrant carbon sources. Enrichment cultures were also used for PCR-based experiments designed to distinguish between the occurrence of Salinispora as spores and vegetative filaments in marine sediments. Enrichment cultures were prepared in 20-ml vials by adding 1.5 g of wet sediment (homogenized and previously frozen) to either 10 ml of seawater enriched with crude chitin (0.1% [wt/vol]), 10 ml of sediment extract (SE; autoclaved supernatant from a 0.5% [wt/vol] sediment-seawater solution), or 10 ml of medium M1low (0.2% starch, 0.08% yeast extract, 0.04% peptone, seawater). Each of the three enrichment conditions was supplemented with either 5 μg of rifampin/ml or 25 μg of novobiocin/ml (final concentrations). Similar enrichments were prepared in the field with 0.5 g sediment and one of the following antibiotics (final concentrations): kanamycin (20 μg/ml), novobiocin (10 μg/ml), vancomycin (5 μg/ml), gentamicin (2 μg/ml), or tetracycline (4 μg/ml). All enrichment cultures were incubated for 4 to 15 weeks at room temperature and observed at ×10 to ×64 magnification using a stereomicroscope. Bacteria that formed visible mycelia were harvested directly from the enrichment cultures by using a sterile pipette, serially washed three times in 10 ml of sterile seawater, and plated on medium M5. Colonies that morphologically resembled the genus Salinispora (19) were repeatedly transferred onto new media until pure cultures were obtained.
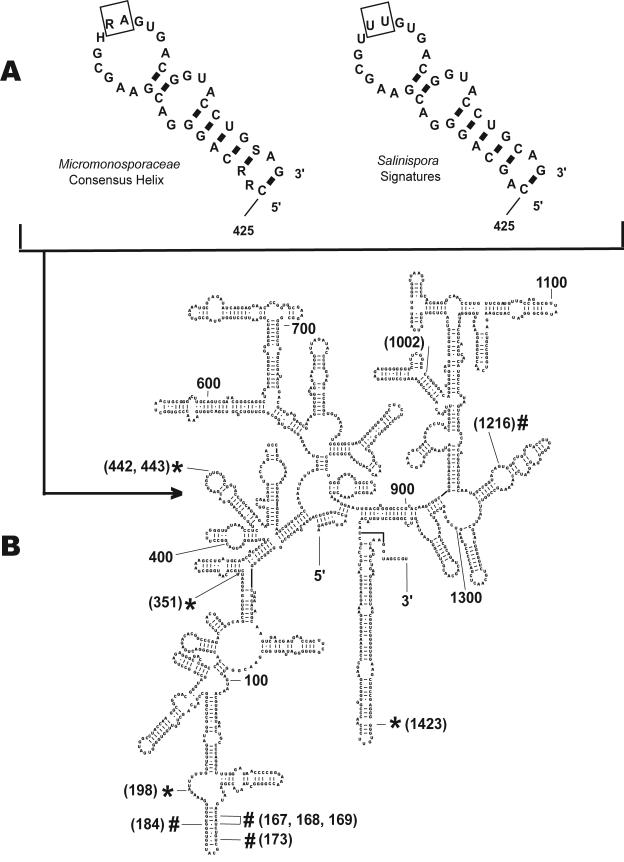
Taxonomic and phylogenetic assignment to the genus Salinispora was verified by the requirement of seawater for growth, successful PCR amplification from a genomic DNA template using Salinispora-specific 16S rRNA gene primers (F468 and RC1492, Table 2), restriction fragment length polymorphism (RFLP) analysis of PCR products and, in some cases, sequence analysis. Almost complete 16S rRNA gene sequences from cultivated isolates were obtained by using the forward primers FC27, F514, and F1114 and the reverse primers R530, R936, and RC1492 (Table 2). All DNA sequencing samples were submitted to the UCSD Cancer Center DNA Sequencing Shared Resource (3100 Genetic Analyzer; PE/Applied Biosystems). Upper- and lower-strand sequence contigs were assembled in MacClade version 4.03 (Sinauer Associates, Inc., Sunderland, MA) and base calling ambiguities resolved by reviewing the sequencing chromatograms in Edit View (version 1.0.1; Applied Biosystems, Foster City, CA). 16S rRNA gene sequences were aligned by using the Ribosomal Database Project (RDPII) Phylip interface (6). Related sequences obtained from an NCBI BLAST (blastn) search were aligned by using MacClade. Phylogenetic analyses were performed by using distance and parsimony criteria implemented in PAUP version 4.0b10 (Sinauer Associates). Bootstrapping was accomplished with 1,000 replicates by using heuristic search methods. Secondary structures of Salinispora 16S rRNA gene sequences (Fig. 1) were assigned by using Streptomyces coelicolor as a reference (5). When portions of the primary sequence differed significantly from that of S. coelicolor, short sections (50 to 100 nucleotides [nt]) were analyzed for the best-predicted secondary structure by using the Mfold program (30) with default settings.
TABLE 2.
Oligonucleotide primers used in this study
| Primera | Sequence (5′-3′) | Specificity | Source or reference |
|---|---|---|---|
| FC27* | AGAGTTTGATCCTGGCTCAG | High G+C gram positive | 18 |
| RC1492* | TACGGCTACCTTGTTACGACTT | High G+C gram positive | 18 |
| F270* | ATGAGCCCGCGGCCTA | Actinomycetales | This study |
| F468* | AGCAGGGACGAAGCGTTT | Salinispora | This study |
| F514 | GTGCCAGCAGCCGCGGTAA | Actinomycetales | 12 |
| F1114 | GCAACGAGCGCAACCC | Actinomycetales | 12 |
| R530 | CCGCGGCTGCTGGCACGTA | Actinomycetales | 12 |
| R936 | GTGCGGGCCCCCGTCAATT | Actinomycetales | 12 |
All primers are named after their respective 16S rRNA gene priming site (E. coli numbering). *, HPLC purified.
FIG.1.
Predicted secondary structures of Salinispora 16S rRNA. (A) Detail of the variable loop containing Salinispora-specific signature nucleotides (U, U) at positions 442 to 443 (467 to 468 by E. coli numbering) used to design the F468 primer. (B) Consensus secondary structure of S. arenicola strain CNH-721 (GenBank accession number AY878316) drawn using Streptomyces coelicolor as a reference (5). Helix loop structures in hypervariable regions were confirmed by using the Mfold program (29). Genus (*)- and species (#)-specific signatures are identified. Position 1002 is the site of the C→T transition observed in the uncultured S. arenicola phylotype III.
Environmental DNA extraction.
Two lysis methods (mechanical and chemical) were used to obtain environmental DNA and differentiate between the occurrence of actinomycetes as spores and vegetative cells in a given sample. Mechanical lysis was performed as follows: marine sediment samples (0.5 g) were placed in bead-beating FastDNA spin kit tubes for soil DNA extraction (Q-biogene, Carlsbad, CA) and macerated by using a FastPrep 120 bead-mill according to the manufacturer's protocol except that two cycles, rather than one, at a setting of 5.5 were performed with cooling on ice between intervals. Triplicate crude environmental DNA preparations from each sample were pooled and purified by using Chroma Spin TE 100 columns (Becton Dickinson Biosciences, Palo Alto, CA.) according to the manufacturer's protocol. Overall, this combination of extraction and purification yielded environmental DNA of high purity and molecular weight (2 to >20 kb). Purified samples were concentrated to 1 mg/ml in TE buffer (50 mM Tris, 10 mM EDTA [pH 8.0]) using Microcon YM-100 filters (Amicon, Beverley, MA) and stored at −20°C. Control experiments were performed to ensure that the mechanical lysis method was capable of extracting DNA from both S. tropica and S. arenicola spore preparations (detailed below). Spore lysis efficiency was determined by phase-contrast microscopy (×1,250), and DNA quality was monitored by gel electrophoresis.
A chemical lysis method to extract environmental DNA from vegetative cells, using conditions determined not to lyse spores, was adapted from previous methods (8). Briefly, triplicate 0.5-g aliquots of homogenized wet sediment from either frozen environmental samples or enrichment cultures were processed separately in 2-ml polyethylene tubes. Sediments were suspended in TE buffer, 1 mg of lysozyme/ml, and 0.2 mg of RNase/ml (all final concentrations) and incubated for 1 h at 37°C. sodium dodecyl sulfate was added to a final concentration of 1% (wt/vol), and samples were incubated for 1 h at 65°C. To clear the lysate of detergent and debris, 200 μl of chloroform and 200 μl of saturated potassium acetate were added, and the tubes were vortexed vigorously for 1 min and centrifuged at 14,000 × g. The aqueous top layer was transferred to a clean tube, and crude nucleic acids were precipitated with 1 volume isopropanol and resuspended in 50 μl of TE buffer. Triplicate crude DNA samples were pooled and purified by using Chroma Spin columns and concentrated by using Microcon filters as detailed above.
To ensure that the chemical lysis method did not disrupt Salinispora spores, the following control experiments were performed. Spores were harvested by using a sterile swab from Salinispora strains grown on M1 plates for 4 to 8 weeks. The spores were suspended in TE buffer and syringe filtered through a sterile cotton plug to remove contaminating vegetative filaments. Purified spores were then concentrated by brief centrifugation, quantified by using a hemocytometer, adjusted to 109 per ml, and then processed by using the chemical lysis method. Using a Pico Green DNA detection method, it was determined that no significant spore DNA was liberated compared to an untreated spore preparation. Pico Green assays were performed by using the microtiter plate protocol supplied by the manufacturer (Molecular Probes, Eugene, OR) and measured by using a Typhoon 8600 laser scanning system (Amersham Pharmacia Biotech, Piscataway, NJ). Similar control experiments performed using the mechanical lysis method were estimated to have lysis efficiencies of 50 to 70% and yielded good quality DNA from spore suspensions.
Seminested PCR and environmental library construction and analysis.
A seminested PCR method was developed to specifically amplify Salinispora 16S rRNA gene sequences from environmental DNA samples. The primary amplification using the forward primer F270, biased toward members of the order Actinomycetales, and the high G+C reverse bacterial primer RC1492 (Table 2) yielded a 1,250-bp product. Reactions contained the following final concentrations: 1× PCR buffer (Perkin-Elmer, Wellesley, MA), 2.5 mM MgCl2, 200 μM concentrations of each deoxynucleotide triphosphates, 0.05 U of AmpliTaq Gold (Perkin-Elmer)/μl, 0.1 μg of bovine serum albumin (Promega, Madison WI)/μl, 0.5 μM concentrations of each primer, and 100 ng of environmental DNA template per 25-μl reaction. Reaction mixtures were thermocycled as follows: there was an initial denaturation of 94°C for 10 min, followed by 23 cycles of denaturation at 94°C (15 s), annealing at 55°C (15 s), and extension at 72°C (1 min). Care was taken to assure that the primary PCR did not reach saturation by quantifying reaction products using the Pico Green assay. It was determined that 23 cycles yielded sufficient product for secondary amplification and was within the linear amplification range of the PCR. All primary reactions intended for library construction were performed in triplicate, and the products were combined, purified by using a QiaQuick PCR clean-up column, and quantified by the Pico Green method.
Using 10 ng of the primary amplification reaction as a template, a secondary amplification, yielding a 1,040-bp product, was carried out in triplicate with the Salinispora-specific forward primer F468 and reverse primer RC1492 as described above (Table 2). Reaction mixtures were thermocycled as follows: an initial denaturation of 94°C for 10 min was followed by 20 cycles of denaturation 94°C (15 s), annealing 65°C (15 s), and extension at 72°C (1 min). Again, to avoid saturation bias, products were quantified by using the Pico Green assay. Negative controls were performed by using 30 thermocycles to aid in the electrophoretic detection of possible contamination.
Clone libraries were constructed from PCR products by using the TA cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. Individual clones were transferred to deep-well, 96-well microtiter plates (each well containing 0.5 ml of LB broth and 50 μg of kanamycin/ml) and grown overnight at 37°C with shaking. Individual plasmid inserts were amplified by direct PCR of clone cell cultures with standard M13 primers and protocols outlined in the Invitrogen TA cloning kit. Amplification products were analyzed by RFLP after digestion with the restriction endonuclease HaeIII (New England Biolabs, Beverly, MA) and analyzed by electrophoresis on a 3% agarose gel containing 0.25 μg of ethidium bromide/ml. Individual clones containing seminested PCR amplified inserts were sequenced by using quantified plasmid template DNA and the M13F and M13R primers by standard methods.
Seminested PCR control library.
To determine Taq polymerase misincorporation rates or other errors introduced by the seminested amplification process, a control library was constructed from a PCR-amplified plasmid DNA template containing the insert of the almost complete 16S rRNA gene from S. tropica isolate CNB-440. The initial plasmid was constructed by using the PCR-amplified 16S rRNA gene (FC27 and RC1492 primers) from strain CNB-440 and the TA cloning kit. Plasmid DNA from a single clone was quantified and sequenced by standard methods. A total of 2 fg (ca. 500 copies) of this plasmid was then processed by using the seminested PCR amplification and library construction methods detailed above. Nine clones were chosen from this library, and the inserts were completely sequenced, aligned with the original clone sequence, and analyzed for errors.
Salinispora RFLP analysis.
To rapidly assess Salinispora diversity, a selective amplification and RFLP screening method was used. Genomic DNA from cultivated isolates was amplified using the Salinispora diagnostic primer set F468, RC1492 as outlined above except that 35 cycles were performed. Amplification products were subsequently digested with the restriction endonuclease HaeIII and analyzed by electrophoresis as indicated above. For the purpose of direct detection and characterization of Salinispora in sediment enrichment cultures, template DNA was amplified by the seminested PCR method with 35 thermocycles in the secondary amplification in order to generate sufficient product for subsequent RFLP analysis.
Intragenomic 16S rRNA gene heterogeneity.
The occurrence of 16S rRNA gene sequence heterogeneity within individual cultivated isolates was assessed by the construction and analysis of clone libraries from nine Salinispora strains. Almost complete 16S rRNA gene sequences (primer set FC27-RC1492) were amplified from each strain using PCR reagent concentrations as described above and 1 μg of genomic DNA template, an initial denaturation of 94°C (10 min), followed by 25 cycles of denaturation 94°C (30 s), annealing 55°C (30 s), and extension at 72°C (1 min). PCR products were cloned and 20 clones were screened from each library using HaeIII RFLP analysis. Select clones from each library were sequenced by using standard M13F and M13R primers, specific for the cloning vector, and primers F514 and R936, specific for the inserts (Table 2).
Chitin hydrolysis assays.
The chitin hydrolysis assay medium consisted of 10 g of colloidal chitin prepared by the method of Makkar and Cross (16), 1 liter of natural seawater, and 20 g of agar. Actinomycete cultures were streaked onto colloidal chitin medium, incubated at 28°C for 2 to 4 weeks, and scored based on the clearing zone diameter: strong (>5 mm), moderate (2 to 5 mm), weak (<2 mm), or no detectable hydrolysis.
RESULTS
Cultured Salinispora diversity.
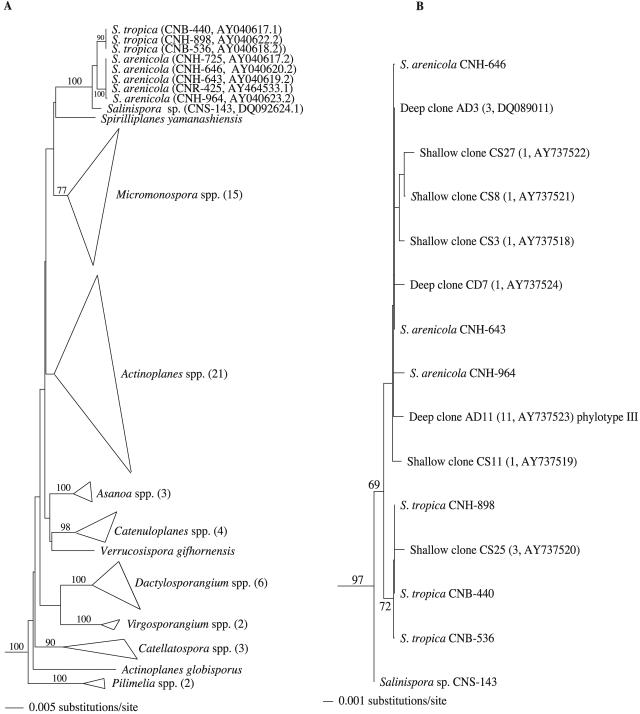
A total of 366 Salinispora strains were cultured from marine sediments collected around the islands of The Bahamas during research expeditions in 2000, 2002, and 2003, including a sample collected at 1,100 m (BA02-36) that exceeded the previous depth record of 570 m (13) for the recovery of this genus. Of these, 105 strains, representing all observed variations in pigmentation, colony size, shape, texture, and level of sporulation, were selected for further study, including strains obtained from various depths and habitats. Seventy-five of these strains originated from the 13 samples listed in Table 1, all of which yielded 2 × 103 to 5 × 103 Salinispora CFU/ml. The PCR-amplified 16S rRNA genes of all 105 strains were analyzed by RFLP using HaeIII restriction endonuclease digestion (Fig. 2), which proved to be an accurate method to delineate between S. tropica and S. arenicola. The RFLP patterns of 102 strains corresponded to S. arenicola, whereas only three strains, originating from samples BA02-35 and BA03-03, displayed the characteristic S. tropica phylotype. Phylogenetic analysis of the almost complete 16S rRNA gene sequences from nine of these isolates corroborated the RFLP findings that no new species-level diversity had been cultured from the Bahamas. The phylogenetic relationships of nine cultured strains, including strains CNS-143 (Palau,) CNH-964 (Sea of Cortez), and CNR-425 (Guam), are shown in Fig. 3A and represent the extant cultured diversity of the genus to date relative to other genera in the Micromonosporaceae. Strain CNS-143 is currently being evaluated to determine whether it represents a new species.
FIG. 2.
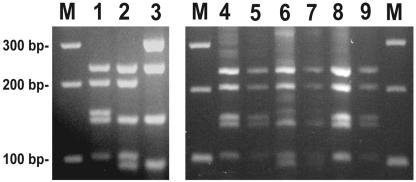
Ethidium bromide-stained 3% agarose gel comparing HaeIII restriction endonuclease digests of Salinispora seminested PCR amplification products. Lanes: M, 100-bp marker; 1, S. tropica cultivated isolate; 2, S. arenicola cultivated isolate; 3, uncultivated phylotype III from sediment BA02-36 environmental library; 4, sample BA02-35 enrichment (sediment extract and rifampin) showing the S. tropica RFLP pattern; 5, sample BA02-35 enrichment (sediment extract and novobiocin) showing the S. tropica RFLP pattern; 6, sample BA02-36 enrichment (sediment extract and rifampin) showing the predominant S. arenicola RFLP pattern; 7, sample BA02-36 enrichment (sediment extract and novobiocin) showing the S. arenicola RFLP pattern; 8, sample BA03-03 enrichment (sediment extract and rifampin) showing the S. tropica RFLP pattern; 9, sample BA03-03 enrichment (sediment extract and novobiocin) showing the S. tropica RFLP pattern.
FIG. 3.
(A) Neighbor-joining phylogenetic dendrogram constructed with nearly full 16S rRNA gene sequences (1,407 unambiguous nucleotides) illustrating cultured Salinispora species diversity including strains isolated from the Red Sea (CNH-725), the Sea of Cortez (CNH-964), Palau (CNS-143), and Guam (CNR-425) in comparison to all formally described genera within the Micromonosporaceae (List of Bacterial Names with Standing in Nomenclature [http://www.bacterio.cict.fr/]) for which sequence data was available from the NCBI Web site. Strain CNS-143 was cultured as part of a separate study and is currently being examined to determine whether it represents a third Salinispora species. Genera other than Salinispora that contain multiple species are represented with a triangle with the number of species included in parentheses. Propionibacterium propionicus, Streptosporangium corrugatum, and Streptomyces lividans were used as outgroups. In both panels A and B, bootstrap values were calculated from 1,000 resamplings and are shown at their respective nodes for values ≥60%. (B) Neighbor-joining phylogenetic dendrogram, obtained after seminested PCR amplification of the 16S rRNA gene, using 995 unambiguous nucleotides illustrating cultured and culture-independent Salinispora diversity. Clones were derived from deep (BA02-36) and shallow (BA00-11) samples with the number of identical clones and accession numbers noted in parentheses. Strain CNS-143 was isolated from Palau and may represent a new species (subject of a separate study). Micromonospora olivasterospora and Dactylosporangium aurantiacum (not shown) were used as outgroups.
Enrichment cultures frequently produced visible aggregates of filamentous bacteria that upon microscopic examination were recognized as actinomycetes by the presence of branching filaments. These actinomycete blooms could be harvested with a sterile pipette and, upon transfer to solid media, yielded an additional 60 actinomycete strains. Based on morphological and RFLP analyses, seven of these strains were identified as S. tropica and 31 were identified as S. arenicola. The remaining isolates were more closely related to Micromonospora or Streptomyces spp., including a new taxon within the Streptomycetaceae (subject of another study). These non-Salinispora actinomycetes were not observed using the original cultivation techniques (heat shock or drying methods), supporting the use of enrichment culture for the recovery of new actinomycete taxa.
The enrichment conditions that yielded Salinispora cultivars (M1low and SE) typically led to the formation of visible actinomycete blooms at the sediment-seawater interface, suggesting that these organisms may have a growth preference in the upper sediment layer. In contrast to many of the M1low and SE enrichments, only one chitin enrichment yielded Salinispora cultivars, whereas all others yielded actinomycetes that were more closely related to Micromonospora or Streptomyces spp. Of the 30 Salinispora cultures tested for chitin hydrolysis, only 5 displayed moderate to weak chitinolytic activity, indicating that the ability to hydrolyze chitin is not a consistent trait among Salinispora strains.
Culture-independent Salinispora diversity.
In preliminary studies, analysis of over 100 clones from two libraries constructed from environmental DNA amplified using the general bacterial primer set FC27-RC1492 or the actinomycete primer set F270-RC1492 revealed no sequences affiliated with the genus Salinispora. Since Salinispora cultivars had been obtained from the sediment samples from which the DNA was extracted, a Salinispora-specific, seminested PCR method was developed to improve the resolution with which this group could be detected. Thirteen environmental DNA samples were processed by using the seminested PCR method (Table 1), and all yielded an initial PCR product with the actinomycete-biased primer set F270-RC1492 (Table 2). However, only two samples (BA00-11 and BA02-36) yielded secondary amplification products using the Salinispora-specific primer set F468-RC1492. Clone libraries were constructed from both of these PCR products (Table 1).
Forty-eight clones from the library created from the shallow (30 m) sediment (BA00-11) were analyzed by RFLP after HaeIII restriction endonuclease digestion. Five cutting patterns or phylotypes were observed with three occurring only once. These three rare phylotypes, upon careful sequence analysis and comparison with Salinispora and other members of the Micromonosporaceae, were found to contain bases that differed within highly conserved regions of the 16S rRNA gene, suggesting Taq polymerase misincorporation events were responsible. These nucleotide substitutions were masked from subsequent analyses. The remaining 45 clones displayed the typical cutting patterns of S. tropica (eight clones, six sequenced) and S. arenicola (37 clones, 14 sequenced). The clone sequences were aligned and representatives incorporated into a phylogenetic tree where they are denoted as “shallow clones” (Fig. 3B). As predicted, all of these clones fell into the S. arenicola or S. tropica phylotypes, differing by one to three nucleotides from previously cultured strains. In one case (CS25), three clones with the identical sequence were observed providing strong evidence that the single nucleotide substitution that distinguishes them from all cultured S. tropica represents new intraspecies diversity.
A second environmental library was created from the deep (1,100 m) sediment sample BA02-36 (Table 1). Fifty-four clones were analyzed by RFLP and seven phylotypes were observed of which four occurred only once. As was the case with the shallow library, the rare cutting patterns could be attributed to polymerase misincorporation events that disrupted or created HaeIII restriction sites. Two phylotypes constituted the majority of the remaining clones with five displaying the typical S. arenicola cutting pattern (four clones sequenced). These S. arenicola clones were either identical in sequence to previously cultured strains (i.e., deep clone AD3) or different by one nucleotide (e.g., CD7). Forty-three clones from the deep sample, however, displayed a unique cutting pattern designated phylotype III (Fig. 2, lane 3). Sequence analysis of 11 phylotype III clones showed one consistent nucleotide difference from cultivated S. arenicola (Fig. 1B). Although subtle, phylotype III represents additional new and as-yet-uncultivated, intraspecific Salinispora diversity. Despite these sequence differences, all of the clones examined fall within the previously described S. arenicola and S. tropica phylotypes (Fig. 3B). Two non-Salinispora clones observed from the deep library shared low (92%) identity (blastn) over a 937-bp region with an uncultured soil Verrucomicrobium.
PCR error analysis.
A library constructed from a PCR-amplified plasmid DNA template was used to estimate the frequency of sequence error introduced by the seminested PCR method. Sequence analysis of nine clones from this control library displayed an overall observed error frequency (f) of 24 of 9,459 bases analyzed or 0.25% (i.e., two or three aberrant bases per clone sequence). This corresponds to a misincorporation frequency per template doubling (m) of 1.2 × 10−4 per bp per cycle, using the formula 2(f/d) = m, where d (43 cycles) is the theoretical doubling (23). These base substitutions appeared to be interspersed randomly throughout the amplified gene sequences and not localized in “hot spots.” Base transitions G↔A or C↔T accounted for ca. 80% of the overall substitutions observed, a finding which agrees with the Taq polymerase transition substitution frequency determined by others (4, 14).
Detection of Salinispora spores and vegetative cells.
All of the 13 sediment samples in Table 1 yielded 2 × 103 to 5 × 103 Salinispora CFU/ml. Using the chemical lysis method, only 2 of these 13 sediment samples yielded a secondary Salinispora-specific amplification product. Since this DNA extraction method does not lyse spores, it can be concluded that Salinispora were present as vegetative filaments in these two samples. It is also implied that the 11 samples that did not yield a secondary amplification product harbored predominantly Salinispora spores. To test this hypothesis, a mechanical lysis method capable of extracting DNA from spores was applied to all 13 samples; however, once again, only the same two samples yielded a secondary amplification product. Given that the CFU/ml for each sample were approximately the same, it appears that spore concentrations of 2 × 103 to 5 × 103/ml are below the detection limits of the mechanical lysis method. Of the 13 sediments, 9 were subjected to enrichment culture, after which all 9 yielded a secondary amplification product using DNA obtained by the chemical lysis method, indicating that spores had germinated in response to these culture conditions.
PCR and RFLP analysis of enrichment cultures.
DNA extracted from the SE and M1low enrichment cultures all yielded a nested PCR product with typical Salinispora RFLP cutting patterns. Even in cases where growth was not evident by microscopic inspection of the cultures, secondary amplification products were obtained by using the chemical lysis method, indicating that Salinispora were actively growing under these conditions. Even though phylotype III was detected in the clone library generated from sample BA02-36, it was not observed in any cultured strains or detected in any of the enrichment cultures inoculated with sediment from this sample and thus members of this phylotype have yet to be cultured.
Typical RFLP results in response to different antibiotic treatments are illustrated in Fig. 2, lanes 4 to 9. Enrichment cultures from sediments BA02-35 (lanes 4 to 5) and BA03-03 (lanes 8 to 9) were the only samples that displayed a clear S. tropica RFLP pattern. These are the same two samples from which S. tropica was directly cultured. All of the remaining samples subjected to enrichment cultivation displayed the S. arenicola RFLP pattern (lanes 6 to 7), while phylotype III (lane 3) was not observed in any of the enrichment cultures. The presence of S. arenicola or S. tropica RFLP patterns appeared to be specific to the sample and not dependent on antibiotic treatment. The rare occurrence of the S. tropica phylotype in the enrichment cultures is consistent with what was detected by direct plating and from environmental clone libraries supporting the observation that this species is relatively uncommon in the samples analyzed.
Intragenomic 16S rRNA gene sequence heterogeneity.
Intragenomic heterogeneity among copies of the 16S rRNA gene is well documented (2), has been shown to occur among members of the actinomycetes (29), and was suspected to be the source of the as-yet-uncultured Salinispora diversity observed in the two environmental clone libraries. To test this possibility, clone libraries were constructed from PCR-amplified 16S rRNA genes from six S. arenicola and three S. tropica cultures. RFLP analyses of 20 clones from each library did not detect the presence of intragenomic heterogeneity or any of the new phylotypes. Further sequencing of two to three clones from each of the nine Salinispora libraries displayed no fine-scale heterogeneity. Overall, this suggests that the observed diversity in the environmental libraries is not due to intragenomic allelic diversity of the 16S rRNA gene.
DISCUSSION
The genus Salinispora is a pan-tropical, marine actinomycete that has proven to be a productive source of biologically active secondary metabolites (10). Despite a widespread distribution in marine sediments, polyphasic taxonomic analyses of cultivated isolates have led thus far to the recognition of only two species (17). To further investigate Salinispora species diversity, new cultivation techniques, along with cultivation-independent methods, were tested to assess the extant diversity and depth distributions of this commercially important marine actinomycete taxon.
The initial cultivation methods used to isolate Salinispora strains relied upon the resistance of spores to heat shock and desiccation (19). Since these relatively harsh treatments may have contributed to the limited cultured diversity observed within the genus, new enrichment cultivation techniques and selective antibiotic treatments were tested. Despite the molecular characterization of 143 Salinispora strains cultured from sediment samples collected throughout the Bahamas, including 38 isolated from enrichment cultures, no new species level diversity was detected. There was, however, new but subtle intraspecific diversity, within both the S. arenicola and S. tropica clades, detected from the environmental libraries, including an abundant new phylotype (III) from a library created from a sample collected from 1,100 m. Despite extensive effort, strains possessing these sequence variations were not cultured.
Culture-independent molecular methods have led to a vastly improved understanding of microbial diversity in the world's oceans (9). It is interesting that in the present study both culture-dependent and culture-independent analyses revealed the same level of Salinispora species diversity. This is unusual since most culture-independent studies yield diversity estimates that are in striking contrast to what can be cultured from the same samples (26). Given that both methods detected the presence of only two Salinispora species in the sediments collected from the Bahamas (a potential third species has been cultured from Palau), the results emphasize that cultivation efforts can be highly successful when a specific taxon is targeted. This was recently demonstrated on a larger scale with the cultivation of members of the ubiquitous marine bacterioplankton clade SAR11 (21). Thus, it is becoming increasingly clear that when appropriate cultivation methods are applied marine bacteria are far more amenable to cultivation than previously believed.
The ability of Salinispora to form spores complicated the culture-independent detection of this taxon in the samples studied. PCR amplification of environmental DNA, extracted by using a chemical lysis method that does not extract DNA from spores, led to the detection of Salinispora in only 2 of 13 sediments. This result was obtained despite the occurrence of approximately equal numbers of Salinispora CFU/ml (2 × 103 to 5 × 103) in all of the samples. Based on these results, it is concluded that Salinispora were present as vegetative filaments in only two of the samples, providing additional evidence that actinomycetes are capable of growth in the marine environment (12, 20) and that actively growing actinomycetes may be uncommon and unevenly distributed relative to spores. Although it is not known what triggers spore germination in marine sediments, all sediment enrichment cultures extracted using the chemical lysis method (Table 1) yielded Salinispora-specific secondary amplification products, indicating that appropriate nutrient conditions are a factor and that, without prior enrichment, spore-forming actinomycetes may be overlooked in molecular analyses of marine bacterial diversity.
The mechanical lysis method was found to be effective at recovering DNA from spore preparations in control experiments; however, this DNA extraction method did not improve our ability to detect Salinispora in the sediments studied. Although at first surprising, it is proposed that spore abundances of 2 × 103 to 5 × 103 CFU/ml remain below the detection limits of this method, an observation that is in agreement with other workers who found that DNA extraction efficiency falls precipitously when spore abundances are ≤103/ml of sediment (8). Similar, confounding results have been reported for actinomycetes in terrestrial soils where cultivation-based studies have shown Streptomyces spp. to be cosmopolitan and dominant, whereas PCR-based molecular analyses have underestimated or failed to detect this important group of microorganisms (18). These results again emphasize the limitations of molecular techniques when working with spore-forming bacteria that spend a majority of their life cycle in a lysis-resistant resting stage interspersed with sporadic but ecologically important periods of growth.
Nested PCR methods have proven effective for monitoring uncultured Actinobacteria in environmental samples (22) and, in the present study, highly specific for the amplification of Salinispora DNA. The signature nucleotides (U, U) at positions 442 to 443 (Fig. 1A, positions 467 to 468, E. coli numbering) were incorporated into the Salinispora-specific primer F468, which yielded Salinispora 16S rRNA gene sequences in all but 2 of 102 clones analyzed. The two aberrant clones showed closest homology to members of the Verrucomicrobium. It is likely that the Actinomycetales biased primer set F270-RC1492, which amplified unacceptable levels of Verrucomicrobium and Acidimicrobium 16S rRNA gene sequences in pilot studies, amplified a significant amount of Verrucomicrobium DNA in the primary amplification step of the nested PCR thus making cross-amplification events more likely in the secondary amplification. Although it is possible that new Salinispora species were missed in the present study, the signature nucleotides targeted by the F468 primer are a defining phylogenetic feature of the genus as currently defined (19) and have been observed in all Salinispora strains analyzed to date.
Although RFLP provided a rapid method to identify Salinispora phylotypes from clone libraries, careful inspection of sequence data revealed that, of the 10 phylotypes originally observed in the two clone libraries, 7 occurred only once and ultimately could be attributed to PCR error. This determination was made if a consensus for a variable position could not be established in multiple clones and if the nucleotide change did not occur in a region of appropriate variability based upon aligned 16S rRNA gene sequence data and secondary structure. Suspect bases, predominantly transitions, that did not meet these criterion were masked from the phylogenetic analyses, resulting in an overall reduction in branch lengths and in the number of new phylotypes predicted by RFLP from eight to one. These changes did not affect the overall tree topology of Fig. 3B. The 11 phylotype III clones sequenced from the deep library all shared a C→T transition at position 1002 (Fig. 1B). Although this base change is a transition, it appears in an area of moderate to high variability among all bacteria (27) and represents the only new Salinispora phylotype that could be identified by RFLP from the present study.
Environmental clone libraries have recently been shown to overestimate diversity unless special precautions are taken to avoid PCR-induced artifacts (1). This appears to be of special concern when Taq polymerase is used (4, 14, 23). Given the suspect Salinispora diversity originally detected in the environmental clone libraries, a control library was constructed, using plasmid DNA template from a well-characterized strain (CNB-440), to assess misincorporation error in the seminested PCR protocol. The overall error frequency per base pair per template doubling (m) was calculated to be 1.2 × 10−4, falling toward the high end of previously reported values for Taq DNA polymerase (4, 14, 23). Misincorporation rates for the shallow and deep libraries were estimated to be 1.5 × 10−4 and 1.7 × 10−4 per bp per cycle, respectively, again within previously determined values. A large number of transitions, as observed in the control library, is known to be a characteristic of Taq polymerase (4, 14, 23), further suggesting that misincorporation events were the major source of error in our environmental libraries. Since neither the primary nor secondary PCR amplifications reached saturation, heteroduplex formation, a known source of clone library error (25), is less likely to explain the error in this particular case.
The obligate marine distribution of the genus Salinispora could be a factor in the limited 16S rRNA gene sequence diversity detected. Compared to their terrestrial relatives Micromonospora, Dactylosporangium, and Actinoplanes, Salinispora spp. inhabit a less variable environment in terms of temperature, salinity, and hydration and thus possibly fewer selective forces driving speciation. Given that habitat heterogeneity has been posited as one of the major determinants of biological diversity (11), the relatively low level of Salinispora species diversity is what would be predicted from a stable marine environment or limited niche suitability. Although difficult to test experimentally, environmental stability may help explain the relatively low level of species diversity detected in this marine actinomycete genus (Fig. 3A). Another factor for lower 16S rRNA gene sequence diversity could be that Salinispora have experienced fewer lateral gene transfer events. Evidence exists that Dactylosporangium and Micromonospora spp. have undergone recent lateral transfers of short gene segments characterized by nonrandom variations outside of hypervariable regions of the 16S rRNA gene that can be identified by variable tree topologies for different regions of the 16S rRNA gene (28). These proposed transfer events can increase the 16S rRNA gene sequence diversity within a given group and are known to cause intragenomic heterogeneity. However, the tree topologies generated from partial and full 16S rRNA gene sequences were always maintained, and intragenomic heterogeneity was not observed in control clone libraries generated from nine cultured Salinispora strains.
S. tropica has proven to be an important source of bioactive secondary metabolites (10), yet this species is rarely cultured relative to S. arenicola with only ca. 20 strains isolated to date. S. tropica was detected by RFLP in only one of the two environmental clone libraries generated (sample BA00-11), where it represented 8 of 45 clones belonging to the Salinispora clade, and in only two of the samples subjected to enrichment culture (BA03-03 and BA02-35), thus corroborating the relative rarity of this species. Since the secondary metabolite profiles of these two species differ significantly (subject of another study), it would be interesting to determine whether the ability to produce various secondary metabolites affects fitness.
Although members of the marine actinomycete genus Salinispora were readily cultured at 2 × 103 to 5 × 103 CFU/ml from all 13 marine sediment samples analyzed in the present study, only 2 of these samples showed a positive result when a targeted, seminested PCR technique was applied. These results emphasize the extra effort that must be made when culture-independent techniques are used to assess the diversity of spore-forming marine bacteria, which undoubtedly remains widely underestimated, as well as the utility of selective cultivation techniques for bacteria that spend a large portion of their life cycle in a resting stage. Although some actinomycetes may occur in marine sediments primarily as spores and these may be distributed to great depths by ocean currents, evidence is presented for actinomycete growth in marine sediments even at the deepest sites sampled (1,100 m). Based on our current understanding of the genus, it is likely that the two approaches: enrichment cultivation and targeted, seminested PCR have revealed the majority of Salinispora species-level diversity within the sediment samples studied. Continued improvements in our ability to culture marine actinomycetes, in combination with sampling new habitats, are essential for a more complete assessment of their diversity and potential as a resource for biotechnology.
Acknowledgments
We gratefully acknowledge the Khaled Bin Sultan Living Oceans Foundation for a fellowship to T.J.M. and J. Pawlik for his invitation to participate in the Bahamas 2000, 2002, and 2003 R/V Seward Johnson expeditions. Additional financial support was provided by the University of California Industry-University Cooperative Research Program (IUCRP, grant BioSTAR 10102). P.R.J. and W.F. are scientific advisors to and stockholders in Nereus Pharmaceuticals, the corporate sponsor of the IUCRP award. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. DNA sequencing was performed by the Molecular Pathology Shared Resource, UCSD Cancer Center, which is funded in part by NCI Cancer Center support grant 5P0CA23100-16.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. C. Ceraj, D. L. Distel, and M. F. Poltz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Poltz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berdy, J. 2005. Bioactive microbial metabolites: a personal view. J. Antibiot. 58:1-26. [DOI] [PubMed] [Google Scholar]
- 4.Bracho, M. A., A. Moya, and E. Barrio. 1998. Contribution of Taq polymerase-induced errors to the estimation of RNA virus diversity. J. Gen. Virol. 79:2921-2928. [DOI] [PubMed] [Google Scholar]
- 5.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BioMed. Central Bioinform. 3. [Online.] [DOI] [PMC free article] [PubMed]
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colquhoun, J. A., J. Mexson, M. Goodfellow, A. C. Ward, K. Horikoshi, and A. T. Bull. 1998. Novel rhodococci and other mycolate actinomycetes from the deep sea. Antonie Leeuwenhoek 74:27-40. [DOI] [PubMed] [Google Scholar]
- 8.Cresswell, N., V. A. Saunders, and E. M. H. Wellington. 1991. Detection and quantification of Streptomyces violaceolatus plasmid DNA in soil. Lett. Appl. Microbiol. 13:193-197. [Google Scholar]
- 9.DeLong, E. F. 1997. Marine microbial diversity: the tip of the iceberg. Trends Biotechnol. 15:203-207. [DOI] [PubMed] [Google Scholar]
- 10.Feling, R. H., G. O. Buchanan, T. J. Mincer, C. A. Kauffman, P. R. Jensen, and W. Fenical. 2003. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinispora. Angew. Chem. Int. Ed. 42:355-357. [DOI] [PubMed] [Google Scholar]
- 11.Horner-Devine, M. C., Carney, K. M., Bohannan, B. J. M. 2004. An ecological perspective on bacterial biodiversity. Proc. R. Soc. Lond. B 271:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, P. R., R. Dwight, and W. Fenical. 1991. Distribution of actinomycetes in near-shore marine sediments. Appl. Environ. Microbiol. 57:1102-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen, P. R., E. Gontang, C. Mafnas, T. J. Mincer, and W. Fenical. 2005. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ. Microbiol. 7:1039-1048. [DOI] [PubMed] [Google Scholar]
- 14.Keohavong, P., and W. G. Thilly. 1989. Fidelity of DNA polymerases in DNA amplification. Proc. Natl. Acad. Sci. USA 86:9253-9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, T. K., M. J. Garson, and J. A. Fuerst. 2005. Marine actinomycetes related to the “Salinospora” group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ. Microbiol. 7:509-518. [DOI] [PubMed] [Google Scholar]
- 16.Makkar, A., and T. Cross. 1982. Actinoplanetes in soil and on plant litter from fresh water habitats. J. Appl. Bacteriol. 52:209-218. [Google Scholar]
- 17.Maldonado, L. A., W. Fenical, P. R. Jensen, C. A. Kauffman, T. J. Mincer, A. C. Ward, and M. Goodfellow. 2005. Salinispora gen. nov., sp. nov., S. arenicola sp. nov., and S. tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 55:1759-1766. [DOI] [PubMed]
- 18.McVeigh, H. P., J. Munro, and T. M. Embley. 1996. Molecular evidence for the presence of novel actinomycete lineages in a temperate forest soil. J. Indust. Microbiol. 17:197-204. [Google Scholar]
- 19.Mincer, T. J., P. R. Jensen, C. A. Kauffman, and W. Fenical. 2002. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 68:5005-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moran, M. A., L. T. Rutherford, and R. E. Hodson. 1995. Evidence for indigenous Streptomyces populations in a marine environment determined by determined with a 16S rRNA probe. Appl. Environ. Microbiol. 61:3695-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the uniquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 22.Rheims, H., and E. Stackebrandt. 1999. Application of nested polymerase chain reaction for the detection of as yet uncultured organisms of the class Actinobacteria in environmental samples. Environ. Microbiol. 1:137-143. [DOI] [PubMed] [Google Scholar]
- 23.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA-polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 24.Takizawa, M., R. R. Colwell, and R. T. Hill. 1993. Isolation and diversity of actinomycetes in the Chesapeake Bay. Appl. Environ. Microbiol. 59:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson, J. R., L. A. Marcelino, and M. F. Polz. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by “reconditioning PCR.” Nucleic Acids Res. 30:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torsvik, V., R. Sorheim, and J. Goksoyr. 1996. Total bacterial diversity in soil and sediment communities: a review. J. Indust. Microbiol. 17:170-178. [Google Scholar]
- 27.Van de peer, Y. S. Chapelle, and R. Dewachter. 1996. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 24:3381-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, Y., and Z. S. Zhang. 2000. Comparative sequence analyses reveal frequent occurrence of short segments containing an abnormally high number of non-random base variations in bacterial rRNA genes. Microbiol. 146:2845-2854. [DOI] [PubMed] [Google Scholar]
- 29.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]